Abstract
Atypical hemolytic uremic syndrome (aHUS) is now well recognized to be a disease characterized by excessive complement activation in the microvasculature. In both the familial and sporadic forms, inherited and acquired abnormalities affecting components of the alternative complement pathway are found in ∼ 60% of patients. These include mutations in the genes encoding both complement regulators (factor H, factor I, membrane cofactor protein, and thrombomodulin) and activators (factors B and C3) and autoantibodies against factor H. Multiple hits are necessary for the disease to manifest, including a trigger, mutations, and at-risk haplotypes in complement genes. The prognosis for aHUS is poor, with most patients developing end-stage renal failure. Renal transplantation in most patients also has a poor prognosis, with frequent loss of the allograft to recurrent disease. However, improving results with combined liver-kidney transplantation and the advent of complement inhibitors such as eculizumab offer hope that the prognosis for aHUS will improve in future years.
Definition of aHUS
The term hemolytic uremic syndrome (HUS) was first used by the Swiss hematologist Conrad Von Gasser in a paper published in 1955.1 In this paper, he described 5 patients with renal failure, thrombocytopenia, and acquired hemolytic anemia. Since then, the term HUS has mainly been used in association with a thrombotic microangiopathy occurring after infection with verocytotoxin (Shiga-like toxin)–producing bacteria, particularly enterohemorrhagic Escherichia coli. Most individuals with this condition (Stx-HUS) recover renal function. Less often, patients present with the features of HUS without any evidence of infection with veryocytotoxin-producing bacteria. In this group of patients, the prognosis is worse, with a significant number developing end-stage renal failure. Over time, the prefix “atypical” has been added to define this group. The nomenclature used to describe those diseases characterized by a thrombotic microangiopathy has been a source of debate for decades. Defining the molecular mechanisms underlying these conditions has in the past decade enabled the development of a mechanistic classification.2 We review herein the genetic basis underlying aHUS and discuss how this has changed patient management.
Complement and aHUS
aHUS can be both familial and sporadic.3 In both the familial and sporadic forms of aHUS, a series of studies undertaken since the late 1990s have established that dysregulation and/or excessive activation of the alternative pathway of complement plays a pivotal role in the pathogenesis of the disease. Both inactivating mutations in genes encoding complement regulators (factor H, factor I, and membrane cofactor protein) and gain-of-function mutations in genes encoding the complement activators (C3 and factor B) have been described.4 Mutations in the gene encoding thrombomodulin, a membrane-bound glycoprotein with anticoagulant properties that modulates complement activation on cell surfaces, have also been described in aHUS.5 The following sections contain descriptions of the genes that may be mutated in aHUS. The protein products of these genes are involved in regulation of the activity of the alternative pathway of complement amplification, C3 convertase. For an overview of the function of each protein within the alternative pathway, see the accompanying article in this section by Atkinson and Liszewski entitled “Too much of a good thing at the site of tissue injury: The instructive example of the complement system predisposing to thrombotic microangiopathy.”
Factor H
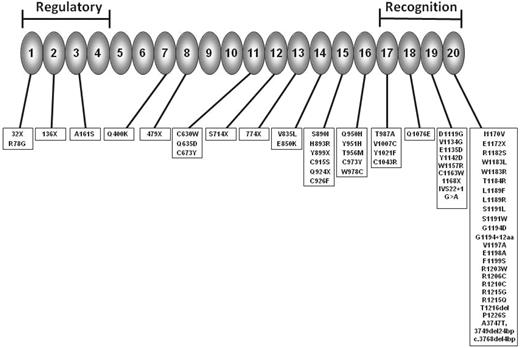
Factor H is a serum protein produced almost exclusively by the liver that acts as the principal regulator of the alternative pathway of complement. Factor H and other proteins within the regulators of complement activation (RCA) cluster at 1q32 share a common basic structure consisting of multiple (contiguous) homologous modules called short consensus repeat (SCR) domains. Factor H has 20 SCRs each comprising ∼ 60 amino acids (Figure 1). The N terminal SCRs form the regulatory domain, whereas the C-terminal SCRs form the recognition domain. Mutations in CFH in aHUS patients have been widely described and are found in ∼ 30% of patients.4 The majority are heterozygous missense mutations that cluster in the C-terminal exons. These mutations commonly do not result in a quantitative deficiency of factor H, but instead result in normal levels of a folded protein with only localized structural perturbations.6 In vitro experiments have demonstrated that these secreted mutants can control complement in the fluid phase, but are unable to bind to and regulate complement on host cells and platelets.7 To test this hypothesis in vivo, mouse models of CFH mutations have been generated. A mouse completely deficient in factor H (Cfh−/−) had uncontrolled turnover of the alternative pathway, with very low C3 levels and the pathological features of membranoproliferative glomerulonephritis in the kidney, not aHUS. In contrast, a mouse lacking the C-terminal end of factor H (Cfh−/−Δ16-20) designed to mimic the mutations commonly seen in aHUS had higher plasma C3 levels than the Cfh−/− mouse, and spontaneously develops aHUS, not membranoproliferative glomerulonephritis.8 Therefore, the mutant factor H seen in aHUS fails to bind to and control complement activation on the glomerular endothelium, basement membrane, and platelets,9 leading to the procoagulant state necessary for aHUS. A minority of CFH mutations are deletions or missense mutations that result in either a severely truncated protein or impaired secretion. This leads to systemic factor H deficiency, which is usually heterozygous.
Factor H consists of 20 SCRs. The N-terminal SCRs form the regulatory domain and the C-terminal the recognition domain. The site of mutations reported in aHUS are shown. (Adapted from Kavanagh and Goodship4 and used with the kind permission of Springer Science and Business Media.)
Factor H consists of 20 SCRs. The N-terminal SCRs form the regulatory domain and the C-terminal the recognition domain. The site of mutations reported in aHUS are shown. (Adapted from Kavanagh and Goodship4 and used with the kind permission of Springer Science and Business Media.)
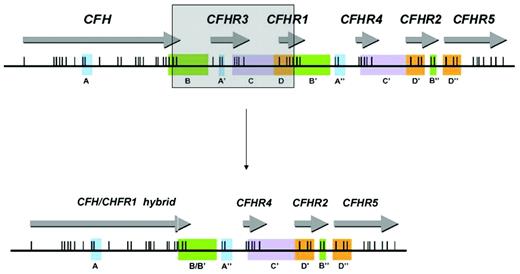
Genomic disorders affecting CFH have also been described in association with aHUS. Complement genes within the RCA cluster at chromosome 1q32 are arranged in tandem within 2 groups. In a centromeric, 360-kb segment lie the genes for factor H (CFH) and 5 factor H–related proteins: CFHR1, CFHR2, CFHR3, CFHR4, and CFHR5. Sequence analysis of this region shows evidence of several large genomic duplications, also known as low copy repeats (LCRs), resulting in a high degree of sequence identity between CFH and the genes for the 5 factor H–related proteins.10 LCRs such as those seen in the RCA cluster are frequently associated with genomic rearrangements.11 These usually result from nonallelic homologous recombination (NAHR) between LCRs. It has been shown that NAHR in this region can lead to the formation of a hybrid gene consisting of the first 21 exons of CFH (encoding SCRs 1-18 of the hybrid gene) and the last 2 exons of CFHR1 (encoding SCRs 19 and 20 of the hybrid gene) (Figure 2).13 This hybrid gene encodes a protein product that is identical to a functionally significant factor H mutant (Ser1191Leu/Val1197Ala) that arises by gene conversion.
The genes encoding factor H and the factor H–related proteins lie in a centromeric segment of the regulators of complement activation cluster at 1q32. Genomic duplications including the different exons of the 6 genes (originally determined by Male et al12 ) are shown in panels A-D by color coding. Exons are indicated as vertical lines. The shaded box shows how a deletion occurring as a result of nonallelic homologous recombination results in a hybrid gene consisting of the first 21 exons of CFH (encoding SCRs 1-18 of the hybrid gene) and the last 2 exons of CFHR1 (encoding SCRs 19 and 20 of the hybrid gene). (Adapted from Venables et al.)
The genes encoding factor H and the factor H–related proteins lie in a centromeric segment of the regulators of complement activation cluster at 1q32. Genomic duplications including the different exons of the 6 genes (originally determined by Male et al12 ) are shown in panels A-D by color coding. Exons are indicated as vertical lines. The shaded box shows how a deletion occurring as a result of nonallelic homologous recombination results in a hybrid gene consisting of the first 21 exons of CFH (encoding SCRs 1-18 of the hybrid gene) and the last 2 exons of CFHR1 (encoding SCRs 19 and 20 of the hybrid gene). (Adapted from Venables et al.)
In addition to inherited defects in CFH, acquired abnormalities affecting factor H function are also seen in the form of inhibitory autoantibodies.14 Factor H autoantibodies are reported in 5%-10% of aHUS patients. Analogous to the genetic defect seen in CFH, these autoantibodies also mainly target the C-terminal end of the protein, thereby impairing complement regulation on host cell surfaces. The development of factor-H autoantibodies in aHUS has a genetic predisposition, being strongly associated with a deletion of CFHR1 and CFHR3, which also arises through NAHR.14 More detailed analysis suggested that the association between the CFHR1/CFHR3 deletion and the presence of autoantibodies in aHUS is probably related to the absence of factor H–related protein 1.14,15 However, this deficiency is not a prerequisite for the formation of autoantibodies, because patients with no evidence of deficiency of either of these factor H–related proteins and high titers of autoantibodies have been described.14
Factor H and factor H–related protein 1 share a high degree of homology with the 2 C-terminal SCRs (4 and 5) of CFHR1, being almost identical to SCRs 19-20 of factor H. It is not surprising, therefore, that autoantibodies to SCR 19-20 of factor H also bind to SCRs 4-5 of factor H–related protein 1.14
MCP
Membrane cofactor protein (MCP, CD46) is widely expressed on the surface of all cells except erythrocytes. The extracellular domain comprises 4 SCRs followed by an alternatively spliced region rich in threonine, serine, and proline (the STP region) and a group of 12 amino acids. This is followed by the transmembrane domain and an alternatively spliced cytoplasmic tail which mediates signalling events. With factor I, MCP degrades C3b and C4b bound to the cell surface. Mutations in the gene (CD46) encoding membrane cofactor protein account for up to 15% of aHUS.5 Most CD46 mutations result in decreased cell-surface expression, and the remaining mutations are produced but are functionally inactive.16
Factor I
Factor I is a soluble regulatory serine protease of the complement system that cleaves 3 peptide bonds in the alpha chain of C3b and 2 bonds in the alpha chain of C4b, thereby inactivating these proteins. Factor H acts as a cofactor for C3b, C4 binding protein for C4b, and membrane cofactor protein for both. Factor I is a heterodimer of ∼ 88 kDa that consists of a noncatalytic heavy chain of 50 kDa that is linked to a 38-kDa catalytic light chain by a disulfide bond. The protein is synthesized as a single chain precursor of 565 amino acids, mainly in the liver. Four basic amino acids are then excised from the precursor before secretion of the heterodimer. Like many of the complement proteins, factor I has a modular structure. The heavy chain contains 2 low-density lipoprotein receptor domains, a CD5 domain, and a module found only in factor I and complement proteins C6 and C7. The gene encoding factor I is located on chromosome 4q25 and spans 63 kb. It comprises 13 exons and there is a strong correlation between the exonic organization of the gene and the modular structure of the protein. The light chain of factor I, which is the serine proteinase region of the molecule, is encoded in 5 exons. The genomic organization of the enzymic part of factor I is similar to that of trypsin. The gene structure is unusual in that the first exon is small (86 bp) and is separated from the rest of the gene by a large first intron of 36 kb. CFI mutations are present in up to 12% of aHUS patients.17 Most mutations result in heterozygous quantitative deficiency of factor I, although some mutants are secreted but lack regulatory capacity.17
Factor B
Factor B is a zymogen that carries the convertase serine protease domain necessary for amplification of the alternative pathway. The convertase for this pathway is assembled in 2 steps. First, C3b associates with factor B, and then the C3bB complex is cleaved by the serum protease factor D at a single site in factor B, producing Ba and Bb fragments. The Ba fragment dissociates from the complex, whereas Bb remains bound to C3b to form the active C3 convertase C3bBb. Dissociation of the 2 components of this complex results in inactivation of the convertase and is promoted by decay accelerating factor, complement receptor 1, and factor H. The gene encoding factor B (CFB) is located on chromosome 6p21.3 and consists of 18 exons coding for a 5-domain glycoprotein. The N-terminal Ba fragment contains 3 complement control protein domains, the C-terminal Bb fragment contains 2 domains, the N-terminal type A domain contains the C3b-binding region, and the C-terminal domain is the site of the serine protease. The frequency of CFB mutations in the cohorts of aHUS patients examined to date is low (< 3%).18 Functional analysis of the mutants has demonstrated either increased generation or enhanced stability of the alternative pathway convertase.
C3
C3 is the pivotal component of the complement system.19 Activation of the classical, lectin, and alternative pathways results in cleavage of C3 to generate C3b and the anaphylatoxin C3a. When C3b is produced, the thioester is cleaved, and then this highly reactive species may bind covalently to targets. Interaction of the zymogen factor B with C3b and subsequent cleavage of factor B by factor D results in formation of the alternative pathway C3 convertase C3bBb. This set of reactions represents an amplification loop. A series of complement regulators including factor H and membrane cofactor protein prevent feedback via this loop by increasing the rate of dissociation of C3bBb and/or by serving as cofactors for the serine protease factor I to cleave C3b. The gene encoding C3 is located on chromosome 19 and consists of 41 exons, 16 of which encode the β chain and 25 the α chain. Mutations in C3 have been described in ∼ 10% of aHUS.5,20 Functional analysis of 5 of the 9 mutations has revealed increased resistance to complement regulation, and in another 2, the mutations resulted in a decreased secretion of C3.20 The mechanism by which impaired C3 secretion results in aHUS is as yet unclear.
Thrombomodulin
Thrombomodulin is a key component of the protein C anticoagulation pathway, which facilitates the activation of protein C by thrombin. Additionally, it enhances thrombin-mediated activation of plasma procarboxypeptidase B (TAFI), an inhibitor of fibrinolysis that also inactivates complement-derived anaphylatoxins C3a and C5a. Thrombomodulin has been shown to down-regulate the alternative pathway of complement by accelerating factor I–mediated inactivation of C3b in the presence of cofactors. Mutations in the gene encoding thrombomodulin have recently been shown to predispose to aHUS. These mutations resulted in a loss of cofactor activity.21
Factors affecting the development of aHUS
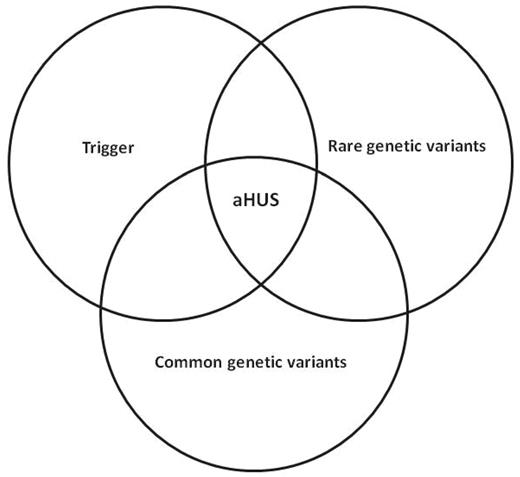
Characteristic of the familial form of aHUS is that ∼ 50% of individuals will not manifest aHUS despite carrying a mutation in one of the aforementioned genes. Two other factors are thought to determine the development of the disease (Figure 3). First, in most patients there is a trigger, with infection and pregnancy being described most frequently.5,22 Second, a further genetic variant (modifier) can increase the risk of developing the disease. This can be in the form of either an additional mutation in one of the aforementioned genes and/or the presence of a common at-risk genetic variant. It is now recognized that ∼ 20% of aHUS patients will have mutations in more than one gene. Common at-risk genetic variants (single nucleotide polymorphisms and haplotype blocks) in CFH, CD46, and CFHR1 have been shown to act as susceptibility factors for the development of the disease.15,23 Therefore, the presence of a rare genetic variant (mutation), a common at-risk genetic variant (single nucleotide polymorphisms and haplotype blocks), and a trigger may be necessary for the disease to be manifest.24
Manifestation of aHUS in an individual may need the presence of a trigger such as pregnancy, a rare genetic variant such as a mutation in a complement gene, and a common genetic variant such as an at-risk haplotype in a complement gene.
Manifestation of aHUS in an individual may need the presence of a trigger such as pregnancy, a rare genetic variant such as a mutation in a complement gene, and a common genetic variant such as an at-risk haplotype in a complement gene.
Investigation and treatment of aHUS
The increased understanding of the molecular mechanisms responsible for the development of aHUS have underpinned the development of national and international guidelines for the investigation and treatment of this disease.25–27 Table 1 shows the diagnostic tests recommended in patients presenting with the clinical features of aHUS.
Currently, plasma exchange and/or plasma infusions is the recommended first-line management for aHUS. Plasma exchange is commonly undertaken daily using 1-2 plasma volumes per session in adults and 50-100 mL/kg in children. Typically, plasma exchange is undertaken daily initially, and then the duration and frequency of treatment is determined by the clinical response. Patients with abnormalities in soluble regulators such as factor H respond better to plasma exchange than patients with abnormalities in the transmembrane regulator MCP.5 Despite treatment with plasma therapy, the majority of patients progress to end-stage renal failure within 3 years of presentation.5 Whereas no problems have been reported with the use of hemodialysis and peritoneal dialysis in aHUS, the use of transplantation has been controversial. The risk of recurrence of HUS in an allograft in all patients is ∼ 50% but is significantly greater (∼ 80%) in patients known to have a mutation in either CFH or CFI.28 In contrast, the outcome for patients known to have only an MCP mutation is favorable.29 Because both factor H and factor I are produced mainly by the liver, combined liver/kidney transplantation is a logical therapeutic option. Whereas the outcome in initial reports of this modality was less favorable,30 more recent reports31–33 suggest that a favorable long-term outcome is possible. Pivotal to this is the use of prophylactic plasma exchange immediately before surgery.25 Another option is to undertake a renal transplantation alone with prophylactic plasma exchange preoperatively, postoperatively, and long-term. Whereas this approach has been used successfully,34 recurrent disease can occur and patients may become intolerant of plasma exchange.
For some time, there has been enthusiasm for the potential use of complement inhibitors in aHUS and it is probable that this will be realized within years. Recent anecdotal case reports of the use of the anti-C5 monoclonal antibody eculizumab in aHUS have been encouraging. In total, there have been 9 published reports.35–43 The patients' age ranged from 18 months to 32 years. In 5 of these patients, eculizumab was used to treat recurrent disease after renal transplantation (4 showed complete recovery and 1 partial). In 3 patients, eculizumab was given for aHUS affecting their native kidneys (2 showed complete recovery and the other partial recovery). The results of trials of eculizumab currently being undertaken in aHUS patients sensitive to and resistant to plasma therapy are eagerly awaited. If these results confirm the efficacy and safety of eculizumab, it is possible that in the future it may be possible to not only prevent new patients from developing end-stage renal failure, but may also enable patients on dialysis to undergo transplantation successfully.
Future directions
Whereas our understanding of the molecular mechanisms underlying the pathogenesis of aHUS has increased dramatically in the last decade and a half, there are still many questions unanswered. In 30% of patients, no abnormalities affecting any of the aforementioned complement genes (or the proteins that they encode) can be found. We envisage that studying families in which there is more than one affected member with techniques such as whole exome sequencing will be particularly informative. We anticipate that additional genomic disorders affecting the genes encoding factor H and the factor H–related proteins will also be found. Whereas it is probable that “new” aHUS genes will be in complement pathways, it is also possible that other pathways such as coagulation may be implicated. Why the presence of factor H autoantibodies in aHUS is associated with homozygous deletion of CHFR1 and CFHR3 is a conundrum that is unresolved. Likewise, the role of factor H autoantibodies in the pathogenesis of the disease remains to be established. In conclusion, aHUS is a paradigm that clearly demonstrates the translational benefits of understanding the molecular mechanisms responsible for the pathogenesis of a disease.
Disclosures
Conflict-of-interest disclosure: D.K. has received research funding from Wellcome Trust and Kidney Research UK. T.H.J.G. has received research funding, honoraria, consultancy fees, and fees as chief investigator and has been affiliated with the speakers' bureau for Alexion Pharmaceuticals. Off-label drug use: Use of eculizumab in atypical hemolytic uremic syndrome.
Correspondence
Professor Tim Goodship, Institute of Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, NE1 3BZ, United Kingdom; Phone: 44-191-241-8632; Fax: 44-191-241-8666; e-mail: tim.goodship@ncl.ac.uk.