Abstract
Several prognostic markers based on genetic, phenotypic, and molecular characteristics of chronic lymphocytic leukemia (CLL) B cells have emerged in the past decade. The clinical utility of these newer prognostic indicators, alone or in combination with each other and other clinical predictive systems, is still being determined. This chapter attempts to define biologic and molecular underpinnings of 3 sets of prognostic indicators in CLL: genetic abnormalities quantified by FISH and/or defined by exploratory sensitive molecular techniques, expression of specific proteins in or on CLL cells (ie, CD38, CD49d, and ZAP-70), and the IGHV mutation status of a CLL clone. Although not demonstrated conclusively, each probably reflects the biologic properties of the leukemic cells of individual CLL patients. This reflection may be direct, indicating a specific property of the CLL cell itself, or indirect, representing how the CLL cell interacts with the host's microenvironment. The new tyrosine kinase inhibitors that are currently in clinical trials support this interpretation. These and other biology-based indicators of patient clinical course and outcome can be used as starting points from which to understand and treat CLL.
Introduction
Over the past decade, several prognostic markers based on genetic, phenotypic, or molecular characteristics of chronic lymphocytic leukemia (CLL) B cells have been brought forth. These have been valuable adjuncts to the time-honored staging systems of Rai and Binet and other parameters that incorporate patient physical findings and clinical laboratory data. The clinical utility of these newer prognostic indicators, alone or in combination with each other and the clinical prognostic systems, is still being analyzed. Indeed, at the last ASH education session on CLL, Richard Furman reviewed this information and the latest guidelines for their use in clinical trials and practice.1 Table 1 lists the prognostic markers currently in clinical use.
A possible difference between the “old” prognostic markers such as staging systems and the “new” prognostic markers such as IGHV mutation status, FISH cytogenetics, and CD38 and ZAP-70 expression is an underlying biological connection to the disease that these new molecular markers reflect. Therefore, the purpose of this chapter is not to readdress in detail the clinical utility of the older versus newer markers, but rather to evaluate the biologic and molecular underpinnings of a few of these in the hope of achieving a better understanding of CLL in general.
This chapter focuses on 3 sets of prognostic indicators: (1) genetic abnormalities quantified by FISH and/or defined by exploratory and more sensitive molecular techniques, (2) expression of specific proteins in or on CLL cells (ie, CD38, CD49d, and ZAP-70), and (3) the IGHV mutation status of a CLL clone. Although it has not yet been demonstrated conclusively, each of these is thought to be a reflection of the biologic properties of individual CLL patients, either directly, indicating a specific property of the CLL cell itself, or indirectly, representing influences of the host's microenvironment on the CLL cell.
Finally, prognostication in patients with CLL should not only address disease progression and overall survival, but also response to therapy.2,3 Therefore, there are important conceptual differences between prognostic factors, which are typically evaluated at the time of diagnosis, and an ongoing evaluation that could lead to more accurate “response predictors” capable of assessing the risks, for example, of targeted therapy. A complete discussion of this distinction is outside of the scope of this manuscript; the reader is referred to Moreno and Montserrat2 and Zenz et al3 for further information.
Genetic abnormalities quantified by FISH and/or defined by exploratory and sensitive molecular techniques
Because adaptation of the FISH technique to the clinical armamentarium has proven valuable in a wide range of disease settings, this approach is probably the most frequently used and widely available for the clinical care of CLL patients. A set of specific defined chromosomal abnormalities has predictive value for patient course and outcome.4 Listed in order of increasing disease severity, these include del(13q), tri12, del(11q), and del(17p).
Deletion at 13q14
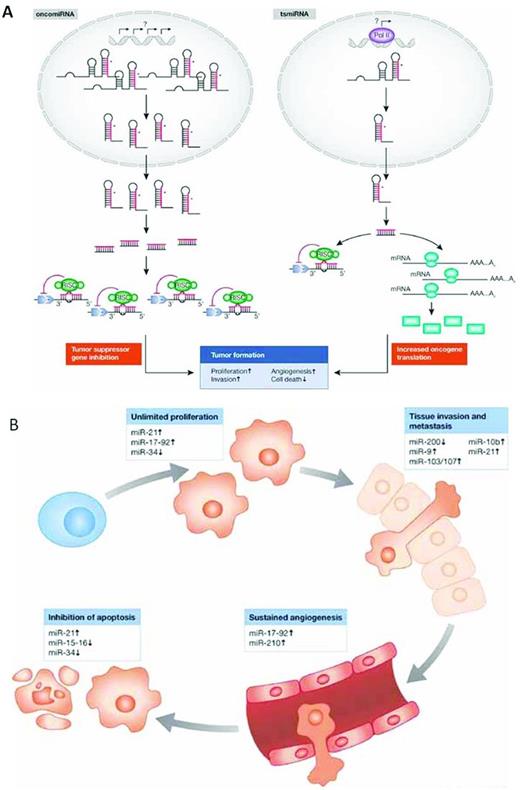
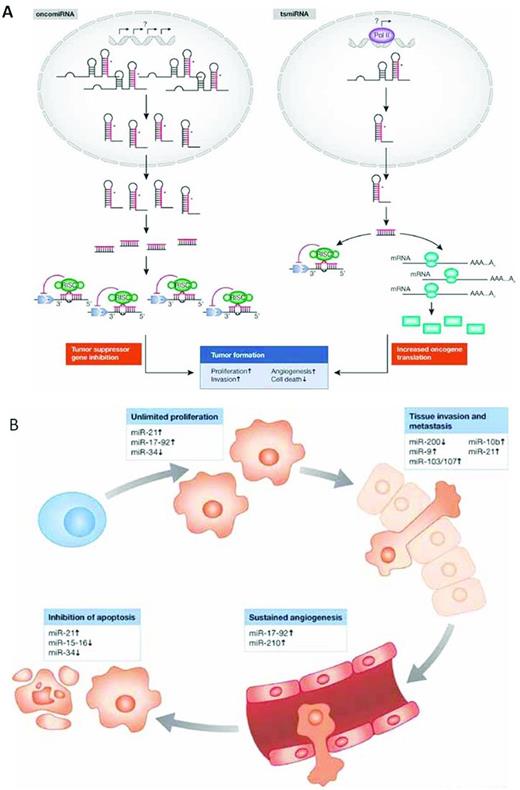
The 13q14 deletion is the most common genetic abnormality in CLL,4 found more often in patients with mutated IGHV genes (M-CLL), a subset with a more favorable clinical outcome.5,6 The critical gene(s) within this site were elucidated when Calin et al identified a relatively small interval within the DLEU2 gene that was lost in the majority of cases and that coded miR15a and miR16-1.7 miRs are small RNA molecules that alter mRNA and protein expression by degrading mRNAs or inhibiting translation to protein. Based on prediction algorithms, miRs act on a large number of target mRNAs, and the experimental validation of these predictions for particular miRs is under way. Nevertheless, because the number and diversity of target mRNAs for any given miR is large, the same miR can lead to distinct and opposing functional outcomes. Depending on the context in which an individual miR is expressed and the expression of members of its target panel, miRs can function as oncogenes or tumor suppressors (Figure 1A). Therefore, the consequences of the deletion of the miR15a/16 cluster on chr13, which leads to up-regulation of BCL2 and enhanced production of Bcl2,8 indicates that these miRs normally function as tumor suppressors.
Functions of miRs in the prevention and development of cancer. (A) miRs as oncogenes or tumor-suppressor genes. (B) miRs target the hallmark features of cancer. Used with permission from Iorio et al.87
Functions of miRs in the prevention and development of cancer. (A) miRs as oncogenes or tumor-suppressor genes. (B) miRs target the hallmark features of cancer. Used with permission from Iorio et al.87
Recently, the miR15a and miR16-1 deletion has been modeled in mice.9,10 After eliminating the miR15a/16-1 cluster or the originally defined minimal region of loss that encompasses DLEU2 or a more extensive deletion that included both of the former plus additional downstream DNA, CD5+ leukemias resembling CLL developed. Tumor development and aggressiveness followed the extent of genetic loss. Therefore, elimination of only the miR15/16 cluster led less often to a CLL-like disease, whereas elimination of the largest region yielded the most penetrant and damaging disease. Remarkably, an “experiment of nature” consistent with the above 2 observations, the spontaneous mutation in the murine miR16 gene of NZB mice,11 can lead to a CLL-like malignancy.
How, then, does this information affect the prognostic value of the 13q14 deletion and the finding that the greater the loss, the worse the impact on survival? First, because the miR15a/16 cluster appears to regulate Bcl2 protein production,8 its loss would make the host CLL cell more resistant to apoptotic deletion. Furthermore, because the findings in the miR15/16–knockout mice suggest that this region also controls proliferation,9,10 a growth advantage of this lesion is also a possibility. The different functional actions of the same miRs in these functional studies are not unexpected because of the range of targets that individual miRs can affect. This also might help to explain the fact that patients with only a monoallelic loss at 13q14 can have a longer life span than those with a normal FISH panel analysis.4 Because del(13q14) occurs more frequently in M-CLL patients, the coding and noncoding gene expression panel of these cells may minimize the pathogenicity of the evolving clone; therefore, an analysis of the outcome of M-CLL and unmutated CLL (U-CLL) patients with monoallelic loss at 13q14 might be informative. Alternatively, as has been suggested by analyses of CLL cohorts12 and mice in which miR15a/16 plus an even greater deletion has been created,9,10 the differences in patient outcome may be due to the loss of other genes on chr13. Furthermore, because in patients the same 13q deletion is not seen in all members of the leukemic clone, the deletion could be associated at times with another abnormality that confounds survival analysis.
Several other miRs have been found to play prognostic roles in CLL and other cancers. In particular, a 13-gene signature including miR-29 and 223 has been identified13 and corroborated,14 and miR34a added to the list15 for CLL. These miR signatures are generally correlated with other prognostic markers such as unmutated IGHV, or in the latter case with del(17p) and loss of p53 function, and with disease progression and refractory disease.
tri12
CLL patients with trisomy 12 (tri12; approximately 15% of CLL patients) have a somewhat shorter survival than those with a normal FISH panel analysis. There appears to be an association between tri12 and the presence of mutations in the NOTCH1 gene, which extend the half-life of the protein. Furthermore, patients with a NOTCH1 mutation have a shorter time to therapy and overall survival,16,17 whereas those with combined tri12 and NOTCH1 mutation fair worse.18,19 Cells from patients with tri12 and concurrent NOTCH1 mutations may be more resistant to apoptosis-induced cell death, leading to a less favorable course.
Deletion at 11q22-23
CLL patients with the 11q22-23 deletion (approximately 15% of CLL patients) often exhibit bulky adenopathy and experience an aggressive clinical course with shorter survival.4 Furthermore, these patients are often U-CLL, which is consistent with their poor clinical outcome.5,6 Nevertheless, these patients may respond to treatment with chemoimmunotherapy, although they inevitably relapse.20
The minimal region of deletion on 11q22.3-23.1 often involves the Radixin (RDX) and Ataxia telangiectasia mutated (ATM) genes.21 Because Atm is crucial for DNA repair, deletion of this gene would likely lead to enhanced clonal aggressiveness and evolution due to the acquisition of novel genomic variants, as suggested by serial genetic analyses.22–24
Of major interest is how deletion at 11q results in bulky adenopathy that can involve the abdomen, an unusual feature in CLL. No revealing information is available regarding this physical sign, and determining its genetic cause might shed light on cell trafficking or interactions of cells with stromal elements in lymph nodes.
del(17p)
CLL patients with the 17p deletion [del(17p); approximately 7% of CLL patients] almost invariably have aggressive clinical courses,4 most likely because of loss of TP53. Furthermore, mutations of TP53 on the other allele occur in approximately 80% of patients and lead to dire outcome.25 This deletion is a prime facilitator of clonal evolution to a more aggressive disease, and the number of leukemic cells with del(17p) often increases in patients who do not respond to therapy or who relapse after therapy.26,27
TP53 abnormalities appear to alter levels of miR34a,15 providing a convergence between the principle that miR abnormalities are important in CLL and the master regulator p53. Lower miR-34a levels due to either deletion or mutation in TP53 alter the repair of DNA lesions and correlate with fludarabine-resistant disease.15
Newer techniques to find genetic abnormalities
Although FISH analyses of these and other chromosomal aberrations have been extremely helpful in predicting clinical course and indicating ongoing evolution of the leukemic clone, the FISH approach is limited by the fact that it can only detect lesions that have already been defined in other patients. For this reason, new and more sensitive techniques that define abnormalities heretofore unrecognized in CLL have great potential and have begun to yield valuable information. Three such techniques are comparative genomic hybridization (CGH), genome-wide analyses of single nucleotide polymorphisms (SNPs), and whole-exome or whole-genome sequencing (“next-generation sequencing” [NGS] or “deep sequencing”). Although promising, such techniques are still investigational, and their application and integration into clinical practice will require a meticulous process of validation.
Prognostic information provided by CGH and SNP analyses
CGH and SNP analyses12,22,23,28–30 have revealed the near universality of genomic aberrations in CLL, even at diagnosis and before therapy. These abnormalities, which are more often losses than gains of DNA, are found more frequently in U-CLL, ZAP-70+, and CD38+ patients. M-CLL patients who eventually require treatment can have as many DNA lesions as U-CLL patients. Because the number and breadth of these abnormalities are correlated with an unfavorable clinical course, the degree of genetic complexity is an indicator of shorter time to treatment and survival.
Whole-exome or whole-genome NGS
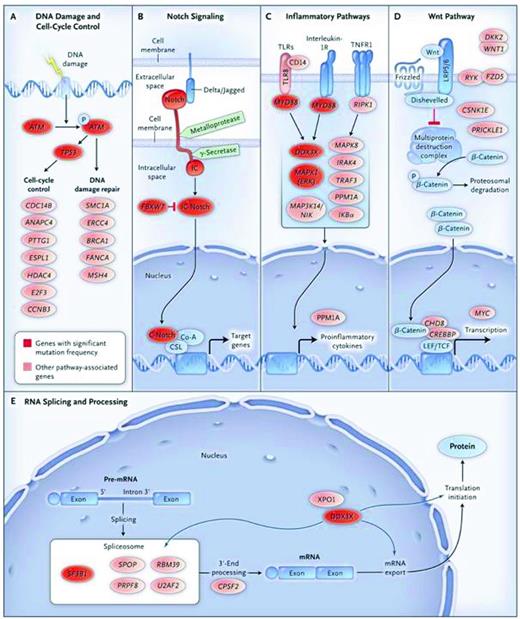
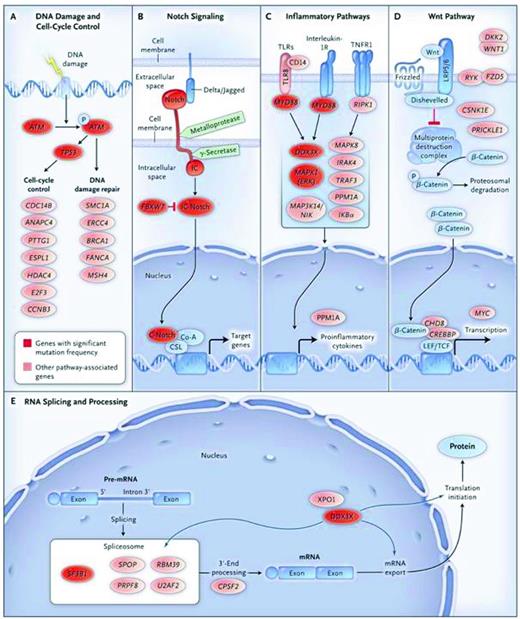
Using primarily whole-exome sequencing of CLL cells, the low but significant level of genomic complexity in CLL has been documented previously.16,17,31,32 Adding to this complexity, 18 recurrently mutated genes have been identified thus far: NOTCH1, Exportin 1 (XPO1), MYD88, Kelch-like 6 (KLH6), TP53, TGM, BIRC3, PLEKHG5, ATM, splicing factor 3, B1 unit (SF3B1), ZMYM3, MAPK1, FBXW7, and DDX3X. These mutations coexist with some of the genetic abnormalities analyzed by FISH: SF3B1 mutations with del(11q), NOTCH1 with tri(12), and MYD88 with del(13q). Furthermore, several of these mutations segregate based on biologic functions: TP53 and ATM link with the cell cycle and DNA repair, NOTCH1 and FBXW7 with inflammation, and SF3B1 and DDX3X with RNA processing (Figure 2). Finally, the mutations likely lead to changes in amino acids and protein structure, strongly suggesting clonal selection of the mutations in vivo. For example, there was a clear relationship between the NOTCH1 mutation and poor patient outcome, resistance to therapy, and transition to Richter's transformation. Therefore, CGH, NGS, and SNP analyses have confirmed an existing and ongoing level of genomic complexity in CLL and revealed new mutations, some of which associate with the more common losses/gains known in the past. It is likely that our understanding of the genetics of CLL, how it relates to CLL cell biology, and its impact on clinical course, outcome, prognostication, and response to specific therapies will increase dramatically in the near future.
Signaling pathways elucidated by NGS in CLL. Nine genes with significant mutation frequencies identified by Wang et al31 fall into 5 core signaling pathways in which the genes play well-established roles: DNA damage repair and cell cycle control (A), Notch signaling (B), inflammatory pathways (C), Wnt signaling (D), and RNA splicing and processing (E). Genes with significant mutation frequencies are shown in red, and genes with mutations that are in a signaling pathway related to CLL are shown in pink. Used with permission from Wang et al.31
Signaling pathways elucidated by NGS in CLL. Nine genes with significant mutation frequencies identified by Wang et al31 fall into 5 core signaling pathways in which the genes play well-established roles: DNA damage repair and cell cycle control (A), Notch signaling (B), inflammatory pathways (C), Wnt signaling (D), and RNA splicing and processing (E). Genes with significant mutation frequencies are shown in red, and genes with mutations that are in a signaling pathway related to CLL are shown in pink. Used with permission from Wang et al.31
Expression of specific proteins in or on CLL cells
Expression of CD38 and CD49d on5 and ZAP-70 in33 CLL cells has proven valuable in predicting outcome in CLL. Like FISH, immunofluorescence is well established in clinical laboratories and therefore the approach is widely used. Although ZAP-70 protein is likely the most accurate of the 3 markers in its prognostic capacity,33 its detection by immunofluorescence and quantification by flow cytometry is more complicated because of intracellular staining and gating approaches that have limited general use to specialty laboratories. Combining these markers in various ways may provide more robust prognosis,34,35 suggesting a network of interactions among these molecules.36 Unlike the aberrations defined by FISH that generally indicate abnormalities intrinsic to CLL leukemic B cells, these markers reflect the ability of CLL cells to respond to signals from the microenvironment.
CD38
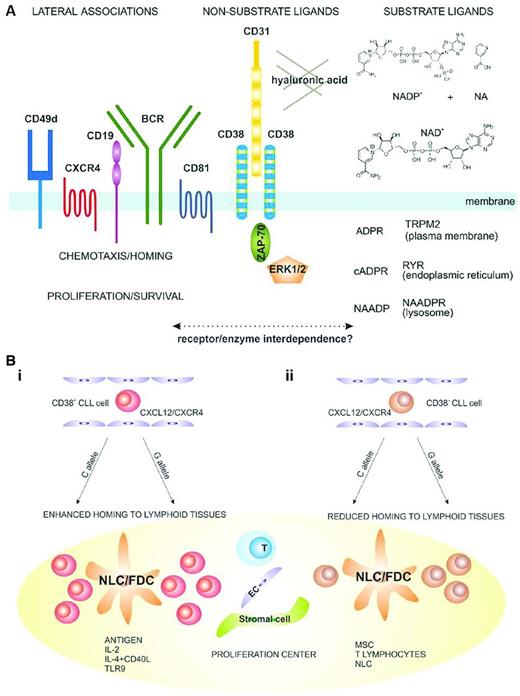
Cell-surface CD38 has a pivotal role in initiating and modulating a series of input signals from the microenvironment (Figure 3A). For several reasons, the percentage of cells within a CLL clone that display CD38 is an indicator of the potential and actual degree of cellular activation of the clone: those with higher numbers (than a defined percentage) are more responsive to activation signals or are activated and are therefore more often more aggressive. For example, CD38+ fractions of CLL clones are enriched in cells expressing Ki-67 and ZAP-70.37 Furthermore, analyses of CLL cells from patients using incorporation into cellular DNA of deuterium (2H) administered in vivo in the form of heavy water (2H2O) suggest that within individual clones CD38+ cells proliferated more recently than CD38− cells.38 CD38+ clones respond more effectively to surface membrane immunoglobulin (smIg) cross-linking,35,39–41 and this process involves ZAP-70. Therefore, CD38 expression is a measure of cell division and a reflection of growth in vivo. The aggressiveness of CD38+ cells appears to be compounded by their ability to migrate and take advantage of interactions with the microenvironment (Figure 3B); ZAP-70 also appears to be involved in this action.42
Structural and functional considerations for the role of CD38 in normal and CLL B cells. (A) Structural and functional characteristics of the human CD38 molecule. CD38 is expressed as an integral surface membrane molecule on B lymphocytes, often in a dimeric conformation. Because it is an ectoenzyme, CD38 may interact with the substrate ligands NAD+ and NADP+, which are converted to the intracellular Ca2+-mobilizing agents cADPR, ADPR, and NAADP. CD38 also interacts with nonsubstrate ligands, including CD31 and hyaluronic acid, which regulate cell-cell and cell-matrix contact. CD38 is preferentially localized in membrane lipid microdomains in close contact with the BCR complex (CD19/CD81) and with molecules regulating homing (CXCR4 and CD49d). CD38 engagement by natural (CD31) or surrogate (agonistic mAb) ligands triggers activation of intracellular signaling pathways that include ZAP-70 and ERK1/2 as major players. These signals increase chemotaxis and proliferation of neoplastic B cells. (B) Proposed model explaining a role of CD38 in the pathogenesis and progression of CLL. (Bi) CD38+ CLL cells (red) are more sensitive to CXCL12 signals and have a higher propensity to home to lymphoid tissues than their CD38− counterparts (brown). (Bii) Once inside lymph node proliferation centers, CLL cells come into contact with accessory cells such as nurse-like, follicular dendritic, stromal, endothelial, mesenchymal, and T cells. The presence of Ag and accessory signals leads to proliferation and potentially acquisition of novel genetic lesions, promoting clonal evolution and disease progression. These events are more apparent in the CD38+ subsets. Used with permission from Malavasi et al.42
Structural and functional considerations for the role of CD38 in normal and CLL B cells. (A) Structural and functional characteristics of the human CD38 molecule. CD38 is expressed as an integral surface membrane molecule on B lymphocytes, often in a dimeric conformation. Because it is an ectoenzyme, CD38 may interact with the substrate ligands NAD+ and NADP+, which are converted to the intracellular Ca2+-mobilizing agents cADPR, ADPR, and NAADP. CD38 also interacts with nonsubstrate ligands, including CD31 and hyaluronic acid, which regulate cell-cell and cell-matrix contact. CD38 is preferentially localized in membrane lipid microdomains in close contact with the BCR complex (CD19/CD81) and with molecules regulating homing (CXCR4 and CD49d). CD38 engagement by natural (CD31) or surrogate (agonistic mAb) ligands triggers activation of intracellular signaling pathways that include ZAP-70 and ERK1/2 as major players. These signals increase chemotaxis and proliferation of neoplastic B cells. (B) Proposed model explaining a role of CD38 in the pathogenesis and progression of CLL. (Bi) CD38+ CLL cells (red) are more sensitive to CXCL12 signals and have a higher propensity to home to lymphoid tissues than their CD38− counterparts (brown). (Bii) Once inside lymph node proliferation centers, CLL cells come into contact with accessory cells such as nurse-like, follicular dendritic, stromal, endothelial, mesenchymal, and T cells. The presence of Ag and accessory signals leads to proliferation and potentially acquisition of novel genetic lesions, promoting clonal evolution and disease progression. These events are more apparent in the CD38+ subsets. Used with permission from Malavasi et al.42
Although, in general, CD38 levels do not move above the threshold that classifies a clone as CD38+ or CD38−, when the levels do transgress the boundary for positivity, this is usually associated with more virulent, progressive disease.42 Depending on the research group, the threshold at which a clone is defined to be negative or positive ranges from 7%-30%. This range may reflect either technical differences between individual laboratories or other, yet to be defined variables; this issue is also relevant for other markers to be discussed below (CD49d and ZAP-70).
The greater proliferative potential of CD38+ clones and cells increases the likelihood that new genomic abnormalities will occur at the time of DNA replication, consistent with finding more cells with 11q and 17p deletions43 and clonal evolution12,22,23,28–30,44 in CD38+ clones and explaining the poor prognosis of patients with clones with higher numbers of CD38+ cells.
CD49d
The percentage of CD49d+ cells, like CD38+ cells, is an independent indicator of prognosis in CLL, with higher levels (≥ 30%) being correlated with shorter survival times. CD49d is an α-integrin subunit (α4) that can pair with CD29 (the β1 subunit) to form a complete integrin (α4β1) that binds fibronectin and VCAM-1. Like other integrins, α4β1 is involved in anchoring cells to tissues via extracellular matrix, which can result in cell survival and can also be a first step in cell migration; the latter function is important for CLL cells and for the prognostic relevance of CD49d expression.
CD49d and CD38 are often expressed on CLL B cells,45–47 and a large macromolecular complex comprising CD49d, CD38, CD44v, and MMP-9 has been identified on U-CLL clones,48 bringing these prognostic markers into a presumptive biologic network.49 Indeed, recent studies have shown that CD49d/CD29 are physically and functionally linked with CD38 on CLL cells.50 This may be especially important in helping both molecules carry out their biologic functions.
ZAP-70
Intracellular expression of the ZAP-70 protein above a certain threshold of cells by immunofluorescence and flow cytometry (≥ 20%) has proven to be an important indicator of time-to-treatment and survival in CLL.33 Although the numbers of CLL cells expressing this protein are correlated with IGHV mutation status and CD38 expression,51,52 ZAP-70 levels are an independent marker of clinical outcome.33 Clinical course is correlated with augmented signaling down the BCR pathway, albeit not necessarily due to its enzymatic actions.53,54 Recent studies suggest that ZAP-70 retards internalization of smIgM and CD79b from the cell membrane, leading to prolonged BCR pathway signaling.53,55 In addition, ZAP-70+ CLL cells are more likely to express adhesion molecules such as CD49d and chemokine receptors, in particular CCR7,55 that promote migration toward a series of chemokines56–58 and inhibit apoptosis.59 The latter suggest that an important component of ZAP-70 expression is trafficking to solid tissue niches where signaling through chemokine receptors and BCRs might promote survival and further proliferation.
Linking to clonal evolution
Based on the prognostic value of CD38, CD49d, and ZAP-70, it appears that interactions with the microenvironment via a series of cell-surface receptors provide important signals for cell proliferation and survival. Indeed, one could envision independent and concerted actions of these 3 molecules and their signaling partners promoting tumorigenesis. Furthermore, ZAP-70 and CD38 are both activation markers of normal B cells, and CLL cells express these markers due to their activation status. At least a portion of this activation is probably due to signaling (BCR and other receptors) and is also possibly dependent upon signals delivered by cytokines. The response to various cytokines by CLL cells, whether due to interactions with other cells in the microenvironment or to signaling, has consequences on clonal expansion and regulation. The biological significance of signaling is especially relevant for the prognostic indicator IGHV mutation status, which is discussed next.
IGHV mutation status
The clinical utility of IGHV mutation status has been well established in multiple clinical trials, with a high correlation between unmutated IGHV status and poor survival and, correspondingly, better prognosis in cases with mutations in IGHV.5,6 The biologic correlates of the prognostic utility of IGHV mutations in CLL have been linked to BCR structure, with the key findings being the differences in the presence or absence of significant numbers of IGHV mutations5,6 ; the use of specific IGHV genes, which is in general,60–62 but not always,63 linked to IGHV mutation status; and the presence of stereotyped BCR structures.64–68 The latter are defined as BCRs having amino acid structures that, in particular at the third complementarity determining region (CDR) of the IGH (HCDR3), are remarkably similar among patients, often because of the use of the same IGHV or IGHV family members linked to identical IGHD and IGHJ genes.64–67
The common link between these 3 BCR structural parameters is believed to be the ability of the CLL precursor cell and the full-fledged CLL cell to bind autoantigens in vivo and then to respond in a manner that supports survival and clonal growth versus anergy/apoptosis.69,70 In normal B lymphocytes, mutations in the IGH variable domain genes (IGHV-D-J) are desired outcomes that permit selection of an altered specific BCR structure and higher-affinity Ag binding. Therefore, it is assumed that CLL B cells with BCRs containing somatic mutations were selected in such a process before leukemic transformation for reactivity with a restricted set of antigenic epitopes. Because patients with IGHV mutations have superior clinical courses, better outcome implies either that their BCRs have better binding to a few autoantigens or no binding to Ags usually encountered by the cell. In the former scenario, anergy might result due to high-affinity BCR-Ag interactions,71 and for the latter scenario, Ag-binding neglect might lead to apoptosis due to a lack of BCR-derived signals69,70 that are needed to keep B lymphocytes alive.
In contrast, BCRs without somatic mutations are likely to have an amino acid structure similar to that encoded by germline variable domain gene segments (IGHV-D-J). Therefore, these receptors would likely bind a wider array of antigenic epitopes,72–75 albeit most likely with lower affinity than those with mutated IGHV-D-Js. Patients without IGHV mutations typically have inferior clinical courses. This poor clinical outcome seems to be correlated with these patients' BCRs binding many autoantigens at variable affinities76 ; therefore, multiple binding potential could result in the survival and growth of the leukemic clone (as opposed to anergy/apoptosis).
Ag binding usually involves the physical association of the Ig heavy chain CDRs (HCDR) of the IGHV-D-J amino acid rearrangement with epitopes of a classical Ag. Furthermore, the CDRs of the Ig light chain IGLVκ-Jκ (LCDRκ) or IGLVλ-Jλ (LCDRλ) rearrangement often increase classical Ag-binding efficiency. In this context, patients with stereotyped BCRs, in whom IGHV-D-J rearrangements and their CDRs are shared, would be expected to be selected for Ag binding. However, because most, albeit not all, CLL clones with stereotyped BCRs do not carry somatic mutations,68 these B cells are either of a type that cannot carry out the IGHV-D-J somatic hypermutation process or they bind Ags that do not turn on the process (eg, autoantigens or T-cell–independent Ags). It is important to keep in mind that, except in the setting of autoimmunity, T lymphocytes, which are usually necessary to initiate somatic hypermutation, are tolerant to autoantigens displayed on B cells or other APCs. Therefore, for patients with stereotyped BCRs, especially those of an unmutated variety, binding of autoantigens or exoantigens (eg, microbial Ags are often carbohydrate in composition), is expected to be T-cell independent.69 Because autoantigens are ubiquitous in the host microenvironment, their binding could occur often, especially because Igs with unmutated IGHV-D-J rearrangements are often able to bind several apparently distinct Ags,72–75 but the consequence would not be IGV gene diversification.
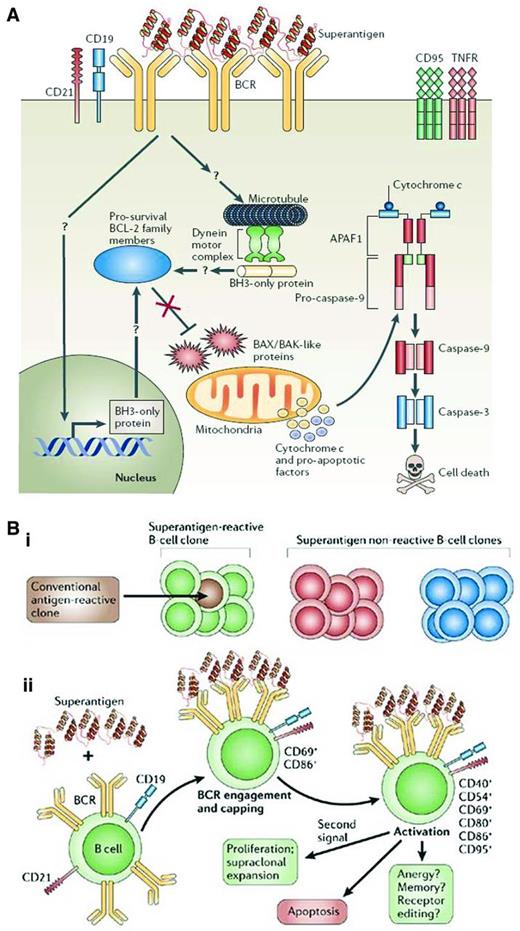
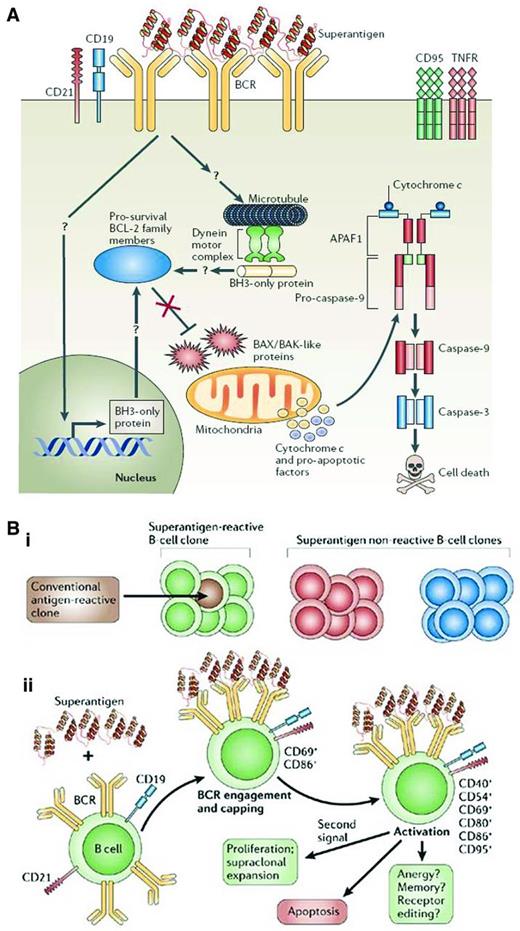
How is IGHV gene bias incorporated into this scenario? Because the best binding of classical Ags involves interactions of the HCDRs, use of the same IGHV gene but not the same IGHD and IGHJ segments that form the third CDR (HCDR3) among patients might not maximize classical Ag binding. In this case, if Ag binding and its consequences are the biologic basis for survival and growth of CLL cells, then one needs to incorporate binding of nonclassical Ags to BCRs of such structure. “Superantigens” are epitopes found most often on molecules from microbes that bind to conserved amino acids in BCRs that are usually found in framework regions of IGHV-D-J amino acid rearrangements (Figure 4A). Unlike classical Ag binding, which occurs between an Ag and the Ab's generally highly variable CDRs, superantigens bind the relatively more constant framework regions. This makes it possible for them to bind a larger variety of Abs, and therefore B lymphocytes, in a nonclonally restricted manner because CDR structure does not affect binding in most instances. Therefore, superantigen engagement for BCRs with common IGHVs but different IGHDs and IGHJs could be relevant for these CLL patients. It is interesting that such IGHV gene biases are seen in both U-CLL and M-CLL patients. Therefore, in M-CLL patients, critical amino acids in the IGHV protein would need to be spared replacement mutations that might alter superantigen-BCR interactions, and for at least some superantigens, this is the case in CLL.77
Consequences of superantigen binding and signaling to B lymphocytes. (A) Superantigen-induced cell death through BCR engagement. Upon engagement by a superantigen, the BCRs are recruited into complexes that include CD19, CD21, and other coreceptors, which results in signaling events for cellular activation. When these interactions occur in the absence of a “second signal,” they can cause a series of intracellular events that could lead to changes in mitochondrial membrane potential and ultimately to the release of cytochrome c and proapoptotic factors that can lead to DNA fragmentation and nuclear fragmentation. (B) Susceptibility and outcome of B-cell superantigen exposure. (Bi) Superantigen can interact with all clones of a particular family (green) no matter their conventional Ag specificity. Conventional Ags, however, can interact with only certain clones having defined binding specificities (brown) and of a particular family (green). (Bii) On initial exposure to a superantigen, the BCRs of all susceptible clones are recruited into complexes, followed by clustering of the CD21 and CD19 coreceptors and the resulting activation of the B cell, associated with up-regulation of CD69 and CD86 expression. Later activation events include up-regulation of CD40, CD54, CD80, and CD95. Migration of B cells to the spleen is observed at early time points. Several outcomes follow, a main one being apoptosis, although with an appropriate second signal such as CD40 ligand or IL-4, survival and proliferation may result. Based on B-cell responses to conventional Ags, differentiation of clones into memory cells, functional inactivation (anergy), or receptor editing may occur. Used with permission from Malavasi et al.42
Consequences of superantigen binding and signaling to B lymphocytes. (A) Superantigen-induced cell death through BCR engagement. Upon engagement by a superantigen, the BCRs are recruited into complexes that include CD19, CD21, and other coreceptors, which results in signaling events for cellular activation. When these interactions occur in the absence of a “second signal,” they can cause a series of intracellular events that could lead to changes in mitochondrial membrane potential and ultimately to the release of cytochrome c and proapoptotic factors that can lead to DNA fragmentation and nuclear fragmentation. (B) Susceptibility and outcome of B-cell superantigen exposure. (Bi) Superantigen can interact with all clones of a particular family (green) no matter their conventional Ag specificity. Conventional Ags, however, can interact with only certain clones having defined binding specificities (brown) and of a particular family (green). (Bii) On initial exposure to a superantigen, the BCRs of all susceptible clones are recruited into complexes, followed by clustering of the CD21 and CD19 coreceptors and the resulting activation of the B cell, associated with up-regulation of CD69 and CD86 expression. Later activation events include up-regulation of CD40, CD54, CD80, and CD95. Migration of B cells to the spleen is observed at early time points. Several outcomes follow, a main one being apoptosis, although with an appropriate second signal such as CD40 ligand or IL-4, survival and proliferation may result. Based on B-cell responses to conventional Ags, differentiation of clones into memory cells, functional inactivation (anergy), or receptor editing may occur. Used with permission from Malavasi et al.42
As a consequence of either classical or superantigen binding to CLL BCRs, the cells could receive signals for survival and growth versus apoptosis (Figure 4B). Because U-CLL cells would have a greater chance of binding Ag(s) than M-CLL cells based on planar versus concave binding clefts, respectively, and because U-CLL cells more readily transduce signals through smIgs than M-CLL clones,35,40,78 BCR signaling would be intimately linked to CLL cell biology, clinical course, and prognostication.69–71 The striking beneficial responses seen recently after inhibiting the Syk, Btk, and PI3Kδ kinases79–81 appear to validate the importance of BCR signaling in initiating and perpetuating CLL. This topic is addressed in a separate chapter by Dr Adrian Wiestner.
Concluding remarks
Simplistically, CD38, CD49d, and ZAP70 can be viewed as dynamic markers the prognostic value of which is a sign of level of aggressiveness at the time of sampling, whereas IGHV mutation status is a static marker (at the clonal level) that is indicative of the origin and maturational events of a B cell before transformation. Absence or presence of IGHV mutations reflects the type of Ag binding that the BCR accomplishes (eg, polyreactive vs oligo/monoreactive, respectively). Ag binding specificity is correlated with outcome because it indicates the breadth of antigenic epitopes that a BCR can engage, the affinity of these interactions, and the likelihood that survival/proliferation signals are delivered to a CLL cell.
The genetic abnormalities that occur in CLL are more difficult to classify in this manner, although their ability to predict clinical course and outcome are probably more akin to CD38, CD49d, and ZAP70, because subclones bearing such lesions can increase over time and lead to clinical deterioration (clonal evolution). It is likely that, like CD38, CD49d, and ZAP70, chromosomal abnormalities are dynamic markers that are correlated with disease progression. This may not have been apparent at first, because most initial studies were conducted in a retrospective manner on patients at all disease stages and tested at different points in their disease course. Now it appears that chromosomal and genetic abnormalities are less numerous in the early stages of the disease and accumulate progressively in the disease,82,83 and this is likely the case for certain gene mutations identified recently by sensitive screening techniques.
It is tempting to try to link the biologic underpinnings of each of the above prognostic markers into a set of common interactions. This is feasible if the signaling nodes are expanded beyond those immediately identified by the specific markers to other known signaling molecules. This may be especially relevant because of the considerable therapeutic efficacy that the tyrosine kinase inhibitors are showing in the clinic, albeit in studies that are still relatively short (approximately 30 months). For example, although Btk and PI3Kδ inhibition dampens BCR signaling, it also alters the consequences of CXCR4 stimulation.84 Likewise, PI3Kδ is a key player in CD38-mediated signal transduction that affects cell trafficking and survival. Furthermore, Btk likely influences the CD38 pathway in humans, because mice with a genetic defect in Btk are unresponsive to CD38 ligation,85 and this unresponsiveness extends to a series of NF-κB proteins.86 These findings illustrate not only the enormity of signaling network connections, but also the difficulty in pinpointing a single interaction as the crucial event in CLL cell biology or prognostication. This becomes even more complex for the broad effects that the genetic abnormalities that occur in CLL introduce; in particular, miR changes can have widespread actions on cell survival and growth (Figure 1B).
In conclusion, it appears that at least the 3 sets of markers discussed herein, and probably others (eg, lymphocyte doubling time and serum levels of β2-microglobulin and thymidine kinase), reflect direct and indirect influences on the initiation, perpetuation, and accumulation of CLL clones, and it is these effects that translate into prognostic value. Viewed in this manner, the identification of biology-based prognostic indicators are starting points from which to understand CLL and modulate its clinical course and outcome.
Acknowledgments
The author thanks Drs Manlio Ferrarini, Sophia Yancopoulis, and Michael Chiorazzi for their comments and suggestions about the text.
Studies from the author's laboratory cited in this review were supported in part by the National Institutes of Health (RO1 CA81554), the Karches Foundation, the Prince Family Foundation, the Nash Family Foundation, the Mildred and Frank Feinberg Foundation, the Marks Foundation, the Jerome Levy Foundation, and the Leon Levy Foundation.
Disclosures
Conflict-of-interest disclosure: The author declares no competing financial interests. Off-label drug use: None disclosed.
Correspondence
Nicholas Chiorazzi, MD, The Feinstein Institute for Medical Research, 350 Community Dr, Manhasset, NY 11030; Phone: 516-562-1090; Fax: 516-562-1011; e-mail: NChizzi@NSHS.edu.