Abstract
Our genome, the 6 billion bp of DNA that contain the blueprint of a human being, has become the focus of intense interest in medicine in the past two decades. Two developments have contributed to this situation: (1) the genetic basis of more and more diseases has been discovered, especially of malignant diseases, and (2) at the same time, our abilities to analyze our genome have increased exponentially through technological breakthroughs. We can expect genomics to become ever more relevant for day-to-day treatment decisions and patient management. It is therefore of great importance for physicians, especially those who are treating patients with malignant diseases, to become familiar with our genome and the technologies that are currently available for genomics analysis. This review provides a brief overview of the organization of our genome, high-throughput sequence analysis methods, and the analysis of leukemia genomes using next-generation sequencing (NGS) technologies.
The human genome
The haploid human genome contains ∼ 3 billion bp of DNA, amounting to 6 billion bp in a diploid nucleus. The nuclear genome is divided into 46 individual double-stranded DNA molecules, which are visible as chromosomes in metaphase cells. Even though almost all of the complete euchromatic sequence of the human genome has been known since 2001,1-3 we are still very far from understanding the function of the majority of this sequence.
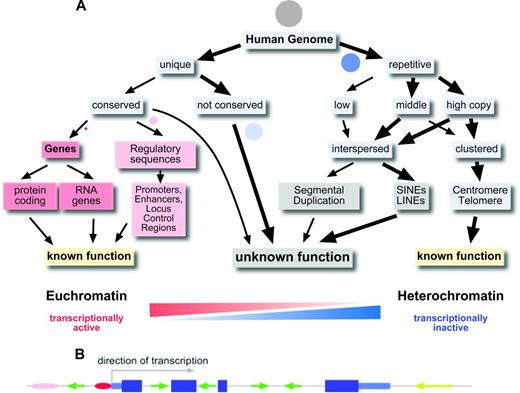
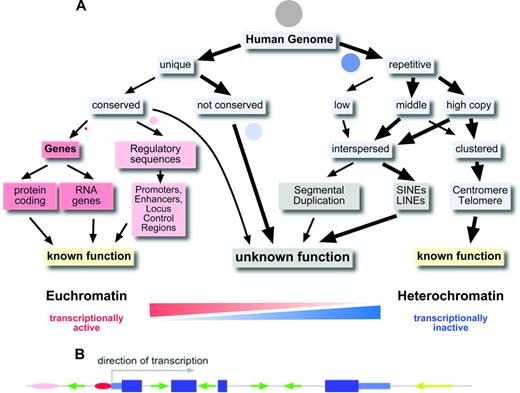
Only 1.5% of our genome, ∼ 45 Mbp, codes for the protein sequences of the classical genes (Figure 1A). There are ∼ 22 000 genes in the human genome, most of them coding for a protein.4 However, it has become apparent that there are also several thousand genes that do not code for proteins, but in which the transcribed and processed RNA itself has a function (eg, miRNA genes, ribosomal RNA genes, and long intergenic noncoding RNAs).4 Some of these RNA genes have unexpected, novel functions, such as the recently described circular RNAs, which regulate the activity of miRNAs.5
The human genome. (A) Organization of the human genome. Circles represent the approximate proportion of the various sequence categories. Grey circle indicates the whole genome. (B) Gene model with exons, introns, and promoter and interspersed repetitive elements. Pink and red ovals indicate enhancer and promoter; dark blue large boxes, coding exons; light blue smaller boxes, 5′ and 3′ untranslated regions; and green and yellow arrows, SINEs and LINEs, respectively.
The human genome. (A) Organization of the human genome. Circles represent the approximate proportion of the various sequence categories. Grey circle indicates the whole genome. (B) Gene model with exons, introns, and promoter and interspersed repetitive elements. Pink and red ovals indicate enhancer and promoter; dark blue large boxes, coding exons; light blue smaller boxes, 5′ and 3′ untranslated regions; and green and yellow arrows, SINEs and LINEs, respectively.
More than 98% of our genome does not code for proteins or is part of functional RNA molecules (Figure 1A). The average human gene codes for a protein of ∼370 amino acids in length that is composed of 7 exons that span ∼ 3 kbp of genomic sequence.3 However, there is a is a huge variation in the size of the proteins for which human genes can code (from ∼ 100 to > 26 000 amino acids), the number of exons a gene has (1-364) and the genomic region a gene can occupy (from < 1 kbp up to 2.2 Mbp; http://en.wikipedia.org/wiki/Human_genome). Up to 8% of our genome, excluding the 1.5% that is protein coding, is highly conserved in evolution and/or contains important regulatory elements such as promoters, enhancers, and locus control regions.4 Some genes are controlled by enhancer elements and locus control regions that can be > 1 Mbp away from the gene.
A prevailing feature of the 90% of the human genome that does not constitute protein coding regions or highly conserved regions is the presence of repetitive elements. Repetitive elements can either occur as clusters of tandem repeats or as interspersed repeats. Overall, > 50% of the human genome can be assigned to repetitive elements (Figure 1A). We know very little about the function of the various repeat elements and the noncoding, nonconserved unique DNA sequences in the human genome. Great efforts are under way to decipher the function of this, sometimes referred to as “junk” DNA.4 The sheer abundance of repetitive elements and noncoding unique DNA sequences in our genome requires that we are aware of these elements when we embark on analyzing changes in the genome that are relevant to hematopoietic malignancies and cancer in general.
Because so little is known about the function of the majority of our DNA sequences, rather than sequencing the complete genome, the transcriptome and the exome have become the focus of interest when analyzing leukemia-associated genetic changes.
Transcriptome
The transcriptome is defined as all of the RNA molecules that are present in a given cell at a given time.6 It is estimated that there are ∼ 300 000 mRNA molecules in a cell. What is really contained in a transcriptome sequence is therefore very much context dependent: on the cell type, the differentiation stage, and also on the way the RNA was isolated and which sequencing library preparation protocols were used. Usually, only the polyadenylated mRNAs are isolated and sequenced. Of the ∼ 22 000 genes in our genome, only ∼ 6000 to 8000 are expressed at significant levels in differentiated cells.6
Exome
The exome is defined as the combined DNA sequence of all exons of protein-coding and RNA genes in the genome. Even though this definition appears to include almost the same sequences as the transcriptome, there are important differences. The transcriptome comprises all RNA molecules in a given cell (everything that is transcribed), so it varies from cell type to cell type. In contrast, the exome is identical for all cells of an organism. In practice, the sequences that are included in an exome will depend on the design of the specific exome capturing kit that is used. The captured sequences usually comprise the sequences from the consensus coding sequence (CCDS) database or an extended set of sequences such as the GENCODE exome target.7 On average, the target of an exome capturing kit is ∼ 50 Mbp in size, or ∼ 1.5% of the whole genome.
Variability of the human genome
Just as every human being is an individual with unique characteristics and talents, so is his or her genome. All of our genomes are “individuals.” This fact has to be borne in mind when we try to identify tumor-specific changes. The variation of the human genome is apparent at all levels: from polymorphic single base pairs up to polymorphic chromosomal features that can be seen in the light microscope.
Single nucleotide polymorphisms
Approximately 1 in every 300 bases in our genome is found to be polymorphic, with an alternative base present in > 1% of the individuals in a population. These so-called single nucleotide polymorphism (SNPs) are so frequent that any 2 individuals will differ at > 3 million SNP locations.8 Although most (> 99%) of these SNPs occur in noncoding regions, there is still a large number of SNPs that affect the coding portion of our genome, and both coding and noncoding SNPs can lead to alterations in the function of proteins.9
Copy number variants
SNPs are easy to detect using sequencing, restriction fragment length polymorphism analyses, and several high-throughput genome analysis tools. However, the variability of our genome is not confined to a single nucleotide at a time. Our genome is not only highly variable at length scales of a few nucleotides (1-5), but also at length scales of several hundred to millions of base pairs. These copy number variations (CNVs) are much more difficult to detect with current methodologies.10 There is an overlap between very low copy number repeats (LCRs), also called segmental duplications, and CNVs. LCRs are often restricted to specific chromosomal regions and can be a few thousand to several hundred thousand base pairs in length. LCRs are estimated to comprise ∼ 5% of the human genome. CNVs in the form of gene duplications can, for example, have important phenotypic consequences such as the increased number of amylase genes found in the genome of the bushmen in southern Africa.11,12
Genome analysis methods
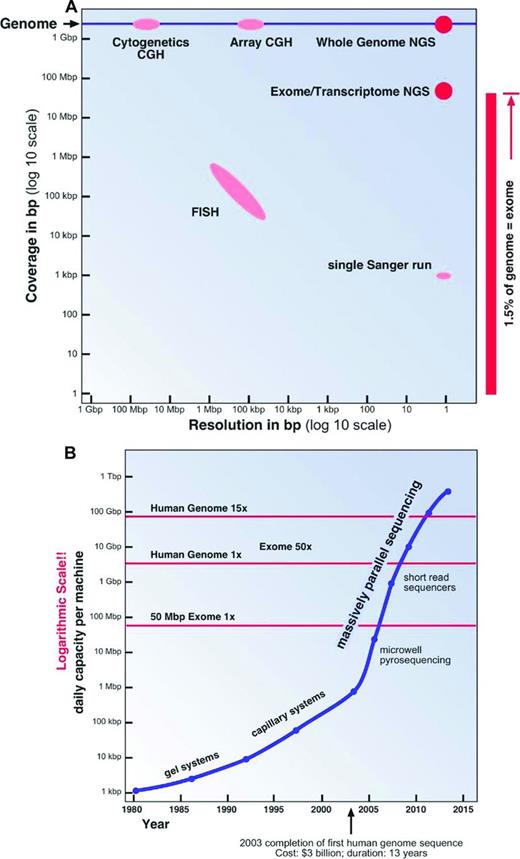
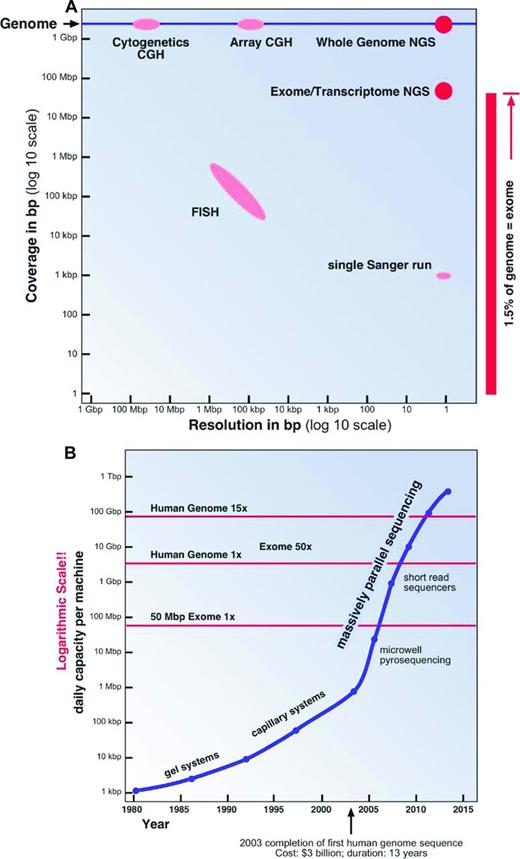
Our technical abilities to analyze the human genome have also shaped the way we perceive the genome and its diversity. Over the past half century, increasingly more sophisticated and powerful genome analysis technologies have been developed. Two important aspects of these technologies have to be considered: resolution and analysis coverage (Figure 2A).
Genome analysis methods. (A) Coverage and resolution. (B) Daily sequencing capacity per machine.
Genome analysis methods. (A) Coverage and resolution. (B) Daily sequencing capacity per machine.
Cytogenetics and molecular cytogenetics and high-throughput array platforms
The most widely used and one of the oldest genome analysis technologies is chromosomal analysis or cytogenetics. In the early 1970s, with the invention of chromosome banding techniques,13 several breakthrough discoveries in the field of leukemia cytogenetics were made; for example, the discovery by Janet Rowley that the t(9;22)(q34;q11) translocation is the cause of the Philadelphia chromosome in chronic myeloid leukemia.14 A chromosomal analysis will visualize the whole genome at a low resolution of ∼ 0.5% to 1% of the genome (15-30 Mbp; Figure 2A). To overcome some of the limitations in resolution and sample requirements of classical cytogenetics, molecular cytogenetics techniques, especially FISH and comparative genomic hybridization, were introduced in the 1990s.15 The utility and resolution of a FISH assay depends on the size of the fluorescently labeled DNA probe used. Therefore, a conventional FISH assay will only interrogate ∼ 0.01% of the genome at a resolution of ∼ 100 Kbp (Figure 2A).
To improve the genomic coverage of molecular cytogenetics techniques, a large number of chip or array platforms have been developed in the last decade; for example, bacterial artificial chromosome comparative genomic hybridization, or oligonucleotide SNP arrays, which interrogate between 3000 and > 500 000 individual genomic loci.16 Only the preselected loci on the chips can be assayed. Therefore, novel SNPs and small indels cannot be “seen” on these platforms (Figure 2A).
Sequencing
DNA sequencing is the genome analysis method with the highest resolution available. However, until very recently, our DNA sequencing technology was only able to cover minuscule stretches of the human genome in a single experiment.
Sanger sequencing.
The first DNA sequencing technology that allowed the reliable sequencing of more than a few dozen base pairs in a single experiment was the dideoxy chain termination method developed by Fred Sanger in the 1970s.17 A typical Sanger DNA sequencing experiment is able to “read” a sequence of ∼ 1000 bp in length, less than one-millionth of the human genome. The DNA fragments to be sequenced have to be cloned or PCR amplified, a labor-intensive process. For this and other reasons, Sanger sequencing is quite an expensive method of sequencing, typically costing several dollars ($1-$10) per 1000 bp. This method was used to complete the sequencing of first human genome at a cost of more than 3 billion dollars over a period of more than 10 years.1-3 It is quite apparent that Sanger sequencing can only be used economically to analyze known mutational hotspots (Figure 2A).
NGS.
Using sequencing to “explore” large genomic regions or even entire human genomes for mutations in cancer and other genetic conditions was only made possible through the invention of NGS technologies in the latter half of the last decade18,19 It was not until just 5 years ago that the NGS technologies had matured sufficiently to allow the almost routine assembly-line sequencing of complete genomes8,20 (Figure 2B).
There are two features that enable the enormous sequence output of all current NGS technologies: (1) a highly simplified workflow to produce a library of clonally amplified DNA fragments that can be directly sequenced, and (2) an extremely high degree of sequence reaction parallelization, which goes hand in hand with the miniaturization of the individual sequencing reaction. In this process, the length of the individual sequence read had to suffer and is, with 100 to 200 bases, much shorter than the individual read length of ∼ 1000 bases of a Sanger sequencing run.
At the moment, there is a fierce competition between different NGS platforms. The mostly widely used NGS platforms are: Roche 454, Illumina, ABI solid, and Ion Torrent, with the Illumina platform currently contributing most of the NGS data worldwide. However, considering the dynamics of the field, this could change rapidly.
For example, currently the Illumina sequencing machine with the highest capacity (HiSeq 2500) is capable of generating ∼600 Gbp of sequences in a single 11-day run.21 The output of such a run is composed of 6 billion 100-bp-long single reads. This is equivalent to 100 diploid human genomes or to 64 human exomes at 50 to 100 × coverage each. The estimated cost for the sequencing reactions only for a human exome at 50 to 100 × coverage will be less than $450 on such a machine, excluding the cost for the library preparation and exome enrichment.
Analysis of cancer genomes using NGS
Although it is relatively affordable now to generate the amount of sequence data that is required to cover an exome or a whole genome at sufficient depth, the process of extracting useful information from this sequence is still a great challenge (Figure 2B). In the following paragraphs, I give a broad overview of this process using exome sequencing of a leukemia sample on the Illumina platform as an example.
Sample preparation and exome capturing
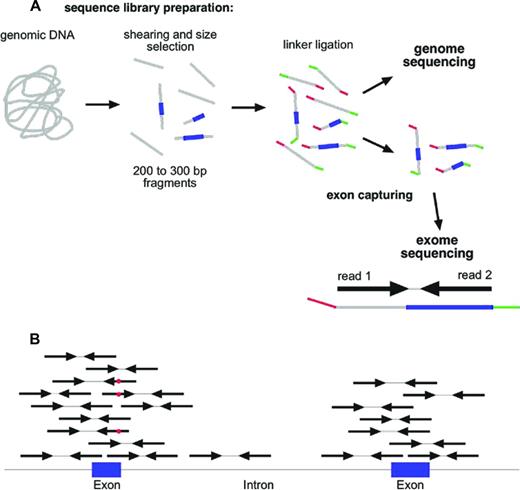
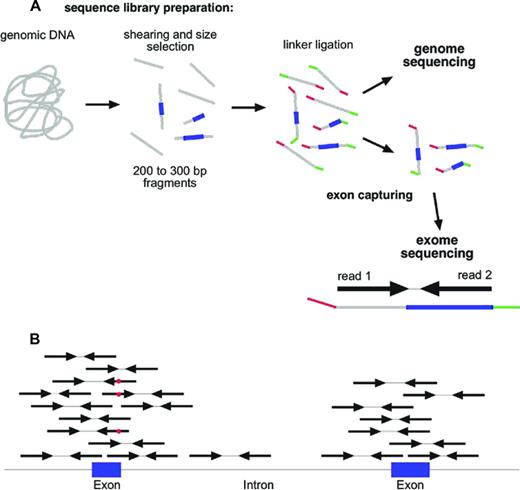
One of the most common strategies in the field of cancer genomics is currently the complete sequencing of the exome (whole exome sequencing [WES]).22 For WES, a sequencing library is prepared from genomic DNA (Figure 3A) and the 1% to 2% of the fragments that represent the exons are captured with oligonucleotide probes (Figure 3A). Exome capturing kits became commercially available ∼ 3 to 4 years ago. Usually, an exome capturing kit has a target region of 50 Mbp.7 The library construction and exome capturing can be completed in 2 to 4 days. The sequencing takes place in a microscope slide-sized device, the flow cell. In the newest machines, up to 1.5 billion fragments can be sequenced in parallel in a single flow cell. Usually, 100 bases are sequenced from either side of the fragments in a so-called paired-end run. Typically, 5 to 10 gigabases of primary sequence are generated from a single exome library, which corresponds to between 25 and 50 million sequenced fragments. This results in a 100- to 200-fold average coverage of every single base in the exome. Although this might appear to be more than should be necessary to discover mutations, it should be noted that the sequence reads are not distributed evenly across all exons and all genes. Even at a 100-fold average coverage, ∼ 10% of the exome will be covered with < 10 reads per base, which makes mutation detection less reliable in these regions.
Whole exome sequencing. (A) NGS library construction and exome capturing. Grey indicates DNA; blue, exons; and green and red, sequencing adaptors. (B) Alignment of reads to genome after exome capturing and sequencing. Red dot in sequencing read indicates a mismatch to reference sequence.
Whole exome sequencing. (A) NGS library construction and exome capturing. Grey indicates DNA; blue, exons; and green and red, sequencing adaptors. (B) Alignment of reads to genome after exome capturing and sequencing. Red dot in sequencing read indicates a mismatch to reference sequence.
Analysis strategies
Low level analysis.
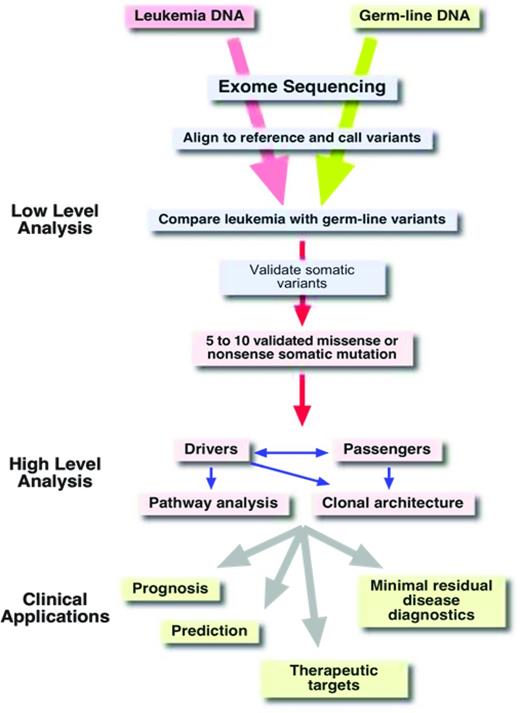
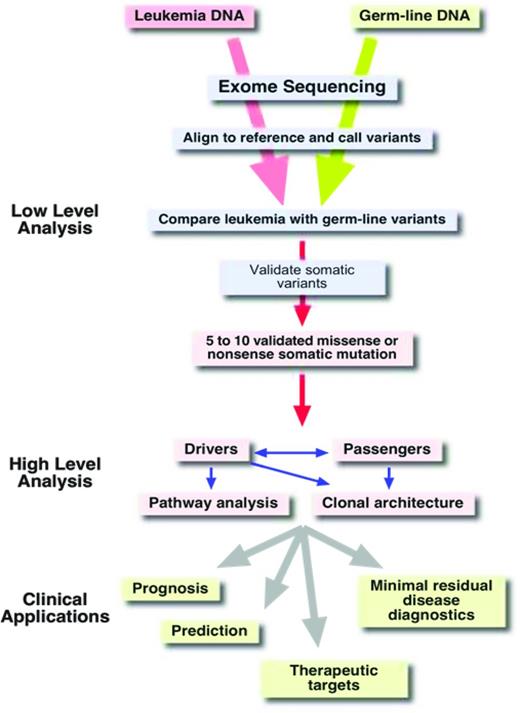
Once the sequence of an exome is available, the next challenge is to extract useful information from this massive amount of data. The first step in the analysis process is to align the millions of reads to a reference genome (Figure 4). This alignment step requires special algorithms (eg, Burrows-Wheeler aligners such as bowtie or bwa) because the familiar BLAST (basic local alignment search tool) searches are too computationally intensive for aligning so many sequences.23 Once alignment files (which can be 100 or more gigabytes in size per exome) have been generated,24 the alignments can be visualized with software tools such as the Integrative Genome Viewer.25 As is illustrated schematically in Figure 3B, the reads from an exome-sequencing experiment usually align to the target, the exons. In addition to the exons themselves, the sequences of the splice donor and acceptor sites, which are adjacent to the exons, are also covered by the sequence reads. Next, deviations from the reference genome, such as single nucleotide variants (SNVs) and small insertions and deletions (indels), are detected in the alignment files using special programs (eg, Varscan26 ; Figure 3B red dots). Typically, an exome sequence will yield ∼ 18 000 to 20 000 SNVs (Figure 4). Of course, most of these SNVs are known SNPs that are annotated in databases. After removing all of the known polymorphisms, ∼ 600 to 1000 SNVs will remain. If we assume that our exome sample was derived from the leukemic blasts of an AML patient, these remaining 600 to 1000 SNVs would then be candidates for somatic mutations that are leukemia specific. However, close to 99% of these remaining SNVs are rare polymorphisms that are not included in the databases yet. To identify these rare polymorphisms, it is greatly preferable to sequence the exome from a nontumor tissue sample of the same individual (ie, a germline reference; Figure 4). After comparing the SNVs from the AML exome sample with the SNVs of the corresponding germline sample, there are usually between 8 and 15 SNVs in the coding regions that are unique to the tumor sample. The effect of these mutations on the coding region is then evaluated using programs such as SNP Effect Predictor.27 Usually, between 5 and 10 mutations will result in an amino acid exchange (missense mutation), a chain termination (nonsense mutation), or an alteration in splicing (splice site mutations). All missense and nonsense mutations that are detected in the NGS data need to be validated by resequencing both the tumor DNA and the germ line reference DNA at the location of the mutation using Sanger sequencing or amplicon deep sequencing.
Small variations in the filter settings that are used for detecting and comparing SNVs in the tumor and germline sample can have a huge effect on the number of candidate somatic mutations that are identified. Because Sanger or amplicon resequencing for mutation validation is very labor intensive, it is critical to tune the filter settings in the analysis pipeline in such a way that the number of false-positive mutation calls is minimized and, at the same time, not too many mutations will go undetected (false negatives).
It should be noted that the number of mutations detected in WES or whole genome sequencing (WGS) experiments in an individual sample that alter the amino acid sequence of genes is between 5 and 10 in AML and in a similar range in chronic lymphocytic leukemia. Certain leukemia subgroups have significantly fewer mutations (eg, childhood leukemias with an MLL rearrangement have only 1 or 2 additional mutations28 ), whereas other entities such as myelomas have a much larger number of mutations (∼ 35 mutations and 21 rearrangements29 ). Solid tumors such as lung or breast cancer have many more mutations.30,31 These varying numbers of mutations in the different tumors probably reflect the time to tumor development (the cells that initiate tumors in older people had more time to acquire more driver and passenger mutations), the strength of the predominant driver mutation (a strong initial driver mutation such as an MLL rearrangement in a childhood leukemia requires fewer additional changes), and the different thresholds for malignant transformation in different tissues (an epithelial cell might require more changes to transform than a hematopoietic cell).
High-level analysis.
Although it is a considerable achievement to condense the 20 Gbp of primary sequence information from 2 exomes down to just 5 to 10 missense or nonsense mutations, the real challenge is to determine which of these mutations are so-called “drivers” of the disease process and which mutations are just along for the ride, the so-called “passenger” mutations. Passenger mutations just happened to have been present in the original transformed cell before it started its clonal expansion.32 In contrast, driver mutations are those mutations that “drive” or are responsible for the malignant phenotype of the leukemia. They are therefore potential targets for therapeutic interventions. There are two ways to identify these important driver mutations: (1) examining whether a mutation is recurring; (2) analyzing the functional consequences of a mutation, for example, by creating an animal model.
Although the second option is the gold standard for identifying driver mutations, it is very time consuming and labor intensive. Therefore, option 1 is more widely used. Looking for the recurrence of a mutation involves the screening of a large number of samples with the same leukemia. However, there is now increasing evidence that, for some driver mutations, the number of samples that have to be screened for recurrence has to be very high (> 200). Recent large-scale genome and exome-sequencing studies of apparently well-defined leukemia entities have almost always found a bewildering genetic heterogeneity and an enormous number of genes that can harbor potential driver mutations.33-35 In a recent study of 200 AML samples, 237 genes were found to be recurringly mutated in 2 or more samples.36 These results suggest that even sequencing the complete genome or exome of 200 samples is not sufficient to identify all potential driver mutations in AML. Several other large sequencing studies of defined leukemia subtypes (eg, chronic lymphocytic leukemia) have provided similar results.33,34 The more genomes and exomes of a given disease entity that are completely sequenced, the higher the chances that certain genes are found to be mutated repeatedly just by chance, even if they are not drivers. This is especially the case for very big genes such as titin (TTN), which represents a very large mutational target with a coding region of > 100 kbp. Therefore, recurrence of mutations in a gene does not necessarily allow the conclusion that these mutations are drivers.
To explore the functional significance of a putative driver mutation, it will become necessary to establish an animal or cell line model. However, even a negative result for a given mutation will not allow the conclusion that this mutation is not a driver mutation. It is becoming more and more apparent that a single driver mutation will, in most cases, not be sufficient to initiate leukemia on its own, but rather that the concerted action of several driver mutations is the basis of most leukemias. Modeling the interplay of several driver mutations is extremely difficult and time consuming with the technologies that are currently at our disposal. We already have evidence that certain driver mutations are dependent on each other and presumably synergize. For example, normal karyotype AML with a biallelic mutation of the CEBPA gene has very specific zinc finger 1 mutations in the transcription factor GATA2.37
Clinical applications
Of course, what we would really like to know is which mutations have an impact on the clinical course of the disease (prognostic significance) and which mutations should guide our treatment decision (predictive significance). Ideally, we would also like to identify mutations that are targetable for therapeutic interventions. The development of algorithms and expert systems that are able to provide this kind of information to the physician after the analysis of NGS data is still in its infancy. For example, it will become necessary to group the many potential driver mutations according to cellular pathways and to predict which drugs might be most suitable for treatment (Figure 4).
WES versus WGS
In the above description of analysis of cancer genomes using NGS, we mainly focused on the information that can be derived from WES experiments. In a WES experiment, one will find missense and nonsense mutations in the coding region, as well as splice site mutations. However, we need to be aware of the fact that the exome constitutes just 1.5% of the genome, albeit the portion of the genome we understand best and that which appears to be the most important functionally. In WGS projects, many more somatic aberrations (∼ 60 times more) can be identified in tumor samples, including not only point mutations and small indels, but also larger genomic rearrangements such as chromosomal translocations, deletions, and duplications.38 However, for practical purposes, at present, only the 1.3% of WGS mutations that affect coding regions, splice sites or certain large genomic rearrangements are considered to be potential drivers and are studied in more detail. Approximately 8% to 9% of the somatic aberrations detected by WGS affect conserved or regulatory genomic elements. Occasionally, such mutations have been shown to constitute driver mutations.39 However, because our knowledge of the exact function of conserved regions and regulatory elements is very limited, we have a very limited capacity to predict or evaluate the functional consequences of these mutations. Close to 90% of the somatic mutations detected by WGS of tumor samples affect nonconserved single copy sequences or repetitive elements. Therefore, although WGS does detect more somatic aberrations than WES in a tumor sample, almost all of these additional aberrations are of unknown significance and can presently not be interpreted properly. It should be noted, however, that one very important class of aberrations, chromosomal translocations, can be detected by WGS but not by WES.
Transcriptome sequencing
Sequencing the transcriptome of a malignant cell can provide very valuable insights and additional information that can also be useful for the interpretation of WGS or WES data. For example, chromosomal translocations that result in the formation of fusion genes can be identified by transcriptome sequencing. In addition, a transcriptome sequence will also provide a “digital” expression profile, which might point to genes that are overexpressed due to a translocation or a mutation affecting a regulatory element. Conversely, mutations present in genes that are expressed at low levels might be overlooked if only the transcriptome of a leukemia is sequenced and the coverage in these genes is not adequate.40
Gene panel sequencing
An alternative to WGS or WES is the deep sequencing of a panel of genes that is known to be recurringly mutated and have prognostic or predictive significance in a given cancer. Custom gene panels have the advantage of allowing high read depths of the genes included in the panel. The disadvantages are the relatively high initial costs in designing a custom gene panel, the fact that adding additional genes to the panel is cumbersome and expensive, and that one is restricted to analyzing the genes in the panel. Certain types of aberrations such as deletions and larger rearrangements cannot readily be detected by a gene panel. When one is considering gene panel sequencing, one also has to keep in mind that the number of potential mutational target genes in most malignancies is very large and still not completely known.
Clonal architecture of leukemia
NGS has also given us a unique insight into the often complicated clonal architecture of leukemias. These analyses showed, for example, that minor clones that were already present at diagnosis can expand after chemotherapy and lead to a relapse.41
Limitations of NGS
It is important to understand that even though NGS is producing an enormous amount of data in a single experiment, we will only find the “pearls” in this vast sea of data if we know exactly what we are looking for and if we understand how the sequence was generated. Therefore, the 10 gigabases of exome sequence from a leukemia patient will not allow us to detect a t(8;21)(q22;q22) translocation, whereas a WGS or transcriptome sequencing experiment would detect such a rearrangement. Even in the complete genome sequence of a tumor, we will not recognize the importance of certain mutations in the noncoding portion of the genome simply because we do not understand the function of 98% of our genome sequence.
Ethical considerations and future directions
The introduction of NGS methodologies into routine tumor diagnostics will also require that several logistical and ethical issues are addressed. NGS will generate massive amounts of data that have to be stored securely to prevent breaches of patient privacy42 and, at the same time, a global data-sharing infrastructure has to be put into place that respects patient privacy and still allows a global data exchange on genotype phenotype correlations. In addition, ethically correct procedures (ie, appropriate consent forms and guidelines) have to be developed to adequately address incidental findings that are bound to occur (eg, what should be done if a BRCA1 germline mutation is found in the course of WES of a leukemia sample?).43
Even though the more than exponential increases in the daily output of sequencing machines has slowed a little in the past year, we have now reached a point where a human genome and certainly several exomes can be sequenced in a few hours (Figure 2B). With this enormous sequencing capacity available at a reasonable cost, we will see many new applications of NGS, such as in minimal residual disease diagnostics to accurately monitor disease burden and NGS as a partial replacement of more traditional genome analysis methods such as cytogenetics and SNP arrays.
Disclosures
Conflict-of-interest disclosure: The author declares no competing financial interests. Off-label drug use: None disclosed.
Correspondence
Stefan K. Bohlander, Department of Molecular Medicine and Pathology, Faculty of Medical and Health Sciences, University of Auckland, 85 Park Road, Grafton, Private Bag 92019, Auckland 1142, New Zealand; Phone: +64-9-923-8348; Fax: +64-9-367-7121; e-mail: s.bohlander@auckland.ac.nz.