Abstract
Chronic lymphocytic leukemia (CLL) is characterized by a relatively small number of recurrent genetic defects. These can be evaluated by clinically available methods such as fluorescent in situ hybridization and targeted sequencing approaches to provide data that can be very helpful in prognostication and planning of treatment. Acquired defects in the p53 pathway, activating mutations of NOTCH1, and dysfunctional mutations of SF3B1 and BIRC3 identify patients with higher risk of progressive disease, poorer responses to conventional chemoimmunotherapy, and shorter survival. Risk stratification using these data can identify patients with aggressive CLL who require careful monitoring and are unlikely to have durable responses to chemoimmunotherapy at disease progression. Patients with defective DNA damage repair mechanisms because of p53 dysfunction should be considered for non-chemotherapy-based regimens including tyrosine kinase inhibitors, BCL2 inhibitors, monoclonal antibodies, and immunological therapies including allogeneic transplantation and chimeric antigen receptor-targeted T cells. Conversely, patients with no high-risk mutations can usually be monitored for a prolonged time and are likely to have durable responses to chemoimmunotherapy at disease progression. New technologies for genetic analysis such as targeted next-generation sequencing have the potential to make these analyses cheaper, faster, and more widely available. Comprehensive genetic analysis of patients both at diagnosis and before treatment for progressive disease could become an integral component of care for CLL.
Learning Objectives
To understand the role of recurrent genetic lesions in the biology of CLL
To use this knowledge for prognostication and selection of therapy
Introduction
Early attempts to use genetic analysis in the management of chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL) were of limited clinical utility because of the difficulty in obtaining metaphase spreads for karyotype analysis, especially in patients with early-stage disease. However, karyotype analysis did identify recurrent genetic defects in CLL patients, leading to the development of probes for their detection by interphase fluorescent in situ hybridization (FISH). FISH analysis provided important insights into the biology of CLL and resulted in considerable improvement in our understanding of the role of these genetic abnormalities in the diagnosis, management, and treatment of patients with CLL.1 However, FISH analysis can only detect large genetic defects recognized by a specific probe. In addition, the sensitivity of FISH can be low in early-stage CLL, especially when patients have a small percentage of clonal CLL cells in their peripheral blood. Genetic analysis of CLL cells has now been expanded using comparative genomic hybridization and single nucleotide polymorphism (SNP) microarrays and gene sequencing using conventional (Sanger) and next-generation sequencing (NGS) techniques. These have proved to be very useful discovery tools. The challenge is to translate this new knowledge into improved patient care.
Microarray SNP analysis and whole exome sequencing have shown that the complexity of the acquired genetic defects in CLL cells (∼10-20 per patient) is low relative to most other malignancies.2-7 This provides an opportunity for focused genetic evaluation of CLL patients. In addition, whole exome sequencing of CLL samples from relatively small numbers of patients has detected novel recurrent genetic defects in CLL that have appreciable clinical implications.5,6,8-10 This review summarizes some of these new data and their impact on the management of patients with CLL.
Acquired genetic lesions in CLL that disrupt physiological pathways
Extensive research has not yet identified a single acquired genetic defect necessary and sufficient to cause CLL. However, acquired genetic defects affecting several important physiological pathways are associated with disease biology, response to treatment, and outcome in subgroups of patients (Figures 1, 2). Detection of these defects is important for determining prognosis in early-stage disease and for choosing therapy in patients with progressive disease. A basic understanding of these pathways and defects could be useful in application of this knowledge to the care of patients with CLL.
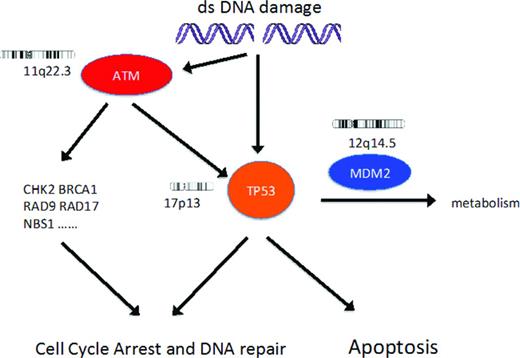
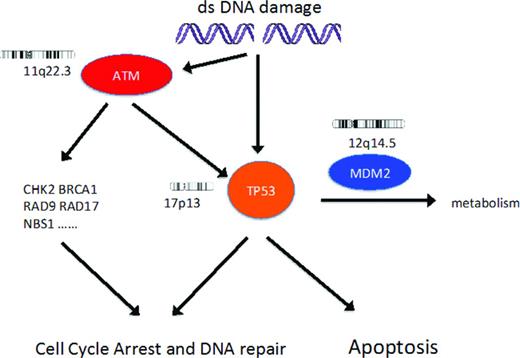
DNA damage response pathway. DNA damage results in activation of ATM and then p53, resulting in cell cycle arrest and then either DNA repair or apoptosis. In patients with CLL, recurrent genetic lesions can result in loss of function of p53 or ATM. The biological effect of the extra copy of the gene coding for MDM2 in CLL patients with trisomy 12 is unknown.
DNA damage response pathway. DNA damage results in activation of ATM and then p53, resulting in cell cycle arrest and then either DNA repair or apoptosis. In patients with CLL, recurrent genetic lesions can result in loss of function of p53 or ATM. The biological effect of the extra copy of the gene coding for MDM2 in CLL patients with trisomy 12 is unknown.
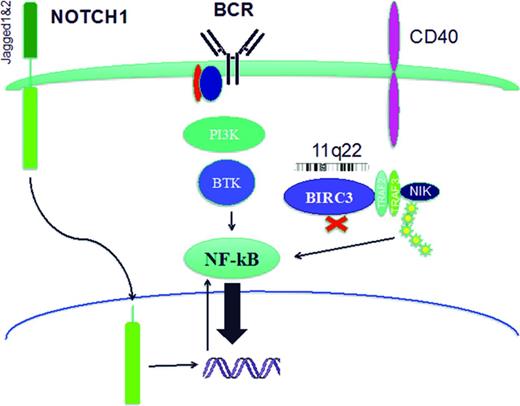
Cell signaling pathways. Cell signaling pathways can be disrupted in CLL cells by activating mutations of NOTCH1 or loss of one allele of BIRC3 by deletion (11q22.2) or mutation. In addition, BCR signaling can be increased in cells with non-mutated or stereotypic IGHV usage. One common feature of these pathways is their ability to activate NF-κB.
Cell signaling pathways. Cell signaling pathways can be disrupted in CLL cells by activating mutations of NOTCH1 or loss of one allele of BIRC3 by deletion (11q22.2) or mutation. In addition, BCR signaling can be increased in cells with non-mutated or stereotypic IGHV usage. One common feature of these pathways is their ability to activate NF-κB.
DNA damage response
Deletions and dysfunctional mutations of genes coding for p53 and ATM (Figure 1) are associated with biologically aggressive and chemotherapy-resistant CLL. Both 17p13 deletion (includes TP53 locus) and 11q22.3 deletion (includes ATM locus) usually affect only one chromosome and the consequences of these deletions usually depends on the functional integrity of the remaining allele of TP53 or ATM, respectively.11 In addition, loss of function of p53 or ATM can occur because of biallelic dysfunctional mutations, mutation in one allele together with copy number neutral loss of heterozygosity, or the generation of a dominant-negative mutant protein.11,12 Clonal CLL cell populations that have both 17p13 deletion and 11q22.3 deletion are rare (1% of all CLL patients, 11% of CLL patients with 17p13 deletion) but have a very poor prognosis.13 There are limited published data on CLL clones with both p53 and ATM dysfunction.
FISH analysis alone thus cannot predict p53 dysfunction in patients with 17p13 deletion or ATM dysfunction in patients with 11q22.3 deletion and is noninformative with regard to p53 and ATM function in patients without these deletions. In contrast, sequencing TP53 and ATM for mutations, together with characterization of the functional importance of detected mutations, can considerably improve the ability to detect defects in the DNA damage response pathway.
p53 dysfunction.
TP53 encodes p53, a pivotal mediator of the DNA damage response pathway controlling cell cycle arrest and apoptosis. Functional p53 is essential for both maintenance of cellular genetic integrity and the cytotoxic effect of DNA-damaging chemotherapy drugs. In patients with CLL, loss of p53 function by deletion and mutation or mutation alone is associated with a very poor response to genotoxic therapy, short median survival, and increased risk of transformation to diffuse large B-cell lymphoma (DLBCL).1,4,11,14-19 Deletion of 17p13 is detected by FISH in ∼5% of CLL patients at diagnosis, 10% of patients requiring initial treatment for progressive disease, and >30% of patients with purine-analog-refractory disease.11 In CLL patients with 17p13 deletion detected at diagnosis, dysfunctional mutations in TP53 are detected in the remaining allele in ∼80% of cases.11 An additional 5% of newly diagnosed CLL patients have dysfunctional TP53 mutations (biallelic, monoallelic with copy number neutral loss of heterozygosity, or dominant-negative monoallelic) in the absence of 17p13 deletion.11 Most TP53 mutations occur in exons 4-9 and frequently disrupt the DNA-binding site.20
ATM dysfunction.
ATM encodes a protein kinase that is activated by DNA damage and has a pivotal role in the DNA damage response mediated in part by the p53 pathway21-23 (Figure 1). Deletion of 11q22.3, including the locus for ATM, occurs in ∼10% of patients with CLL at diagnosis, 20% of patients with progressive disease needing treatment for the first time, and >20% of patients with relapsed/refractory disease.11 Approximately 30%–40% of CLL patients with 11q22.3 deletion and progressive disease have a dysfunctional (missense or a nonsense) mutation in the remaining ATM allele, resulting in more aggressive disease, poorer treatment responses, and shorter overall survival similar to that of CLL patients with dysfunctional p53.24-26 In contrast, patients with 11q22.3 deletion who retain a wild-type ATM usually have less aggressive disease. Patients with a monoallelic dysfunctional ATM mutation (that does not result in production of a ATM protein with dominant-negative activity) together with a wild-type ATM allele have an intact p53 pathway response,27,28 and no significant increase in the risk of poorer response to treatment or shorter overall survival.25 These data suggested that deletion of genes on chromosome 11q22 other than ATM could contribute to the adverse consequences of in 11q22 deletion in CLL patients who do not have a dysfunctional mutation in the remaining ATM allele.25,28
Signaling pathways
B-cell survival and proliferation are dependent on signaling from activated surface receptors mediated by multiple intracellular pathways, many of which activate the transcription factor NF-κB (Figure 2). In CLL patients, activation of these pathways can result in more aggressive disease and poorer prognosis.
NOTCH1.
NOTCH1 is a single-pass type I transmembrane receptor expressed as a noncovalently linked heterodimer (Figure 2).29 The intracellular component of NOTCH1 is a transcription factor that regulates the expression of many genes, including those that encode NF-κB and c-MYC.29 This intracellular component of NOTCH1 is released from the transmembrane heterodimer by γ-secretases after ligand binding and translocates to the nucleus, where it forms a short-lived transcriptional complex until it is phosphorylated on its PEST domain and inactivated.29 NOTCH1 is known to have a physiological role in the development of marginal zone B cells.29 Although NOTCH1 is not expressed by circulating B cells from healthy donors, CLL cells constitutively express NOTCH1 and NOTCH2 and their ligands Jagged1 and Jagged2.30,31
Activating mutations of NOTCH1 are observed at low frequency in CLL patients at diagnosis (∼10%), but at a considerably higher frequency in patients with chemotherapy-refractory CLL (20%) and patients who have transformation of their CLL to diffuse large B-cell lymphoma (∼30%).3,8,19,32-35 The majority of mutations occur in the PEST domain, with the most frequent (67–80%) being a 2-bp frameshift deletion (c.7544_7545delCT).3,8,32,33 More than 90% of the mutations detected in CLL truncate the PEST domain. This decreases NOTCH1 degradation, causing an accumulation of the active isoform resulting in increased NOTCH1 pathway signaling activity with multiple consequences including increased canonical and noncanonical NF-κB pathway activity.3,8,30-33,36,37
In patients with CLL, NOTCH1 mutation has been reported to be associated with decreased overall survival in at least 2 studies (UK LRF CLL4 trial,35 observational study32 ), but was not an independent marker of survival in patients in the CLL8 study.19 In CLL patients, NOTCH1 mutations are associated with a marked increase (∼20-fold) in the risk of transformation to DLBCL.8
BIRC3.
The Baculoviral IAP repeat containing 3 genes (BIRC3, c-IAP2; locus 11q22.2) codes for a multifunctional protein that regulates the ubiquitin-dependent pathways that modulate NF-κB activation38 (Figure 2). The BIRC3 locus is 6 Mb centromeric to the ATM locus and >80% of patients with CLL and 11q22 deletion have also lost one allele of BIRC3.10,26 In addition, BIRC3 can be mutated in CLL cells at multiple loci that usually cause truncation of the BIRC3 protein and loss of E3 ubiquitin ligase activity.10 Loss of one allele of BIRC3 by deletion or mutation in patients with CLL decreases protein expression and increases constitutive noncanonical NF-κB activation10 (Figure 2).
BIRC3 abnormalities (deletion and/or mutation) have not yet been reported in clinical monoclonal B-cell lymphocytosis and occur at low frequency (4%) in CLL patients at diagnosis.10 In contrast, BIRC3 abnormalities occur in 24% of patients with purine-analog-refractory CLL with equivalent frequency of deletion or mutation, which is usually monoallelic. In CLL patients, BIRC3 abnormalities were associated with a significant increase in the risk of death,10 but the independent significance of BIRC3 deletion/mutation in patients with 11q22.3 deletion (loss of one copy of ATM) is uncertain.26 BIRC3 abnormalities do not appear to increase the risk of clonal evolution to DLBCL in patients with CLL.10
BCR.
The BCR signaling required for B-cell survival and proliferation is mediated in part by NF-κB7 (Figure 2). BCR signaling can be modulated by IGHV somatic hypermutation and stereotypy, both of which can be measured in patients with CLL. However, there are no known acquired recurrent mutations that affect the BCR signaling pathway directly.
RNA editing
Splicing factor 3b, subunit 1 (SF3B1, locus 2q33.1) encodes a protein component of the ribonucleoprotein complex (spliceosome) that processes RNA transcripts into mature mRNA.39 Recurrent mutations of SF3B1 in CLL were detected by whole-genome-sequencing techniques. Multiple SF3B1 mutations have been described in CLL patients and are most frequently missense lesions in the HEAT repeats that result in proteins with abnormal splicing activity that modify rather than completely disrupt SF3B1 activity.5,6,9,11 However, the exact mechanisms by which SF3B1 mutations alter CLL biology remain unclear.
SF3B1 mutations occur in ∼5%-15% of patients with CLL at diagnosis, increasing to ∼15%-20% in patients with fludarabine-refractory CLL.6,9,19,35 The frequency of SF3B1 mutations is not increased in CLL patients with clonally related transformation to DLBCL (6%).9 Patients with SF3B1 mutations have decreased progression-free survival after therapy with chemoimmunotherapy19 and multivariate analysis has shown an association with decreased overall survival.6,9,35
Clonal evolution
Acquisition of new mutations, or expansion of small subclones harboring additional mutations, can be associated with major changes in the biology of CLL. This clonal evolution was detectable by FISH in 27% of initially untreated CLL patients followed for at least 5 years.40 The mechanism, risk factors, and clinical implications of clonal evolution are not yet fully elucidated. The ability to understand clonal evolution using FISH is limited by the restricted probe sets and the inability to detect subclones comprising <5% of the CLL cells. Clearly, better methods of evaluating patients for small but potentially important subclones of CLL cells that contain deleterious mutations are needed.
Detecting small subclones in CLL
In purified CLL cells, the resolution of subclone detection is ∼20% for array technology (comparative genomic hybridization or SNP) and 10% for conventional sequencing. Deep NGS methods can detect subclones comprising <1% of the CLL population7,34,41,42 and provide a tool for determining the genetic complexity of CLL, mechanisms of clonal evolution, and the prognostic importance of early detection of small subclones containing deleterious mutations.
Detection of subclones with deleterious genetic defects before initiation of treatment for CLL predict both shorter time to next treatment and clonal evolution with disease progression.7 Rossi et al have shown that, in patients with newly diagnosed CLL, TP53 sequencing by ultradeep NGS identified considerably more mutations than conventional sequencing (15% vs 9%) almost exclusively because of detection of patients with smaller clonal subpopulations within their CLL cells.34 Detection of these smaller subclonal populations with TP53 mutations had the same adverse prognostic implications as detection of large subclones by conventional methods, demonstrating that the size of clone did not influence the effect of the mutation.34 This suggests that small-TP53-mutated subclones detected at diagnosis expand under the selective pressure of treatment with chemotherapy containing regimens to become the dominant clone at relapse.34
Deep NGS for NOTCH1 has also showed that ∼25% of detected mutations were in subclones below the 10% threshold of conventional sequencing.3,33,41 These small subclones were frequently detected (∼12%) in patients with earlier-stage CLL and monoclonal B-cell lymphocytosis. CLL patients with small subclones of NOTCH1-mutated cells had an equivalent poor prognosis and increased risk of transformation to DLBCL as patients with large subclones.
Detection of small subclones of CLL cells with deleterious mutations in patients with CLL has important clinical implications. These patients have an adverse prognosis and require counseling and more careful monitoring of their disease. In addition, these patients could be considered for early intervention clinical trials testing the efficacy and safety of therapies targeting their specific mutation as these drugs become available. When these patients require therapy, they should be considered for treatment with non-DNA-damaging drugs to decrease the risk of expansion of their subclonal population and transformation to more aggressive disease, including DLBCL.
Patient evaluation and management
At diagnosis of CLL
Patients with suspected CLL require an accurate diagnosis. A definitive diagnosis of CLL can usually be made by flow cytometry, but does require a lymphoid tissue biopsy when the immunophenotype is not diagnostic.43 Confirming the diagnosis of CLL is especially important for the correct interpretation of the results of genetic analysis, which are diagnosis specific in B-cell malignancies. Conversely, genetic analysis can largely exclude the diagnosis of mantle cell lymphoma, but cannot be used to make the diagnosis of CLL.
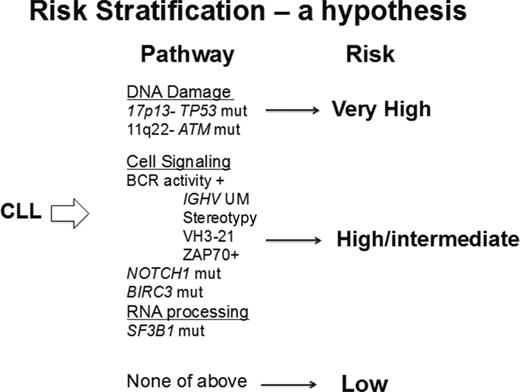
Prognostication at diagnosis for patients with CLL is now possible and of increasing clinical value. The standard of care genetic analysis available to many clinicians is FISH and the hierarchical classification of genetic defects remains a useful prognostic tool.1 Prognostic precision can be increased by more comprehensive analysis of genetic defects, including sequencing of TP53 and ATM, and may be further improved by the addition of BIRC3, SF3B1, and NOTCH1 to the profile (Figure 3). Currently, ATM sequencing is generally not available because of technical difficulties in conventional (Sanger) sequencing of this large gene. Outside of well-designed clinical trials of targeted therapy for earlier treatment of high-risk patients, these data do not provide a justification for early initiation of therapy. However, these data can be very valuable for planning follow-up, determining management strategy, and assisting patients in adapting to their new diagnosis. Patients with no adverse prognostic markers (and especially those with 13q14 deletion as the only genetic defect on FISH analysis) are likely to have stable disease and may not require therapy for a prolonged period of time. However, they are at increased risk of infection, second malignancies, and clonal evolution of their CLL and could benefit from an active monitoring program designed to decrease the risk and consequences of these complications. In contrast, patients with higher-risk disease need to be monitored more frequently (eg, 3 month follow-up) and educated to recognize the clinical features of CLL progression. Those patients with NOTCH1 mutations need to be more carefully monitored for transformation to DLBCL.
Molecular risk stratification in CLL. Patients with CLL could be evaluated by sensitive next-generation sequencing for the mutations and deletions known to confer more aggressive disease and a poorer prognosis. The proposed risk stratification provides a hypothesis for analysis in a prospectively studied population.
Molecular risk stratification in CLL. Patients with CLL could be evaluated by sensitive next-generation sequencing for the mutations and deletions known to confer more aggressive disease and a poorer prognosis. The proposed risk stratification provides a hypothesis for analysis in a prospectively studied population.
At disease progression
FISH analysis for 17p13 deletion should be done before the initiation of therapy for progressive CLL and is required in all patients being considered for chemotherapy containing alkylating agents or purine analogs. Because of the risk of clonal evolution, FISH testing should be repeated in any patient who has had a prior test that did not show 17p13 deletion. This is especially important in patients with an accelerating clinical course and those with treatment-refractory CLL. Because conventional sequencing of TP53 exons 4-9 appreciably improves the ability to predict p53 dysfunction, this test is recommended for all patients on clinical trials, those who could be candidates for chemoimmunotherapy, and those being considered for allogeneic transplantation.44
Patients with progressive CLL and p53 dysfunction have a very high risk of poor response to chemoimmunotherapy and short survival. Patients with p53 dysfunction usually respond to therapy with BCR pathway inhibitors (as reviewed by Dr. Adrian Wiestner elsewhere in this publication). However, the effect of p53 pathway dysfunction on the duration of response of CLL patients to BCR pathway inhibitors is still uncertain. A recent report that CLL patients with 17p13 deletion are overrepresented among patients acquiring resistance to ibrutinib is of particular concern.45 CLL patients with p53 dysfunction should thus be referred to a specialized treatment center for management whenever possible. When treatment is required, they should be considered for inclusion in clinical trials of nonchemotherapy combination regimens and immunotherapy using reduced-intensity conditioning allogeneic transplantation or chimeric antigen receptor T cells.
Patients with relapsed/refractory CLL that do not have detectable defects predictive of p53 dysfunction could benefit from mutation analysis of ATM, BIRC3, NOTCH1, and SF3B1 before retreatment with chemoimmunotherapy regimens if these tests are available. Patients with predicted loss of ATM function are less likely to benefit from chemotherapy-based regimens and should be considered for therapies that are considered to be more effective in CLL patients with loss of p53 function.25,26 The detection of a NOTCH1 mutation in the CLL8 trial of initial therapy of progressive CLL was associated with no clinical benefit from addition of rituximab to fludarabine + cyclophosphamide (FC),19 but the application of this finding to clinical practice is not yet fully defined. Preclinical in vitro studies suggest that CLL patients with activating NOTCH1 mutations could benefit from therapy with γ-secretase inhibitors that prevent NOTCH1 activation.37 Because CLL patients with NOTCH1 mutations are at considerably increased risk of transformation to DLBCL, this diagnosis needs to be considered when evaluating disease progression. Patients with SF3B1 mutations have a shorter progression-free survival after therapy with fludarabine + cyclophosphamide + rituximab (FCR) or FC19 and these patients should be considered for inclusion in appropriate clinical trials whenever possible. The role of the detection of BIRC3 deletions/mutations in therapy decisions is not yet defined.
Conclusion
We are experiencing a paradigm shift in the evaluation and treatment of patients with CLL. Genome-wide and targeted genetic analyses are unraveling the genetic pathologies of CLL. These data are being used to develop tests that provide accurate prognostic data, guide treatment, and identify new targets for drug development. The immediate challenge is to develop targeted NGS methods to provide affordable and accessible data on the genetic defects of importance in CLL patients for prognostication at diagnosis and to assist in therapy decisions at disease progression. Ongoing research is likely to provide novel data on both the genetics and epigenetics of CLL that can be used to develop more effective and safer targeted therapy. This could lead to early risk stratification with risk-based management and eventually to multitargeted and possibly curative combination therapy.
Disclosures
Conflict-of-interest disclosures: C.S.Z. has received research funding from Genzyme, Biothera, Novartis, GlaxoSmithKline, and Genetech. W.R.B. declares no competing financial interests. Off-label drug use: Pentostatin for the treatment of CLL, ibrutinib and ofatumumab for the initial treatment of CLL.
Correspondence
Clive S. Zent, Wilmot Cancer Center, University of Rochester Medical Center, 601 Elmwood Ave, Box 704, Rochester, NY 14642; Phone: 585-273-3258; Fax: 585-276-0350; e-mail: clive_zent@urmc.rochester.edu.