Abstract
As shown with gene expression profiling (GEP), the development and progression of follicular lymphoma (FL) involves complex interactions between neoplastic B cells and the surrounding microenvironment. GEP further reveals that the tumor microenvironment may predict survival in patients with FL and influence the response to therapy and the risk of transformation. Here, we briefly review GEP technology and summarize the role of the tumor microenvironment in FL diagnosis, prognosis, and transformation. Genes expressed by infiltrating T cells and macrophages appear to be the most important predictors of survival, clinical behavior, and outcome. These findings provide a basis for future studies into the pathogenesis and pathophysiology of FL and may ultimately provide guidance in the choice of therapy and the identification of potential therapeutic targets.
Learning Objective
To perceive the strong interconnections revealed by GEP between immunologic and neoplastic cells in the prognosis and transformation of FL
Introduction
Follicular lymphoma (FL) accounts for 20% of all lymphomas worldwide, with highest incidence in the Western hemisphere. FL arises by malignant transformation of normal germinal center B cells and therefore carries the characteristic features of that population: centrocytic/centroblastic morphology; usually some component of nodular growth, expression of pan-B cell markers, along with CD10, BCL6, and LM02; and somatic hypermutation of the immunoglobulin variable chain genes. In addition, FL nearly always harbor the t(14;18)(q32;q21), resulting in overexpression of the anti-apoptotic protein B-cell lymphoma 2 (BCL2), which is absent in normal germinal center B cells.1,2 The FL clinical course is primarily related to disease burden at the time of diagnosis and is generally considered indolent; however, a subset of cases may show early progression. Approximately 25% of cases will transform to aggressive disease, typically diffuse large B-cell lymphoma (DLBCL) with a very poor prognosis.3 Management strategies include watch and wait, immunochemotherapy, and new targeted treatment options.4 The Follicular Lymphoma International Prognostic Index (FLIPI) clinical score reflects the variability in patient outcome but not all patients fit this model.5 Features of the tumor, such as the frequency of large cells and evidence of a late germinal center differentiation, are also related to prognosis.6-8 To better stratify patient risk, major laboratory efforts using high-throughput technologies are focused on identifying tumor-associated features to predict patient outcome. Although it is well recognized that FL samples contain large numbers of T cells of various subtypes, intact dendritic cell meshworks, and macrophages, the enormous contribution of the microenvironment to the disease process was not fully appreciated until the advent of gene expression profiling (GEP). We briefly review GEP technology and then summarize the role of the tumor microenvironment in FL diagnosis, prognosis, and transformation.
GEP
GEP is the collective term for several techniques, all of which simultaneously quantify the mRNA expression levels of nearly all of the expressed genes in the human genome. Although, in the past, mRNA levels were evaluated one gene at a time through Northern blots and later through qRT-PCR, investigators can now evaluate the entire transcriptome in a single experiment. With appropriate statistical and bioinformatic analyses, these techniques identify important pathways and interactions between pathways in the biology of lymphoma.
Initially, results from GEP were viewed with some skepticism. Results were only partially confirmed with Northern blots, qRT-PCR, or immunohistochemistry (IHC) for the associated proteins. Due to expense, studies were rarely performed in duplicate and nonoverlapping results from various groups called into question the reproducibility of the assay. However, as in all scientific endeavors, the details of the experiments are critical to understanding differences in results. A few examples of these variables are listed in Table 1.
Despite the many variables in how GEP studies are conducted, major themes in FL are evident including diagnostic, prognostic, and transformation-associated signatures. Early on, the predominant contribution of the microenvironment to all of these areas was clearly defined and will serve as the focus of the next 3 sections.
Diagnosis
In 2003, in one of the first studies using the earliest GEP technology of 2-color competitive microarrays, investigators described a group of genes also known as the “diagnostic signature.” These genes characterized FL as arising from germinal center B cells, which was distinct from the gene set of naive or memory B cells.9 Similar results showing a germinal center B-cell-like signature were reported using a short oligonucleotide array platform on FL cases sampled by fine needle aspiration.10 Both studies also identified a significant, but not complete, diagnostic signature that overlaps with the germinal center subtype of DLBCL (GCB-DLBCL). One study noted that some FL with higher histological grade, distinguished histologically by more frequent centroblasts, fell into an “intermediate” category, with GEP signatures falling between FL and GBC-DLBCL.10 These results highlighted the biologic continuity from low-grade FL to higher-grade FL to GCB-DLBCL that had long been recognized morphologically.
Low-grade versus aggressive or transformed FL were further compared in another larger study using competitive arrays.11 The cases included paired low-grade (FL1 or FL2) and high-grade (FL3b or DLBCL) biopsies from the same patient. In addition, they studied a group of cases with “ambiguous morphologic features” that were difficult to classify with conventional morphologic techniques (either scored as borderline between FL grade 2 and 3a or the dominant presence of small, centroblast-like cells that fell outside of the consensus criteria for classical “large transformed cells” (centroblasts)). Unsupervised cluster analysis demonstrated the relative homogeneity of all grades of FL. Supervised clustering of cases with low-grade versus high-grade morphology showed distinct GEP signatures for the different grades of the disease. Genes up-regulated in the aggressive phase of the disease were associated with cell cycle control, DNA synthesis, increased metabolism, and several signaling pathways. In contrast, tumor cells from indolent disease highly expressed genes derived from the reactive T-cell infiltrate and macrophages. Although T-cell genes were increased in the indolent cases, T-cell and macrophage activation markers, including several chemokine receptors, were up-regulated at transformation. These findings were confirmed in a separate validation cohort. In the difficult or morphologically “ambiguous” case series, 18 of 19 cases were correctly classified into molecular categories of indolent or aggressive disease, which matched the subsequent patient experience.
In these initial studies, the diagnostic signatures of FL were based on the germinal center nature of the B cells. Once morphologic grade and patient outcome were considered, the T cells and macrophages were identified as the most important factor in separating cases into indolent versus aggressive disease. The cells within the microenvironment are the primary focus of the following sections on prognosis and transformation.
Prognosis
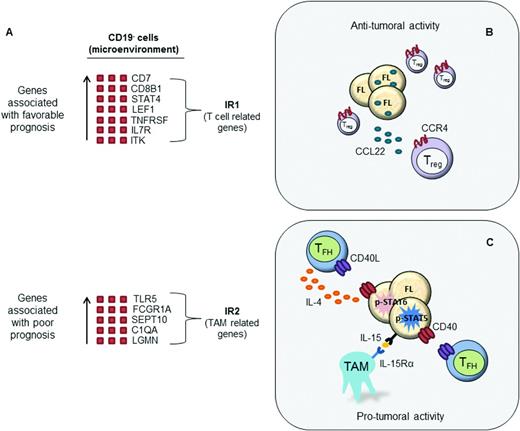
In a landmark study performed on whole tumor biopsies from 191 untreated FL patients, Dave et al defined 2 gene expression signatures that strongly correlated with patient prognosis.12 The 2 gene signatures were termed immune response signature 1 (IR1) and immune response signature 2 (IR2) because they include many genes expressed in T cells, macrophages, and other immune cells. The IR1 signature is enriched for genes expressed in T cells (CD7, CD8B1, ITK, LEF1, and STAT4) and is associated with a favorable clinical outcome. Conversely, the IR2 signature is composed of genes expressed in macrophages and follicular dendritic cells (TLR5, FCGR1A, SEPT10, LGMN, and C3AR1) and is strongly associated with poor clinical prognosis. Interestingly, analysis of purified cell populations revealed that these gene signatures are based on molecular features of nonmalignant, tumor-infiltrating cells and not on the specific genetic characteristics of tumor B cells, suggesting an important interplay between the host immune system and the tumor cells.
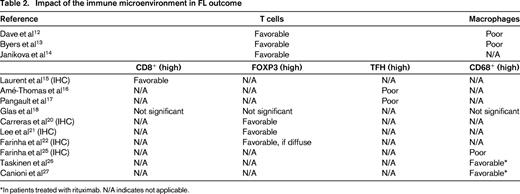
Subsequent studies have since confirmed that immune cells in the tumor microenvironment play an important prognostic role in FL.13,14 Byers et al used poly A RT-PCR to validate 35 “indicator” genes previously shown by Dave12 and Glas11 to have prognostic significance.13 The results confirmed the role of the host immune response in determining outcome for FL and specifically demonstrated that the number of tumor-associated macrophages and T cells are prognostically useful. Another study used custom-designed DNA microarrays containing published genes of prognostic value in lymphomas and found that T-cell-related genes and genes involved in proliferation could predict outcome in patients with FL.14 Specifically, they showed that patients with a poor prognosis exhibited increased expression of proliferation genes and/or decreased expression of T-cell-related genes.
The association between the composition of the immune microenvironment in FL tumors and prognosis led to several GEP studies that characterized the immune cells present in the tumor specimens and the prognostic significance of individual cell populations (Figure 1).15-18 These studies focused on various T-cell subsets, including CD8+ and CD4+ cells, as well as macrophages and other immune cells. The results suggest that these cells affect antitumor immunity and patient outcome in FL. In the case of CD8+ T cells, a recently published IHC and microscopy study correlated an increased amount of intratumoral CD8+ T cells with longer overall survival and disease-specific survival in FL.15 The role of the CD4+ T-cell population, however, is more complex because there are several different populations with distinct functions, including regulatory T (Treg), and T helper (TH) cells.
The tumor microenvironment in FL. (A) Gene expression profiling in FL identified 2 immune response signatures (IR1 and IR2) that can be used to predict clinical outcome. IR1 is associated with genes expressed by non-neoplastic T cells, whereas IR2 is associated with genes expressed by macrophages. Subsequent studies have analyzed specific cell populations within the tumor microenvironment. (B) Increased frequency of Treg cells in the tumor microenvironment is generally associated with a favorable prognosis in FL, particularly when dispersed throughout the tumor. CCL22 secreted from lymphoma B cells induces the migration of CCR4+ Treg cells and may in part account for the increased number of Treg cells in FL. (C) TFH cells overexpress IL-4, which induces signal transducer and activator of transcription 6 (STAT6) activation in neighboring malignant B cells. In addition, TAMs overexpress IL-15, which triggers STAT5-dependent FL B-cell activation. TFH cooperate with this IL-15-mediated signal through CD40L-dependent transcriptional induction of STAT5. The transcriptional targets of STAT proteins play roles in cell cycle progression and cell survival. Therefore, TFH and TAMs in the tumor microenvironment are generally associated with poor prognosis in FL.
The tumor microenvironment in FL. (A) Gene expression profiling in FL identified 2 immune response signatures (IR1 and IR2) that can be used to predict clinical outcome. IR1 is associated with genes expressed by non-neoplastic T cells, whereas IR2 is associated with genes expressed by macrophages. Subsequent studies have analyzed specific cell populations within the tumor microenvironment. (B) Increased frequency of Treg cells in the tumor microenvironment is generally associated with a favorable prognosis in FL, particularly when dispersed throughout the tumor. CCL22 secreted from lymphoma B cells induces the migration of CCR4+ Treg cells and may in part account for the increased number of Treg cells in FL. (C) TFH cells overexpress IL-4, which induces signal transducer and activator of transcription 6 (STAT6) activation in neighboring malignant B cells. In addition, TAMs overexpress IL-15, which triggers STAT5-dependent FL B-cell activation. TFH cooperate with this IL-15-mediated signal through CD40L-dependent transcriptional induction of STAT5. The transcriptional targets of STAT proteins play roles in cell cycle progression and cell survival. Therefore, TFH and TAMs in the tumor microenvironment are generally associated with poor prognosis in FL.
Treg cells are a small subset of CD4+ T cells expressing CD25 regulated by the forkhead/winged helix transcription factor family member p3 (FOXP3); therefore, FOXP3 is often used as a marker for Treg cells.19 Treg cells are involved in the suppression of the immune response. Elevated numbers of Treg cells are frequently observed in FL, resulting in suppressed antitumor immunity. Several GEP and IHC studies investigated whether the number and location of Treg cells alters patient response to therapy in FL.18,20-22 Rather than a numerical change, these studies determined that differences in the spatial distribution of FOXP3+ cells correlated with patient outcome. FOXP3+ T cells were primarily distributed around the follicle (rather than within) in biopsies from patients with longer survival times.22
The TH family consists of 4 major lineages, including TH1, TH2, TH17, and follicular T helper (TFH) cells. These cells secrete cytokines such as IFN-γ, TGF-β, and a variety of interleukins (IL2, IL4, IL12, and IL22).19 Until recently, most of the research on TH cells in FL emphasized the balance between TH1 and TH2 cells. It is generally believed that the TH2 immune response favors tumor growth and TH1 elicits a more favorable antitumor immune response. Using RT-PCR, Jones et al found that both TH1 and TH2 cytokines were expressed at high levels in a cohort of 44 B-cell non-Hodgkin lymphoma patients.23 They also observed that patients with a favorable prognosis had elevated IL4 levels, a cytokine predominantly expressed by TH2 cells. However, in another study, high serum levels of IL12, a major cytokine in TH1 immunity, were associated with a poor prognosis.24 Therefore, the data on the role of TH1 and TH2 cells appear inconsistent and more studies are necessary to draw any conclusions. Similarly, the data on TH17 cells are limited in FL.
Conversely, the prognostic role of TFH cells in FL has recently become a major research focus. TFH cells highly express CXCR5, ICOS (inducible costimulator), CD200, PD-1 (programmed death-1), and BCL6.19 A recent study found that several genes, including IL4 and CD40LG, were overexpressed in the CD4+CXCR5+ICOS+ T-cell population.16 The IL4 gene encodes for IL4, a TH2 cytokine, and CD40LG for the B-cell surface molecule CD40L. Functional studies revealed that these 2 molecules help support FL cell survival, because treatment with anti-CD40L and anti-IL4α neutralizing antibodies significantly inhibited malignant B-cell survival.16 Another investigation validated the importance of IL4 signaling in FL cells.17 The results of that study showed that TFH cells in the tumor microenvironment express IL4 at very high levels and suggested that increased IL4 may contribute to FL pathogenesis and represents a novel therapeutic target.
GEP studies demonstrate that genes expressed by tumor-associated macrophages (TAMs) correlate with poor prognosis in patients with FL.12 These findings have been validated using IHC; unfortunately, the data are inconsistent. Although several studies showed that increased numbers of CD68+ TAMs are associated with adverse outcomes in response to chemotherapy,25-27 no correlation was observed in other investigations.18 Interestingly, the addition of rituximab, a monoclonal antibody directed against the B-cell antigen CD20, to the standard treatment for FL changed the adverse prognostic effect of TAMs in the Taskinen and Canioni studies.26,27 These data highlight the need to investigate further whether TAMs in the tumor microenvironment can be used to predict patient prognosis. They also suggest that discrepancies among datasets could be related to differing treatments, thus highlighting the importance of reevaluating previously identified prognostic factors in the post-rituximab era.
To date, there are only a few studies addressing whether gene expression in FL can predict a patient's outcome in response to rituximab in combination with other chemotherapies. Bohen et al identified several genes that accurately predict whether a patient will respond to rituximab.28 Many of the genes that were highly expressed in tissue from patients who failed to respond to rituximab are involved in mediating the cellular immune response and in inflammation, such as cytokine, tumor necrosis factor, and TCR signaling proteins. Once again, the observed patterns of gene expression reflect all the cells in the tumor, not only the tumor cells themselves. In a separate study, Harjunpää et al found 3 genes, EPHA1, SMAD1, and MARCO, the expression of which correlated with patient response to rituximab + cyclophosphamide + hydroxydaunorubicin + vincristine + prednisone/prednisolone (R-CHOP) treatment in FL.29 EPHA1 is a receptor tyrosine kinase expressed on CD4+ T lymphocytes; SMAD1 is a transcription factor involved in TGF-β signaling and a mediator of growth arrest and apoptosis; and MARCO is a receptor on macrophages that is induced in response to inflammatory stimuli. High EPHA1 expression and low SMAD1 and MARCO expression were associated with better outcomes, such as increased progression-free survival, in FL patients treated with R-CHOP. Consistent with previous studies showing that the microenvironment is critical, IHC analyses showed that these proteins were not expressed in malignant lymphocytes, but rather in the surrounding microenvironment, including vascular endothelia and granulocytes.29
Overall, GEP studies in FL demonstrate that the tumor microenvironment is an important determinant of outcome (reviewed in Table 2). Although there is some variation, genes expressed by infiltrating T cells and macrophages appear to be among the most important predictors of survival, clinical behavior, and outcome. Generally, elevated numbers of T cells are associated with a favorable prognosis, whereas increased numbers of macrophages are correlated with progression and poor prognosis in patients with FL. The importance of the immune microenvironment in FL has led to the development of novel therapeutic strategies targeting the immune system, including immunomodulatory drugs (ie, lenalidomide) and immune checkpoint inhibitors (ie, ipilimumab and pidilizumab).30-32 The following chapter by Nowakowski et al will discuss the clinical status of therapies targeting the immune microenvironment in FL.
Transformation
Several studies have evaluated GEP changes in paired samples from patients before and after transformation. Interestingly 2 main themes emerge. First, there appears to be more than one collective set of changes by which low-grade FL transforms into aggressive disease. Second, at least some portion of the transformative process includes alterations in the microenvironment.
One of the first studies examining mechanisms of transformation used competitive microarray analysis and, later, array comparative genomic hybridization to examine genomic and expressional changes. Looking only at genes that varied by 3-fold between the case sets, the investigators found that MYC and 91 of its downstream genes were altered (either up-regulated or down-regulated) in 9 of the 12 transformed biopsies. Therefore, although MYC often plays an important role in tumor transformation, not all transformed cases have an increased MYC signature. These results suggest that there are additional undetected mechanisms for transformation.33,34 Similarly, in a study 2 years later also using competitive microarrays, MYC expression was again identified as increased in some, but not all, transformed paired cases.35 In a network-based reanalysis of published data, a third group described an embryonic stem cell (ESC)-like signature, including MYC and its target genes, which correlated with histologic transformation. The importance of this observation was demonstrated in a transgenic mouse model demonstrating that the ESC program significantly correlated with transcriptional programs maintaining tumor phenotype. That study further described a decrease in a “stromal signature,” particularly genes that were targets of TGF-β, which together with ESC signatures predicted both transformation and survival.36
Other investigators evaluated mechanisms of FL transformation using a short oligonucleotide technique. Similar to the previously cited studies, these investigators also reported transformation as a heterogenous process with stepwise alternative pathways. That study, however, was the earliest to identify that, among other pathways, there was decreased expression of T-cell-, dendritic-cell-, and stromal-cell-related genes in transformed samples.37 Another study, this time using competitive microarray, studied 3 (unpaired) cohorts: cases that did not progress, cases that did progress, and transformed FL. In that study, immunologic cell-related genes were overrepresented in the list of differentially expressed genes. More particularly, cases that rapidly transformed exhibited higher levels of these genes (similar or exceeding benign follicular hyperplasia) than nontransforming cases, which were postulated to resemble nonactivated lymphoid tissue.18
All of the preceding studies used homogenized whole lymph node/biopsy specimens. However, in a more detailed recent study, the investigators assessed the GEP of purified CD4+ and CD8+ T cells from FL cases. They examined mRNA changes in these populations and related the results to subsequent transformation. Interestingly, the investigators identified altered expression in 109 genes, including 95 up-regulated but only 13 down-regulated genes in the lymphoma-associated T cells compared with T cells from reactive tonsil controls. They identified pro-melanin-concentrating hormone (PMCH) and E-Twenty-Six (ETS) variant-1 (ETV1), among others, which were induced by the FL cells through transwell plates, suggesting that a soluble factor(s) from the FL induced changes in the infiltrating T cells. PMCH is proteolytically cleaved into melanin-concentrating hormone (MCH), which has immunomodulatory effects. The investigators postulated that FL B cells may increase T-cell production of MCH, which binds to surrounding tumor-associated macrophages, altering their behavior. Actin 1 was among the most down-regulated genes in transformed FL. Migration-tracking studies demonstrated altered mobility of both sorted CD4+ and CD8+ T cells from transformed FL compared with tonsil controls, indicating a possible mechanism for impaired tumor immunosurveillance.38
The emphasis of this review is on the role of the microenvironment in transformed FL. However, for completeness and in keeping with the concept multiple mechanisms contributing to transformation, several studies identified alterations in pathways in addition to infiltrating immune cells and MYC-related genes and should be mentioned. These include decreased expression of genes related to evasion of apoptosis and increased levels of genes involved with tissue invasion, metastasis, proliferation, metabolism, and a subset of transcription factors, as well as up-regulation of the MAPK and NFκB pathways.9,35,37,39
Summary
GEP studies have correlated several genes with diagnostic characteristics, progression, response to therapy, and transformation in FL. Although genes involved in apoptosis, proliferation, and metastasis may predict the course of the disease and patient outcome, genes associated with infiltrating immune cells in the tumor microenvironment appear to be the most accurate predictors of clinical behavior and response to treatment in FL. The interplay between the tumor and the surrounding cells of the immune system is supported by several ancillary studies specifically showing that T cells and macrophages in the tumor microenvironment are important factors of patient prognosis and survival. At present, the mechanisms of these alterations in the benign-tumor-infiltrating immune cells and the best way to measure these changes are under investigation. In addition, most published GEP data on FL are based on RNA extracted from fresh-frozen biopsies; however, fresh/frozen material is often not available. The feasibility of using RNA derived from routinely processed formalin-fixed paraffin embedded biopsies, which is the only material for analysis for many patients, was recently shown in DLBCL.40-42 In the future, one of the major tasks will be to translate the FL gene signatures into robust clinical tests for daily practice. Technological platforms for focused GEP that use short probes, tolerate degraded nucleic acid template, and do not require enzymatic activity for assay success may be particularly promising.
Disclosures
Conflict-of-interest disclosure: The authors declare no competing financial interests. Off-label drug use: None disclosed.
Correspondence
Lisa M. Rimsza, M.D., Department of Pathology, University of Arizona, 1501 N. Campbell, Room 5208A, PO Box 245043, Tucson, AZ 85724. Phone: 520-626-8396; Fax: 520-626-6081; e-mail: lrimsza@email.arizona.edu.