Abstract
The outcome for children with acute lymphoblastic leukemia (ALL) has improved dramatically with current therapy resulting in an event free survival exceeding 75% for most patients. However significant challenges remain including developing better methods to predict which patients can be cured with less toxic treatment and which ones will benefit from augmented therapy. In addition, 25% of patients fail therapy and novel treatments that are focused on undermining specifically the leukemic process are needed urgently.
In Section I, Dr. Carroll reviews current approaches to risk classification and proposes a system that incorporates well-established clinical parameters, genetic lesions of the blast as well as early response parameters. He then provides an overview of emerging technologies in genomics and proteomics and how they might lead to more rational, biologically based classification systems.
In Section II, Drs. Mary Relling and Stella Davies describe emerging findings that relate to host features that influence outcome, the role of inherited germline variation. They highlight technical breakthroughs in assessing germline differences among patients. Polymorphisms of drug metabolizing genes have been shown to influence toxicity and the best example is the gene thiopurine methyltransferase (TPMT) a key enzyme in the metabolism of 6-mercaptopurine. Polymorphisms are associated with decreased activity that is also associated with increased toxicity. The role of polymorphisms in other genes whose products play an important role in drug metabolism as well as cytokine genes are discussed.
In Sections III and IV, Drs. James Downing and Cheryl Willman review their findings using gene expression profiling to classify ALL. Both authors outline challenges in applying this methodology to analysis of clinical samples. Dr. Willman describes her laboratory’s examination of infant leukemia and precursor B-ALL where unsupervised approaches have led to the identification of inherent biologic groups not predicted by conventional morphologic, immunophenotypic and cytogenetic variables. Dr. Downing describes his results from a pediatric ALL expression database using over 327 diagnostic samples, with 80% of the dataset consisting of samples from patients treated on a single institutional protocol. Seven distinct leukemia subtypes were identified representing known leukemia subtypes including: BCR-ABL, E2A-PBX1, TEL-AML1, rearrangements in the MLL gene, hyperdiploid karyotype (i.e., > 50 chromosomes), and T-ALL as well as a new leukemia subtype. A subset of genes have been identified whose expression appears to be predictive of outcome but independent verification is needed before this type of analysis can be integrated into treatment assignment.
Chemotherapeutic agents kill cancer cells by activating apoptosis, or programmed cell death. In Section V, Dr. John Reed describes major apoptotic pathways and the specific role of key proteins in this response. The expression level of some of these proteins, such as BCL2, BAX, and caspase 3, has been shown to be predictive of ultimate outcome in hematopoietic tumors. New therapeutic approaches that modulate the apoptotic pathway are now available and Dr. Reed highlights those that may be applicable to the treatment of childhood ALL.
I. New Approaches to Risk-Adapted Therapy for Childhood Acute Lymphoblastic Leukemia
Deepa Bhojwani, MD, Dong-Joon Min, PhD, Elizabeth Raetz, MD, and William L. Carroll, MD*
Mount Sinai and New York University Schools of Medicine, Combined Division of Pediatric Oncology, One Gustave L. Levy Place, Box 1208, New York NY 10029-6574
Supported by a Director’s Challenge grant from the National Cancer Institute U01 CA88361
Acknowledgments; The authors gratefully acknowledge the help of Dr. Harland Sather in reviewing outcomes on CCG studies.
The prognosis for children with acute lymphoblastic leukemia (ALL) has improved dramatically over the past four decades. Breakthroughs in therapy have been achieved in a stepwise fashion through carefully controlled, cooperative group clinical protocols, the hallmark of care within the childhood cancer community. Contemporary therapy has focused on intensification using established agents rather than the introduction of new drugs. Despite these improvements, many children are being overtreated, while subgroups of children still do poorly despite recent therapeutic advances. Risk-adapted therapy tailors treatment based on the predicted risk of relapse—augmenting therapy for those whose tumors require this approach while avoiding the more toxic side effects of augmented therapy in children who can be cured with treatment of standard intensity. Currently, risk-adapted therapy is used for almost all pediatric tumors.
Treatment outcome is dependent not only on the therapy applied, but importantly, also on the underlying biology of the tumor and the host. A detailed discussion of host factors that factor into treatment response is provided in Section II. Each of these variables must be factored into initial treatment decisions, as well as later refinements based on initial response, and several biological features. This review will discuss the most important variables that are currently used to design therapy for children with ALL as well as emerging data from transcript and protein profiling that might be applied to risk assignment in the future. It is recognized that with improvements in therapy, certain variables might lose their prognostic value; therefore, risk assignment plans should be routinely reassessed. Finally an optimal system should allow for comparison of the outcomes of similar, or identical patients, treated on different protocols.
Clinical Features Predictive of Risk
The two most important factors predictive of outcome are age and presenting white blood cell count (WBC) at diagnosis.1,2 The National Cancer Institute (NCI)/ Rome criteria stratify patients into subsets based on age 1 to 9.99 years and WBC < 50,000/μL (standard risk), or age ≥ 10 years and/or WBC ≥ 50,000/μL (higher risk).3 These variables have consistently emerged as independent predictors of outcome in almost all therapeutic studies. Age and WBC are continuous variables, and discrete thresholds used for risk stratification are somewhat arbitrary. However, the obvious advantage of this system is that these variables can be measured reliably in almost all circumstances, and therefore these criteria can be applied to children worldwide. The good outcomes characteristically observed in younger children are partially associated with favorable genetic features of the blast, which are frequently present in these patients. Infants less than one year of age continue to fare poorly, and the unfavorable biology of MLL translocations, which occur in 70% of these patients, accounts for much of these inferior outcomes.
WBC is reflective of tumor burden, although the underlying biological mechanisms that account for the adverse outcomes associated with an elevated WBC are uncertain. Other features associated with high tumor burden, such as hepatosplenomegaly and mediastinal mass, are also associated with a greater risk of relapse. Investigators from the Berlin-Frankfurt-Munster (BFM) cooperative clinical trials consortium incorporate peripheral blast count and liver and spleen size into a single variable that can be used in risk-based classification.
Gender and immunophenotype are other features that have consistently shown to be associated with outcome. Girls have a superior event free survival (EFS) compared to boys, even when they are treated with less therapy. Although the magnitude of this difference may be less apparent on recent studies, intensified therapy has failed to abrogate this difference. Blast immunophenotype has also been shown to have prognostic significance, although the impact of this variable has lessened with improvements in therapy. Coexpression of myeloid antigens on lymphoid blasts (My+ ALL) has been reported to be a poor prognostic feature, but studies now show that outcome of My+ ALL is indistinguishable from that of typical B-precursor ALL.4 A T-cell immunophenotype has also been associated with inferior EFS rates, perhaps in part secondary to the presence of additional adverse prognostic features such as older age, mediastinal mass, and lymphadenopathy.5 However, the rationale for stratifying T-cell patients onto unique protocols is currently based on the distinctive pharmacological properties of T-cell blasts, namely, their relative sensitivity to agents such as asparaginase and 506U and relative insensitivity to lower doses of methotrexate.
The presence of central nervous system (CNS) disease at diagnosis is also an adverse prognostic factor despite intensification of therapy with additional intrathecal therapy and CNS irradiation. The presence of blasts on the cytospin in the absence of an elevated cerebral spinal fluid (CSF) WBC (so called “CNS 2” status) or a traumatic lumbar puncture, as defined as a red blood cell count (RBC) > 10/μL with blasts (TLP+), is also associated with a poorer outcome.6 Evidence suggests that the adverse prognostic significance of CNS 2 status might be overcome with additional intrathecal chemotherapy, and the more recent use of dexamethasone-based regimens might also be beneficial in this regard.7
Genetic and Molecular Characteristics of Leukemia Cells
ALL blasts routinely contain somatically acquired genetic abnormalities that provide insight into pathogenesis and strongly influence prognosis. Approximately one third of cases of ALL show an increase in the modal chromosome number (e.g., hyperdiploid, > 47 chromosomes, and “high” hyperdiploid, > 50 chromosomes) blasts make up a unique biologic subset associated with increased in vitro apoptosis and sensitivity to a variety of chemotherapeutic agents.8,9 Very good outcomes are characteristically observed in patients whose blasts harbor these features (EFS 75%–90%). The good outcome seen in hyperdiploidy is attributed to the favorable prognostic impact of trisomies of chromosomes 4, 10, and 17 (triple trisomies). Patients with triple trisomy ALL have a 7-year EFS > 90%. In contrast, hypodiploid blasts, with fewer than 45 chromosomes, are a negative prognostic feature. While the outcome of patients with 45 chromosome blasts is no different from those patients with a normal karyotype, the EFS for those with 33 to 44, and less than 28 chromosomes, is 40% (± 18%) and 25% (± 22 %), respectively.10
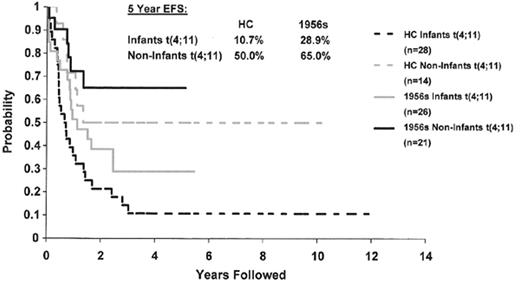
Almost one third of ALL blasts show chromosomal translocations in the absence of changes in chromosome number. Four major translocations have been observed, and each defines a unique biological subset of patients. The t(1;19)(q23;q13) is a hallmark of some pre-B (cytoplasmic μ+) ALL, and is characterized by fusion of the E2A and PBX genes.11 Despite the adverse prognostic impact of this translocation in older studies, recent intensification of therapy has resulted in an improved survival for these children. Translocations between the mixed lineage leukemia (MLL) gene at 11q23 and over 30 different partner chromosomes characterize 6% of ALL cases. MLL translocations, most commonly t(4;11)(q21;q23), are seen in the vast majority of infant patients with ALL. A recent, large series demonstrates that any rearrangement of 11q23 is associated with a worse prognosis (e.g., 20% to 25%).12 In older children the negative impact of 11q23 is less powerful although it still defines a higher risk subgroup (Figure 1 ). The Philadelphia chromosome is a byproduct of a t(9;22)(q34;q11), and this abnormality is observed in 3% to 5% of children but up to 20% of adults with ALL. The resulting BCR-ABL fusion transcript has altered tyrosine kinase activity, which is responsible for its transforming potency. Ph+ ALL is the most difficult to treat of all childhood leukemias. Although certain subgroups of patients have an EFS of 50%, such as those with lower WBCs and a rapid response to initial chemotherapy, as well as those who undergo matched-related stem cell transplant, new options are clearly needed.13 The use of the kinase inhibitor imatinib mesylate has shown some transient effectiveness in relapsed Ph+ ALL but recurrence occurs in almost all cases. However, administration in conjunction with chemotherapy is currently being evaluated in ALL.
The most common translocation is the t(12;21)(p13;q22), which is recognized in up to 25% of B-precursor ALL using molecular screening techniques.14 This translocation fuses the TEL locus at 12p12 (also called ETV6) with the AML1 gene (also called CBFA2 or RUNX1). The resulting fusion transcript is a transcription factor and functions as a corepressor at AML-1 target genes.15 Many studies have demonstrated that patients with the TEL/AML1 translocation do extremely well. However, results from the BFM group showed that the incidence of this translocation in relapsed cases was identical to that seen at initial diagnosis. Relapses tend to occur late, and salvage has been extremely good.16 In contrast, a study from the Dana-Farber Cancer Institute showed very few cases of TEL-AML1+ at relapse.17 Differences in therapy might account for this discrepancy. In vitro studies show a unique sensitivity of TEL-AML1 blasts to asparaginase and protocols that contain augmented therapy with this agent might prove particularly useful for this subgroup of patients.18 This concept will be formally tested in the upcoming Children’s Oncology Group clinical trial for “low risk” ALL. Intriguing results from studies on identical twins indicate that the TEL-AML1 translocations can occur many years before emergence of leukemia, indicating that subsequent genetic events, including deletion of the alternative TEL allele, are necessary for full malignant transformation.19 Interestingly, studies of late relapse TEL-AML1+ ALL show that the recurrence is actually development of another de novo ALL that was generated from an identical premalignant stem cell. This may account for the responsiveness of late-relapsing TEL-AML1+ ALL to retrieval therapy.20
Early Response to Therapy
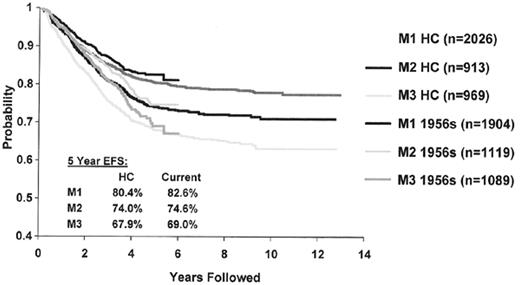
In vivo response to the therapy would be predicted to be one of the most useful predictors of outcome and studies in ALL and other tumors show this to be the case. BFM investigators have shown that patients whose peripheral blast count drops below 1000 blasts/μL have an EFS of 61% after 1 week of prednisone and a single intrathecal dose of methotrexate compared to 38% for those with a higher level of blasts in the peripheral circulation.21 Investigators from St Jude Children’s Research Hospital have shown that the presence of peripheral blood blasts after 1 week of conventional induction chemotherapy is also an adverse prognostic feature.22 Alternatively CCG investigators have looked at blast content in the bone marrow at day 7 and/or 14 of induction.23,24 Since more than 90% of children achieve an M1 status at day 14, the predictive value of day 7 blast content has been investigated more recently. Overall, 52%, 23%, and 25% of children had an M1, M2, or M3 marrow status at day 7 and the associated EFS was 80% (± 1%), 74% (± 2%) and 68% (± 2%).25 The predictive value of marrow blast content, as determined by morphology, continues to be robust in contemporary clinical trials despite issues related to hypocellularity and possible variability in interpretation between individual physicians (Figure 2 ). These results are particularly noteworthy since augmented therapy given in response to a slow early response (SER) can significantly improve outcome. In CCG 1882 day 7 SER patients who received additional therapy with more intensive methotrexate and asparaginase had a 5-year EFS of 75% (± 3.8%) compared to 55% (± 4.5%) for those who received “standard” therapy.26
All of the above measurements of early response rely on morphological recognition of tumor cells. In reality, the bulk of tumor burden is below this limit of detection. Techniques are now available that can now detect 1 tumor cell in a background of 1000 to more than 1 million normal cells, and the assessment of minimal residual disease (MRD) has refined further the evaluation of tumor response. The choice of the particular technique depends on the question being considered and the clinical context in which the sample is being evaluated.
Flow cytometric MRD analysis relies on the detection of surface phenotypes unique to leukemia cells and not present on normal hematopoietic cells. This technique can be applied to the great majority of both B-precursor and T-cell cases, is relatively inexpensive, can be done quickly, and is therefore amenable to the analysis of a large number of samples and has a sensitivity of 10−3 to 10−4.27 Molecular techniques are more sensitive in general but are also labor intensive, and therefore expensive. Analyses of antigen receptor rearrangements have been used frequently in MRD analysis. Since individual T-cell receptor and immunoglobulin genes undergo a unique clonal rearrangement, they can be used as specific targets for residual tumor detection.28 Consensus primers can be used to amplify junctional sequences and the length of the product will allow discrimination from a background of normal cells. The sensitivity of this approach is roughly 10−3. To provide for greater sensitivity, the initial product can be sequenced and allele specific primers can be designed (sensitivity 10−5). Fusion genes resulting from chromosomal translocations provide the optimal target, but this approach is applicable in only one-third of cases.29
A number of studies have demonstrated the value of MRD assessment.28–,32 Most have looked at disease levels at the end of induction and correlated these values with EFS. Regardless of the technique used, the following broad conclusions can be made. Patients with no detectable MRD at end induction have an exceedingly good outcome (EFS > 90% at 3 years). Those children with a high MRD (≥ 10−2) have a poor prognosis (3-year EFS approximately 25%). Patients with intermediate levels (10−4 to 10−3) make up one-third to one-half of all patients depending on the technique used. These patients can be further subdivided based on analysis of a second time point. For example, in the study by Coustan-Smith, of 32 patients who were MRD+ by flow cytometry at the end of remission induction who then became MRD− at week 14, only one relapsed. In comparison, 10 relapses occurred among 18 patients who remained MRD+ at the second time point.32 These same investigators examined the prognostic significance of early clearance of blasts at day 19.31 Lack of detectable leukemic cells at this point was more closely associated with relapse-free survival than lack of detectable blasts at the end of induction (day 46 in the St. Jude studies). Thus, this approach may define a subgroup at extremely low risk of relapse that could be candidates for reduction in therapy. Not surprisingly, the level of MRD prior to transplant correlates with the effectiveness of this modality.
Children’s Oncology Group Risk Groups
The recent merger of the Pediatric Oncology Group and the Children’s Cancer Group into the single new Children’s Oncology Group provided an opportunity to reassess therapeutic stratification since each group used similar but somewhat distinct approaches. A number of variables have been analyzed, and those selected for incorporation into a new classification system were those that maintained prognostic significance using data from both groups or where data from one group were supported by additional published information from additional clinical trials. According to this proposed scheme, patients will be assigned to one of four initial treatment groups at the time of diagnosis, based upon their age, presenting white blood cell count, and immunophenotype: T-cell, infant, high risk B-precursor, and standard risk B-precursor ALL. Patients with B-precursor ALL will undergo further refinement in treatment stratification at the end of induction based on molecular features of the blast, as well as response to therapy as assessed by bone marrow morphology at day 8/15 and 29, and MRD at day 29. At the end of induction, all B-precursor ALL patients will be further classified into low risk, standard risk, high risk or very high risk groups.
Classification of Leukemia by Gene Expression
The active transcription of a subset of all potential genes and the relative abundance of their transcripts contribute to the static and dynamic profile of a cell. Results using microarrays or “DNA chips” indicate promising applications in molecular classification of tumors, definition of distinct subgroups with prognostic significance, and delineation of new therapeutic targets.33
A number of studies using microarrays to classify various tumors have been published.34–,36 In 1999, Golub et al demonstrated the feasibility of using microarrays to accurately distinguish subtypes of leukemia using a set of genes as class predictors.37 Moos et al also described gene expression signatures that could discriminate between the following categories: AML vs. ALL, T vs. B lineage ALL and TEL/AML + ALL.38 Using cross validation, the predicted correct classification rate was 75%–100%. Additionally, genes were selected to differentiate risk groups by NCI/Rome criteria, but the prediction rate was lower, 61%–65% (Figure 3; see Appendix, page 611). This admixing of standard-risk and high-risk patients was not unexpected, possibly reflecting gaps in the traditional classification system. In fact, 3 of the standard risk patients who were misclassified as high risk showed slow initial marrow response at day 7; one patient died from early marrow relapse.
A large scale study by Yeoh et al, using over 327 pediatric ALL samples, identified seven distinct ALL subtypes: T-ALL, E2A-PBX1, BCR-ABL, TEL/AML1, MLL rearranged, hyperdiploid (> 50 chromosomes) and a new novel subtype.39 Recently, a subset of the above samples was reanalyzed using high-density microarrays (39,000 transcripts).40 An excellent discussion on the use of microarrays in pediatric acute leukemias and important insights provided by these analyses is presented by Dr. James Downing in Section III.
In an illustration of using microarrays for class discovery, Armstrong et al41 proposed that mixed-lineage leukemia (MLL) is a distinct clinical entity from ALL and AML. Patients with MLL do not respond well to conventional therapy. They were able to demonstrate that the MLL translocation specified a unique gene expression profile differentially expressing FLT3 and certain HOX genes that may be important for leukemogenesis and hematopoiesis. Likewise, infant leukemia has a poor prognosis, and 70% of the infants have translocations of the MLL gene. Mosquera-Caro et al performed microarrays on 126 infant leukemia samples.42 They identified three discrete biologic groups of infant leukemia and hypothesized that genes expressed in these groups represented distinct etiologic pathways. Interestingly, MLL samples were represented in all subgroups. Prediction of treatment failure was more accurate when modeled on these clusters, rather than on traditional classification methods. Dr. Cheryl Willman will update these studies from her laboratory in Section IV.
Microarrays have the potential to be used in the clinic as frontline diagnostic and risk assignment tools. Before that occurs it is crucial that microarray data be validated in independent experiments and in different laboratories using standard formats to collect, transfer, and archive data.43 The feasibility of microarrays is currently being tested in large clinical trials and to be practical tools, chips must also be cost-effective. Finally, refinements in methods for increasing sensitivity by improved amplification and labeling techniques are ongoing, and it remains to be established whether whole-genome chips vs. custom chips will be used for selected indications.
Cell Death Pathways in Acute Leukemia
Chemotherapy and irradiation trigger apoptosis in tumor cells and an understanding of the biochemical pathways involved in apoptosis provides an opportunity to classify tumors based on their response to common induction regimens. Multiple distinct signaling pathways regulate apoptosis, but two major cell death pathways have been implicated in hematological malignancies: the mitochondrial pathway and the death receptor pathway (Figure 4 ).44 Both of these pathways ultimately activate members of the caspase family of proteins that are responsible for executing the terminal phases of apoptosis. p53 protein levels rise in response to various cellular stresses including chemotherapy. p53 induces the loss of mitochondrial membrane potential with subsequent release of cytochrome c, which forms a complex, the “apoptosome,” with the adapter molecule Apaf-1, ATP, and caspase-9.45,46 This complex, in turn, activates caspase-3.47
Another proximal pathway of cell death involves death receptor signaling at the cell surface. Binding of CD95-L and other tumor necrosis factor (TNF) family ligands to their death inducing receptors, CD95/APO-1/FAS or TNF-α and TRAIL respectively, leads to receptor trimerization and the recruitment of adapter molecules.48 These molecules include FADD/MORT-1 that in turn lead to recruitment and activation of caspase-8.49 This initiator caspase also cleaves and activates downstream caspases, including caspase-3.50 Although generally described as being distinct, these two proximal pathways are interconnected. For example, caspase-8 cleaves the pro-apoptotic protein BID, which results in translocation to the mitochondria and release of cytochrome c.51 Finally members of the Bcl-2 protein family play pivotal roles in the decision and execution phases of apoptosis in the mitochondrial pathway.52 To date, 24 Bcl-2 family members have been identified as either pro- (e.g., Bax, Bak, Bcl-XS, Bid, Bad, and Noxa) or anti- (e.g., Bcl-2 and Bcl-XL) apoptotic proteins.53 Bcl-2 proteins form homo- and heterodimeric complexes to regulate mitochondrial channel formation and subsequent release of cytochrome c from the mitochondria.
Several studies have examined the prognostic significance of apoptotic protein expression in leukemia. Defects in the p53 pathway are distinctly rare in childhood malignancies including ALL, where mutations are detected in < 5% of cases at the time of initial diagnosis.54 However, relapsed blasts may harbor mutations of p53 gene much more commonly.55 Further, ALL blasts at relapse have been noted to express high levels of the Mdm-2 protein, which abrogates p53 signaling.56
Liu et al evaluated changes in apoptotic proteins expression that occur in response to chemotherapy in 33 children with acute leukemia just prior to and 1, 6 and 24 hours following the administration of multiagent chemotherapy.57 They found great heterogeneity in the patterns of apoptotic protein expression in the initial response to chemotherapy among individual patient samples. Importantly, no increases in p53, p21 or MDM-2 protein expression were seen in leukemic blasts from the standard risk patients whose initial treatment consisted of the non-p53-dependent drugs, vincristine and prednisone. In the subgroup of children who received at least one p53 dependent drug, patients could be segregated into two groups, one group that showed up-regulation of p53 protein and its target p21, and another group that showed no increase following therapy, thus identifying at least two distinct pathways leading to apoptosis. Dr. John Reed will elaborate further on the apoptotic pathway in ALL in this session. Application of new approaches to analyze protein levels globally (the “proteome”) is likely to reveal patterns of apoptotic protein expression predictive of long-term outcome.
Summary and Conclusions
Current approaches to risk-based therapy rely on predictive algorithms that use a combination of clinical and biological variables related to the patient and leukemic clone. Further refinement depends on assessment of tumor regression measured by morphology and quantitative MRD detection. Despite improvements in risk-assignment, patients are cured or fail therapy through unknown mechanisms. Global analyses of transcript and protein expression patterns in samples will no doubt refine predictive classification systems and yield insight into mechanisms of tumor regression and resistance.
II. Pharmacogenetics of Acute Lymphoblastic Leukemia
Mary V. Relling, PharmD,* and Stella Davies, MD
Departments of Pharmaceutical Sciences, St. Jude Children’s Research Hospital, 332 N Lauderdale Street, Memphis TN 38101, and University of Tennessee, Memphis, TN
This work was supported by NCI CA 51001, CA 78224, CA21765, and the NIH/NIGMS Pharmacogenetics Research Network and Database (U01 GM61393, U01GM61374 http://pharmgkb.org) from the National Institutes of Health; by a Center of Excellence grant from the State of Tennessee; and by American Lebanese Syrian Associated Charities (ALSAC).
The contribution of acquired genetic changes in acute lymphoblastic leukemia (ALL) blasts to the long-term outcome of treatment has been widely studied, and genetic subtype of ALL blasts (e.g., presence of the TEL/AML1 translocation, MLL rearrangements, hyperdiploidy) is well accepted as one of the features that is used to “individualize” therapy.1 Although many of the mechanisms by which these acquired changes affect prognosis and response to therapy are unknown, their strong prognostic significance has led to use of these somatically acquired genetic variations to intensify (e.g., for MLL rearrangements) or to deintensify (e.g., TEL/AML1) therapy. Much less attention has been given to the role of germline, inherited genetic variation to the outcome of ALL therapy.
It has been known that inheritance affects interindividual variability in response to specific drugs for almost 50 years.2–,4 Driven by phenotypic variation, the field of pharmacogenetics first developed in the absence of molecular biology. Pharmacogenetics is the study of how interindividual genetic variability affects interindividual differences in drug response. Based on a phenotype-to-genotype approach, it is understandable that the first important examples of pharmacogenetics were monogenic, relatively penetrant traits, and molecular biology eventually defined the molecular genetic basis of phenotypic variability.5
Pharmacogenetics can now be conducted using a genotype-to-phenotype approach. The private initiative to sequence “the” human genome involved the sequencing of germline DNA from 5 individuals.6 Related initiatives, from the Single Nucleotide Polymorphism (SNP) Consortium and other groups,7 indicate that there is no justification for the article “the” when referencing our genomes, and that each of us may differ from other human individuals on average every 300–1500 nucleotides.8 These interindividual differences in human genomes may have important functional consequences, and partly account for the ways in which individuals differ from one another in the risk of disease (e.g., in the risk of cancer) and in probability of favorable versus unfavorable outcomes for treatment of cancer (e.g., relapse versus remission; adverse effects versus none). With the technical improvements in assessing genomic variation, a genotype-to-phenotype approach may facilitate the elucidation of effects of multigenic variation on drug-induced phenotypes. Thus, there is increased interest in determining which of the millions of human genetic variations are functionally important, and which, if any, may be important for individualizing therapy for a number of diseases.
Childhood ALL represents a disease that theoretically can benefit tremendously from individualizing dosages. Medications alone can cure the disease, otherwise uniformly fatal, in over 75% of patients; the medications have a narrow therapeutic range, with death from drug toxicity or second tumors being a significant cause of mortality (in addition to relapse); drug-induced adverse events can be dose-limiting in many cases; dose intensity is an important determinant of outcome; there is significant interpatient variability in systemic exposure to most of the antileukemic agents that have been examined; and there is proof of principle that adjusting dosages based on drug clearance improves ALL outcomes.9 Therefore, genetic variants that affect the probability of cure versus adverse effects of antileukemic agents are likely to have an important impact on ALL outcomes. Differences in outcomes may be influenced by population polymorphisms in genes that influence the disposition of chemotherapy drugs (pharmacokinetics), or influence the response to these drugs (pharmacodynamics). It should also be noted that germline genetic variation may influence the probability of or the nature of the acquired genetic changes in ALL, thus influencing directly or indirectly the intrinsic sensitivity of the blasts.
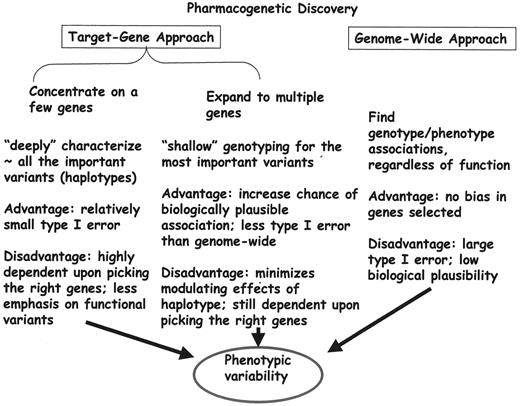
Approaches to establishing genotype/phenotype associations (Figure 5 ) include genome-wide approaches and target gene approaches, in which a small number of genes are very “deeply” sequenced or a somewhat larger number of functionally important genotypes are assessed, haplotypes determined, and associations with phenotypes explored. Several genetic polymorphisms have been studied in childhood ALL.
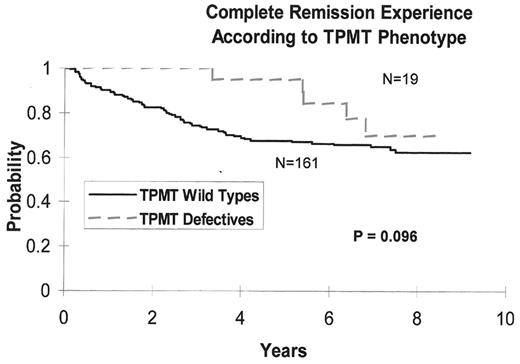
One of the key medications for treatment of ALL is 6-mercaptopurine (6MP). Thiopurine methyltransferase (TPMT) is a key enzyme in the metabolism of 6MP. TPMT activity is inherited as an autosomal codominant trait, and activity is polymorphic in all tissues and in all large populations studied to date. About 1 in 300 people are TPMT-deficient, and approximately 10% inherit intermediate TPMT activity due to heterozygosity at the TPMT locus.10–,13 We and others have shown that, in over 90% of the cases, defective TPMT activity is due to inheritance of TPMT alleles containing at least 1 of 3 single nucleotide polymorphisms (SNPs).14,15 These SNPs have been shown to lead to enhanced protein degradation as the mechanism underlying low TPMT activity.16 Individuals with both alleles carrying inactivating mutations (homozygous mutant) cannot methylate (inactivate) 6MP base, accumulate extremely high levels of active thioguanine nucleotides, and thus have unacceptable, life-threatening toxicity from normal doses of 6MP. The fate of TPMT heterozygotes was less clearly defined. In an analysis of 182 children (St Jude Children’s Research Hospital Protocol Total XII) receiving antimetabolite based therapy for ALL, we examined in detail the impact of 6-MP dosing and metabolism on outcome of treatment for ALL.17,18 We showed that the cumulative incidence of 6-mercaptopurine dose reductions for myelosuppression was highest among patients homozygous for TPMT deficiency (100% of patients), intermediate among heterozygous patients (35%), and lowest among wild-type patients (7%) (P < .001)—indicating that heterozygosity (present in 10% of the population) will have an impact on the optimal dose of 6MP. Importantly, in a further analysis of the same patient population, we showed that a higher dose intensity of 6-mercaptopurine was associated with improved event-free survival. In agreement with this, we also demonstrated a trend toward better survival in patients with at least 1 defective TPMT allele (who would be expected to have greater efficacy if treated with an equivalent or lesser dose of 6-mercaptopurine) compared with wild-type cases (see Figure 6 ).
This same polymorphism has been linked to the occurrence of drug-induced second cancers among children with ALL. The incidence of malignant brain tumors is increased as much as 6-30 times in survivors of ALL, occurring almost exclusively in those who have received craniospinal irradiation.19 In an analysis of patients enrolled on the St Jude Total XII treatment protocol, we reported a 12.8% incidence of brain tumors in irradiated patients.20 Importantly, the incidence of brain tumors was significantly impacted by TPMT genotype (42% versus 8.3% in defective and wild-type TPMT genotypes respectively; P = .0077). Among children with ALL, defective TPMT has also been associated with the risk of topoisomerase II inhibitor–induced secondary myeloid malignancies by 2 independent groups.21,22 Follow-up laboratory studies indicate that thioguanine incorporation into an oligonucleotide DNA substrate, which would be higher in TPMT defective patients, affects the avidity of topoisomerase II–stabilized DNA cleavage, with and without etoposide present.23 Moreover, thioguanine substitution for guanine in DNA creates a structural modification in DNA,24 which affects the interactions of multiple DNA-directed enzymes.25,26 Thus, there are multiple mechanisms whereby a pharmacogenetic polymorphism in TPMT could affect the disposition of 1 antileukemic drug (6MP), which could then in turn have profound effects on the adverse effects of other elements of therapy (e.g., topoisomerase II inhibitors, cranial irradiation).
Thymidylate synthetase (TS) catalyzes the intracellular conversion of deoxyuridylate monophosphate to deoxythymidylate monophosphate, which makes it an essential enzyme in proliferating cells.27 Thymidylate synthase is the target of several anticancer drugs, including the widely used antileukemic agent, methotrexate.28–,30 TS expression has been related to a germline polymorphism in the number of tandem-repeats in its enhancer, with the triple-repeat associated with increased expression of thymidylate synthase, which has been linked to poor antitumor response to the TS inhibitor 5-fluorouracil in adult gastrointestinal tumors.31 The TS enhancer polymorphism has been studied in children with ALL32 and found to be associated with outcome in 205 children with ALL treated with a number of different methotrexate regimens. Individuals who were homozygous for the triple repeat had a poorer outlook than those with other genotypes (odds ratio 4.1, 95% Confidence Interval [CI] 1.9-9.0, P = .001). In a follow-up of this study, Lauten et al reported a case control study of the frequency of thymidylate synthase polymorphisms in 40 children who relapsed and 40 children with ALL successfully treated on Berlin-Frankfurt-Munster (BFM) protocols.33 This study found that the thymidylate synthase polymorphism had no impact on relapse of disease. The discrepant results from these 2 studies could be because of heterogeneity in the ALL cases, as both included subsets of patients enrolled on the respective treatment protocols, and the effect of thymidylate synthase polymorphism might be specific to particular molecular and immunophenotypic subsets of ALL. In addition, these studies included cases treated quite differently and it is possible that the polymorphism is important only in a specific therapeutic context. For example, higher doses of methotrexate were used in the BFM studies than in the treatment protocols used in the study of Krajinovic et al,32 and it is possible that the use of high doses of methotrexate can overcome the adverse impact of the TS polymorphism.
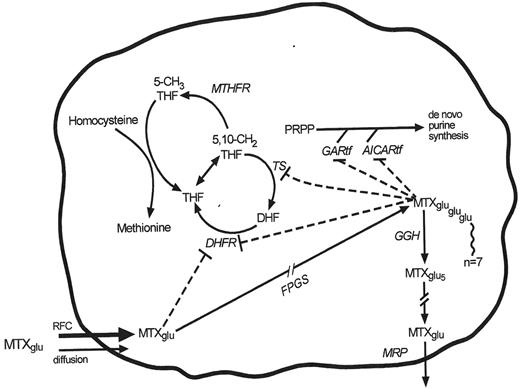
Methotrexate plays a central role in treatment of ALL, and several studies have indicated the importance of achieving high intracellular concentrations of this drug.34–,37 Resistance to methotrexate can be a result of altered cellular uptake,38 and in vivo accumulation of methotrexate is related to the expression of the reduced folate carrier (RFC1).39 A common G(80)A polymorphism has been described in the RFC1 gene, which encodes the major transporter for MTX influx into ALL blasts. In a study of 204 children with ALL treated on a heterogeneous group of treatment regimens, children with the A allele variant had worse event-free survival than patients with the GG genotype (P = .04).40 However, patients homozygous for the A allele had higher levels of MTX (P = .004) than the other genotype groups. Thus, the role of this RFC polymorphism remains unclear, and may interact with other folate-related polymorphisms. A simplified diagram (Figure 7 ) of methotrexate-related targets and enzymes illustrates the fact that multiple gene polymorphisms might interact to affect the pharmacodynamics of this critically important agent for ALL.
The enzyme 5,10-methylenetetrahydrofolate reductase (MTHFR), which catalyzes the reduction of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate, is crucial to folate metabolism. A common polymorphism, a C-to-T substitution at nucleotide 677 (replacing alanine with valine),41 reduces the activity of MTHFR but results in a much less severe phenotype than the rare mutations that cause severe MTHFR deficiency. Approximately 10% of Caucasians and 1.0% of African Americans are homozygous for the lower-activity allele.42 The 677T/T genotype has been associated with hyperhomocysteinemia,43,44 especially in patients with low folate45,46; a lesser effect of a second common SNP at 1298 (A > C) of MTHFR has also been demonstrated.47 A higher incidence of gastrointestinal or hepatic toxicity following chronic, low-dose methotrexate has been noted among patients with the 677T allele48,49 although our preliminary analysis in a relatively small group (53) of children with ALL did not link MTHFR genotypes with MTX-associated neurotoxicity.50 Thus, the role of MTHFR genotypes on MTX-related toxicity and efficacy is still somewhat unclear, and may depend upon the context (high-dose, low-dose, chronicity) of methotrexate therapy in the ALL trials of interest. Common polymorphisms have also been demonstrated in cystathionine beta synthase51,52 and dihydrofolate reductase.53
Conjugation of electrophilic compounds to glutathione, mediated by the family of glutathione S-transferase enzymes (GSTs), is an important detoxifying pathway for mutagens such as organophosphates (including pesticides), alkylating agents, epoxides, and polycyclic aromatic hydrocarbons.54,55 The glutathione S-transferase mu(μ)1 (GSTM1) and the glutathione S-transferase theta 1 (GSTT1) genes are polymorphic in humans, and the phenotypic absence of enzyme activity is due to a homozygous inherited deletion of the gene.56 As we and others have shown, the frequency of the null phenotype varies by race, with approximately 50% of whites and 28% of blacks having the GSTM1 null phenotype.57 The frequency of the GSTT1 null phenotype is 15% in whites and 24% in blacks. It has been suggested that GST expression may play an important role in the outcome of therapy of leukemia as GSTs detoxify many of the drugs used to treat leukemia, and are involved more generally in “protecting the genome” from electrophilic oxidative damage.58 In an immunohistochemical study of 71 cases of childhood ALL, ALL blast samples from 44 were negative for μ class GST; of these, 39 (82%) remained in remission. Of 27 patients who were positive for μ class GST, only 14 (52%) remained in remission,59 so that expression of μ class GST predicts a 3-fold increased risk of relapse (95% CI 1.25-7.26). Pharmacogenetic studies from St Jude Children’s Research Hospital in 197 children with ALL demonstrated that the null genotype for GSTM1, GSTT1, or both was not found to be a prognostic factor for disease-free survival or probability of hematologic remission.60 Central nervous system relapse tended to be less common in those with the GSTM1 null genotype (P = .054), with similar borderline significant findings reported by the BFM group in a case-control design.61
In an investigation from Children’s Cancer Group (CCG), we analyzed GST genotypes in 710 children with ALL.62 Stratification of cases by age at diagnosis, sex, white blood cell count at diagnosis, B or T lineage, or cytogenetics revealed no differences in genotype frequencies. There were no differences in treatment outcomes according to GST genotype. Varying results from these clinical epidemiologic studies could again be because only subsets of patients enrolled on the respective treatment protocols were genotyped, and that the treatment regimens may differ in their dependence on glutathione conjugation.
Cytokines modify the proliferation and activation of normal hematopoietic cells, and can stimulate or inhibit growth in hematological malignancies also. Plasma levels of tumor necrosis factor (TNF) and interleukin 10 (IL-10) have been associated with therapy outcome in hematological malignancies and are influenced by genetic variation due to germline polymorphisms within the TNF and IL-10 genes. TNF and IL-10 genetic polymorphisms might therefore also influence clinical outcome in childhood ALL. In 214 childhood ALL patients,63 patients with a high-risk TNF haplotype were older than patients with low-risk haplotype (P = .024). No statistically significant associations were found between TNF haplotype and sex, white blood cell (WBC) counts, central nervous system involvement, immunophenotype, response to chemotherapy, and event-free survival. In contrast, Lauten et al64 analyzed the association of TNF and IL-10 polymorphisms with response to initial treatment and risk of relapse in 135 children with ALL, treated according to BFM protocols. BFM trials use clearance of peripheral blood blasts in response to an 8-day course of prednisone for treatment stratification, and have shown rapid clearance of blasts to be a powerful prognostic indicator. The data showed that prednisone poor response was less frequent in patients with the IL10GG genotype, whereas no association of the risk of relapse and IL-10 genotype was found. In the total study group, patients expressing the TNF2 allele showed neither a statistically significant general association with prednisone response nor with risk of relapse compared to subjects homozygous for the TNF1 allele. Nevertheless, there was a higher risk of relapse in poor prednisone responders expressing the TNF2 allele compared to poor prednisone responders not expressing the TNF2 allele. The authors concluded that IL-10 genotype might influence prednisone response in patients with childhood ALL, whereas TNF genotype was associated with the risk of relapse in high risk ALL patients. The authors note that the number of cases in this study was small (n = 135) and the cases heterogeneous, and that further investigation in a larger more homogeneous population is necessary.
Infections remain a serious and common complication of ALL therapy, and (after relapse) are the second most common cause of death among children with ALL. Infection risk may be increased due to polymorphisms involved in the pharmacokinetics of myelosuppressive antileukemic agents (resulting in abnormally high systemic exposure to active drug), or to polymorphisms in genes whose products are involved in protective immunity from pathogens. Polymorphisms in TNFα, IL-10, and mannose binding protein have been linked to the risk of infection in other populations,65– 69 but haven’t been fully evaluated in children with ALL.
Many prior studies suffer from relatively small sizes, the possibility of selection bias because only subsets of patients have been studied, lack of multivariate analyses including other known prognostic factors, and lack of accounting for race and population substructure. The ability to genotype at multiple polymorphic loci, many of which display remarkable racial/ethnic diversity in the frequencies of variant alleles, complicates the use of multivariate analyses. For example, perhaps some of the inferior outcome in blacks compared to whites, which has been reported by several groups,70–,72 is in fact due to different polymorphic allele frequencies. Thus, adjusting or stratifying for race might obscure an important relationship between allele frequency and outcome. Additional analyses are necessary to determine the association of prognostically important, acquired genetic abnormalities in the ALL blasts (e.g., TEL-AML1, t(4;11), t(9;22) etc) with the frequency of specific germline genetic polymorphisms.73,74 As is true for the associations of race with genotypes, associations of acquired molecular defects with germline genotype frequencies will greatly complicate the handling of data in genotype/outcome analyses. The optimal methods for analyzing large genotype/phenotype association studies have not yet been demonstrated.
III. Expression Profiling in Acute Leukemia
James R. Downing, MD*
St. Jude Children’s Research Hospital, Department of Pathology, Room #D4047, 332 N Lauderdale Street, Memphis TN 38105
This work was supported in part by National Cancer Institute grants P01 CA71907-06, CA-21765, and by the American Lebanese and Syrian Associated Charities (ALSAC) of SJCRH.
In most contemporary treatment protocols the different genetic subtypes of pediatric acute leukemia are treated using so-called risk adapted therapy—that is, therapy in which the intensity of treatment is tailored to a patient’s relative risk of relapse. Critical to the success of this approach is the accurate assignment of individual patients to specific risk groups. Unfortunately, this is a difficult and expensive process requiring a variety of laboratory studies including morphology, immunophenotyping, cytogenetics, and molecular diagnostics. With the recent development of expression microarrays it should now be possible to take a genome-wide approach to leukemia classification. This approach not only offers the potential of an efficient diagnostic platform for identifying the known prognostic subtypes of leukemia, but should also help us to identify specific gene signatures that will allow us to more accurately identify those individual patients who are at a high risk of relapse. In addition, this approach offers the potential of providing unique insights into the altered biology underlying the growth of the leukemic cells. However, before this methodology can be applied in the clinical setting significant developmental work remains to be done. Importantly, a number of methodological issues must be considered in both the design and analysis of these studies. In this lecture, I will address some of the more important methodological issues and will then summarize the gene expression data that has been generated in my own laboratory on pediatric acute leukemias.
Methodological Considerations
Expression microarray platforms, either cDNA- or oligonucleotide-based, result in the collection of expression values for a large number of genes, varying from several hundred up to 33,000 genes depending on the specific microarray platform being used. For leukemias, analysis is typically performed on leukemic cells isolated from either a diagnostic bone marrow aspirate or a peripheral blood sample.1– 6 Typically, the leukemic cells are partially purified away from more mature hematopoietic cells by density gradient centrifugation prior to analysis. The leukemic cells are then either processed immediately to isolate total RNA, or frozen as viable cell suspensions and the RNA isolated at a later time.
A number of variables affect the expression profile obtained from a clinical sample. These include, but are not limited to, the percentage of leukemic cells, the time between obtaining the sample from a patient and either freezing or isolating RNA, the quality of RNA extracted, and the methods used for labeling the RNA and detecting the hybridized signals. One of the most important variables is the percentage of leukemic blasts within the sample. Since our goal is to obtain the expression profile of the leukemic cells, we strive to ensure that the sample being analyzed consists of a majority of leukemic blasts. For our initial exploratory studies in pediatric acute lymphocytic leukemia (ALL) our criteria for inclusion in the study has been to restrict our analysis to samples that contain a minimum of 70% lymphoblasts. A second critical variable is the time between obtaining the sample and isolating RNA. Experimental data have demonstrated changes in the expression profile of freshly isolated leukemic blasts compared to those placed on ice or stored at room temperature for extended periods of time. The longer a sample sits prior to RNA extraction the greater is the change in the expression profile. Moreover, the extent of change in the expression profile can vary significantly between leukemia subtypes. This is a confounding variable that for many retrospective studies cannot be controlled. Thus, it is important to know that it exists and to ensure that the interpretation of the results of an expression profiling study takes this into account. It is also important to use an RNA extraction procedure that provides high-quality RNA and to rigorously assess not only the quality and purity of the RNA, but also the efficiency of labeling and hybridization to the microarray. Last, variation in expression profiling can result from a variety of technical issues. Therefore, to minimize these variations it is important to assess the reproducibility of data acquisition throughout an experiment. This can easily be done by analyzing replicate samples at multiple points during the experiment.
Acute Lymphoblastic Leukemia
A number of prognostically important genetic subtypes of pediatric ALL have been identified using a combination of immunophenotyping, cytogenetics, and molecular diagnostics. These include B-lineage leukemias that contain t(9;22)[BCR-ABL], t(1;19)[E2A-PBX1], t(12;21)[TEL-AML1], rearrangements in the MLL gene on chromosome 11, band q23, or a hyperdiploid karyotype (i.e., > 50 chromosomes), and T-lineage leukemias (T-ALL).7–,9 Expression profiling is well suited to assist in identifying these leukemia subtypes. In studies performed by a number of different laboratories, expression profiles have been obtained on a large number of diagnostic pediatric ALL samples.1–,6,10–,13 Specifically, in our own studies, we generated a pediatric ALL expression database from the analysis of over 327 diagnostic samples using the Affymetrix U95 microarray, an array that contains probe sets for approximately 10,000 genes.4 This patient cohort represents a largely unbiased selection of patients, with 80% of the dataset consisting of samples from patients treated on a single institutional protocol. The composition of the dataset being analyzed is a very important variable, since changes in the composition can significantly influence the interpretation of the data. For the identification of leukemia subtype specific expression signatures, it is optimal to have a dataset that approaches the normal distribution of leukemia subtypes seen in the clinical setting. In addition, the overall event-free survival rate of the patients in the dataset should not differ significantly from that seen in clinical practice. Our dataset was designed to try and achieve these goals. More recently, we have extended our analysis of this dataset by analyzing a subset of these cases (n = 132) using the higher density Affymetrix microarrays U133A & B, which contain probe sets for ~33,000 genes.6 The data from these 2 studies are available at: http://www.stjuderesearch.org/data/ALL1 and http://www.stjuderesearch.org/data/ALL3.
When these datasets were analyzed using an unsupervised clustering algorithm with all genes that pass a variation filter, 7 distinct leukemia subtypes were identified. Remarkably, 6 of these represented the known prognostically important leukemia subtypes including: BCR-ABL, E2A-PBX1, TEL-AML1, rearrangements in the MLL gene, hyperdiploid karyotype (i.e., > 50 chromosomes), and T-ALL. In addition, 14 cases were identified that lacked any of these other genetic lesions but had a common expression profile, suggestion that these cases may represent a new leukemia subtype.
Class specific genes can be selected using a variety of different statistical approaches, including t-statistics, a chi-squared metric, weighted average, etc.14–,16 An important aspect to the identification of class discriminating genes is to control for false positive gene correlations, a problem that can frequently occur because the number of genes on the microarray greatly exceeds the number of leukemia samples being analyzed. A variety of mathematical approaches have now been developed to assess the false discovery rate.17 Using these approaches, we can now obtain lists of discriminating genes with less than one false positive class discriminating gene per list. In pediatric ALL, the number of class discriminating genes identified using the Affymetrix U133A and B microarrays varied markedly from group to group, with more than 2000 discriminating genes identified for T-ALL, between 700 and 1000 for E2A-PBX1; TEL-AML1, MLL chimeric genes, and hyperdiploid with > 50 chromosomes, and less than 200 class discriminating genes identified for BCR-ABL.6
To formally assess if the identified genes could be used to accurately diagnose the various subtypes of ALL, we turned to the use of computer-assisted supervised learning algorithms.18 In this analysis, discriminating genes are initially used to build a class assignment algorithm using a subset of the cases defined as the training set. In the training set, the diagnostic classification of each case is known and is used to train the performance of the expression-based classification algorithm. Through a reiterative process of error minimalization using cross-validation, different weights are assigned to the discriminating genes so that in the end an algorithm is built that provides the greatest degree of accuracy on the training set. The performance of the algorithm is then assessed using the blinded test set, which consists of the remaining cases. Using this approach, our data demonstrated that the single platform of expression profiling was able to accurately diagnose the various subtypes of pediatric ALL with an overall accuracy of 96%. Remarkably, this level of accuracy is comparable to that achieved at the best institutions using a combination of contemporary diagnostic approaches. This suggests that microarray-based gene expression profiling may provide a viable approach to the front-line diagnosis of pediatric ALL.
Importantly, the number of genes required to diagnose all of the different leukemia subtypes in a parallel format can be as few as 20. This small number of genes raises the possibility that using these class defining genes may not require a high throughput microarray-based approach, but might instead be accomplished using standard diagnostic methods such as automated real-time reverse transcription polymerase chain reaction (RT-PCR) or multiparameter flow cytometry.
The analysis of the class discriminating genes can also provide new insights into the pathogenesis of the different leukemia subtypes. It is important to recognize, however, that the lists of leukemia subtype discriminating genes can be quite large, and therefore, many competing interpretations can be proposed for the importance of different groups of differentially expressed genes. Thus, to translate these lists into biological information, it is essential to develop testable hypotheses from these gene lists, and then perform direct experiments to either validate or disprove these hypotheses. An example of a testable hypothesis that we have generated from our data is based on the aberrant expression of C-MER in E2A-PBX1 leukemias, which encodes the MERTK receptor tyrosine kinase.4 MERTK is not normally expressed in HSCs and when over-expressed in these cells leads to their transformation. Thus, these data raise the possibility that transformation initiated by E2A-PBX1 may require the aberrant expression of the MERTK, a testable hypothesis. If this can be experimentally proven, then MERTK would represent a good therapeutic target against which a specific tyrosine kinase inhibitor could be developed.
Acute Myeloid Leukemia
Like ALL, one of the most important prognostic factors in acute myeloid leukemia (AML) is the presence or absence of specific karyotype abnormalities.19 Based on this information, AMLs are typically categorized into one of three prognostic groups: favorable, including t(15;17), t(8;21), and inv(16); intermediate, which have normal karyotypes; and unfavorable, which include −5/del(5q), −7/del(7q), inv(3)/t(3;3), +8, and complex karyotypes.9,19 Work from a number of different laboratories has identified unique expression signatures for the 3 major subtypes of favorable risk AML— t(15;17), t(8;21), and inv(16).20–,23 Importantly, as few as 13 genes were shown to be sufficient for the accurate diagnosis of these AML subtypes in a relatively small dataset.20 However, determining whether these expression profiles will allow accurate diagnosis in a clinical setting will require evaluating their performance on a large number of cases including a broader range of AML subtypes.
We have recently completed the analysis of 130 pediatric AML samples using the Affymetrix U133A microarray. As expected, class discriminating genes were identified for each of the major prognostic subtypes of pediatric AML, including t(15;17)[PML-RARα], t(8;21)[AML1-ETO], inv(16)[CBFβ-MYH11], MLL gene rearrangement, and cases with FAB-M7 morphology. When subsets of these genes were used in supervised learning algorithms, an overall diagnostic accuracy of > 95% was achieved. Moreover, we were able to use the expression signatures generated from the pediatric samples to accurately diagnose adult de novo AMLs with the same genetic lesions. Thus, these gene signatures should prove valuable in the diagnosis of AML.
The class discriminating genes again provide a view into the molecular pathology of these leukemias and a number of testable hypotheses can be generated. An immediately apparent problem, however, is that an almost unlimited number of different hypotheses can be generated. Thus, what is needed is some way to prioritize these hypotheses so that those most likely to provide insights can be identified. The approach we have taken is to use mouse models of specific genetic subtypes of leukemias, and to do cross-species comparisons between the human and murine leukemia specific gene expression profiles. For example, we are using murine models of core-binding factor leukemias (TEL-AML1, AML1-ETO, and CBFβ-MYH11) and obtaining expression profiles from hematopoietic cells from the preleukemic phase through to overt leukemia. The expression profiles are then compared to those obtained from the identical genetic subtype of human leukemia. This approach should provide a method for prioritizing genes whose altered expression is likely to be functionally relevant.
Prognosis Prediction by Expression Profiling
The data presented above demonstrate that expression profiling can provide prognostic information by accurately identifying known prognostically important subtypes of both ALL and AML. What remains to be proven, however, is whether expression profiling can also provide independent prognostic information. Studies on a variety of other types of cancer including breast, colon, prostate, and melanoma suggest that this should be possible. In fact, early work on ALL suggests that expression signatures can be identified within specific genetic subtypes of ALL that predict whether a patient will have a high risk of relapsing.2–,4,10 These data, however, should be considered preliminary. For these types of studies to be considered “validated,” it will be essential to first make sure that the data has been checked on a blinded test set. Beyond this, it will also be necessary to show that the expression signatures accurately predict prognosis in an independent dataset that has been generated in a second laboratory. The latter requirement is necessary to make sure that no unrecognized confounding variables are inappropriately influencing the interpretation of the data. Last, for a prognosisassociated expression profile to be of clinical value it will need to be determined if it is specific for a particular therapeutic regimen, or alternatively, predicts prognosis irrespective of the specific therapy being used. Although a number of groups are pursuing these types of studies, it is likely to be years before this type of analysis moves into the clinic.
Summary
Gene expression profiling is yielding a view of the leukemia cells that is not only providing insights into pathogenesis, but is also providing new diagnostic markers and therapeutic targets. In the not too distant future, this information should begin to have a major impact on the way we diagnose and treat leukemia patients. Although considerable work remains to be done before these predictions are realized, our ability to acquire and appropriately analyze this type of data continues to mature at a rapid pace. Thus, the fruits of gene expression profiling should soon help us to accurately identify specific leukemia subtypes, and to select therapies targeted to the underlying molecular lesions or their altered downstream consequences.
IV. Discovery of Novel Biologic Clusters, Molecular Classification Schemes, and Genes Predictive of Outcome in Pediatric Acute Lymphoblastic Leukemia Using Gene Expression Profiling
Cheryl L. Willman, MD*
University of New Mexico School of Medicine, UNM Cancer Research and Treatment Center, 2325 Camino de Salud, NE, Room 101, Albuquerque NM 87131
Over the past three decades, remarkable advances have been made in the treatment of acute lymphoblastic leukemia (ALL) in children. Yet significant challenges remain. Although the use of modern combination chemotherapy and post-induction therapeutic intensification now yield long-term remissions in nearly 75% of children affected by ALL, 25% ultimately relapse with disease that is highly refractory to current therapy.1 Conversely, another 25% of children with ALL who now receive dose intensification are likely “overtreated” and may well be cured using less intensive regimens resulting in fewer toxicities and long-term side effects. Thus, a major challenge for the treatment of children with ALL in the next decade is to improve and refine ALL diagnosis and risk classification schemes in order to precisely tailor therapeutic approaches to the biology of the tumor and the genotype of the host.
Current risk classification schemes in pediatric ALL use clinical and laboratory parameters such as patient age, initial white blood cell count (WBC), and the presence of specific ALL-associated cytogenetic or molecular genetic abnormalities to stratify patients into groups at increasing risk for relapse or treatment failure.1–,10 NCI risk criteria are first applied to all children with ALL, dividing them into “NCI standard risk” (age 1.00–9.99 years, WBC < 50,000) and “NCI high risk” (age > 10 years, WBC > 50,000) categories based on age and initial WBC at disease presentation. In addition to these more general NCI criteria, classic cytogenetic analysis or molecular genetic detection of more frequently recurring cytogenetic abnormalities has been used to stratify B precursor ALL patients more precisely into low, standard, high, and very high risk categories. These chromosomal aberrations primarily involve structural rearrangements (translocations) or numerical imbalances (hyperdiploidy—now assessed as specific chromosome trisomies, or hypodiploidy) (Table 1 ). Alternatively, the rate of disappearance of both B precursor and T ALL leukemic cells during induction chemotherapy (assessed morphologically or by other quantitative measures of residual disease) has also been used as an assessment of early therapeutic response and as a means of targeting children for therapeutic intensification.11–,17 In new risk classification schemes employing all of these factors in the Children’s Oncology Group, children with B precursor ALL with “low-risk” disease (22% of all B precursor ALL cases) are defined as having standard NCI risk criteria, the presence of low risk cytogenetic abnormalities (t(12;21)/TEL;AML1 or trisomies of chromosomes 4, 10, and 17), and a rapid early clearance of bone marrow blasts during induction chemotherapy. Children with “standard risk” disease (50% of ALL cases) are NCI standard risk without “low-risk” or unfavorable cytogenetic features, or are children with low-risk cytogenetic features who have NCI high-risk criteria or slow clearance of blasts during induction. Although therapeutic intensification has yielded significant improvements in outcome in these two risk groups, it is likely that a significant number of these children are currently “overtreated” and could be cured with less intensive regimens resulting in fewer toxicities and long-term side effects. Conversely, a significant number of children even in these good-risk categories still relapse and a precise means to prospectively identify them has remained elusive. “Standard-risk” disease in particular is highly heterogeneous both in clinical and molecular genetic features. Nearly 30% of children with ALL have “high-risk” disease, defined by NCI high-risk criteria and the presence of specific cytogenetic abnormalities (Table 1 ); again, precise measures to distinguish children more prone to relapse in this heterogeneous group have not been established. Finally, in a minority (approximately 3%) of children with B precursor ALL, a very poor outcome has been associated with certain “poor prognosis” cytogenetic abnormalities (t(9;22), hypodiploid DNA content < 45 chromosomes). While T ALL cases have not been traditionally divided into distinct risk groupings similar to B ALL, recent gene expression profiling studies published by others18 indicate that distinct intrinsic biologic clusters of T ALL cases can be defined.
Thus, despite the refinement of risk classification schemes employing cytogenetics and the rate of clearance of leukemic blasts or other measures of minimal residual disease, current diagnosis and risk classification schemes in pediatric ALL remain imprecise. Children with ALL more prone to relapse who require more intensive approaches and children with low-risk disease who could be cured with less-intensive therapies are not adequately predicted by current classification schemes and are distributed among all currently defined risk groups and a precise means to prospectively identify such children has remained elusive. As striking differences in therapeutic response and outcome may still be observed in ALL patients with the same cytogenetic profile or within the same risk classification group, it is likely that other molecular genetic abnormalities and functional activation or inactivation of critical cellular pathways (cell signaling, cell cycle regulation, adhesion, DNA repair, apoptosis, drug resistance) in leukemic cells also impact disease biology and therapeutic response. Thus, many investigators in this field are engaged in applying large-scale genomic technologies that measure global patterns of gene expression in leukemic cells to acquire systematic gene expression profiles and sets of genes that can be used for improved diagnosis and risk classification in pediatric ALL and for the prediction of therapeutic response or resistance in individual patients.
Funded under the NCI Director’s Challenge Program: Toward a Molecular Classification of Tumors (NCI CA88361: Molecular Taxonomy of Adult and Pediatric Acute Leukemia; PI: CL Willman, Co-PI: WM Carroll; http://dc.nci.nih.gov), our investigative team has recently completed comprehensive gene expression profiling in two large statistically designed, retrospective cohorts of pediatric ALL patients, designed by Dr. Jon Schuster, registered to clinical trials previously coordinated by the Pediatric Oncology Group (POG): (1) a cohort of 127 infant leukemias; and (2) a case control study of 254 pediatric B-precursor and T-cell ALL cases. Using both unsupervised learning tools and novel data visualization techniques to discover intrinsic biologic clusters of ALL and supervised machine learning algorithms and statistical methods to model gene expression profiles associated with clinical characteristics, cytogenetics, and therapeutic response, we have made a number of novel and potentially important discoveries. We have identified novel intrinsic biologic clusters of ALL and novel genes that are strongly predictive of outcome. These discoveries are providing us with new tools and approaches to refine and improve molecular diagnosis and risk classification in pediatric ALL that will be implemented and tested prospectively in the context of Children’s Oncology Group (COG) clinical trials within the next 5 years.
Gene Expression Studies in Infant Acute Leukemia: Novel Biologic Clusters and Genes Predictive of Outcome
Over the past three years, many investigative teams have developed reproducible methods for leukemia blast purification, RNA isolation, linear amplification, and hybridization to oligonucleotide and printed cDNA microarrays. Our approach is a modification of a double amplification method originally developed by Ihor Lemischka and colleagues from Princeton University (protocols available at the NCI Director’s Challenge website http://dc/nci/nih/gov). Using Affymetrix (U-95A.v2) oligonucleotide arrays, we have obtained gene expression profiles from ALL patient cohorts in the KUGR (Keck University of New Mexico [UNM] Genomics Resource) housed in the UNM Cancer Research Facility (http://hsc.unm.edu/som/micro/genomics). We have used powerful, multidimensional unsupervised learning algorithms and data visualization tools (VxInsight, principal component analysis)19,20 for class discovery and for the identification of intrinsic biologic clusters of pediatric leukemia. Supervised computer learning methods (primarily Bayesian analysis of gene expression networks, support vector machines [SVM], and neuro fuzzy logic)22– 24 were used to identify genes and groups of genes that were significantly associated with various parameters (outcome, specific cytogenetic abnormalities, etc) by our collaborators at UNM and Sandia National Laboratory (http://www.cs.sandia.gov/projects/VxInsight.html).
Infant Leukemia Cohort Studies
In the 2 POG infant trials, 142 retrospective cases (9407 for infant ALL; 9421 for infant AML) were initially chosen for analysis in our infant leukemia cohort. Infants as defined were < 365 days in age and had overall extremely poor survival rates (< 25%). Of the 142 cases, 127 were ultimately retained in the study; 15 cases were excluded from the final analysis due to poor quality total RNA, cRNA amplification, or hybridization. Of the final 127 cases analyzed, 79 were considered traditional ALL by morphology and immunophenotyping and 48 were considered AML. Of the 127 cases, 59 had rearrangements of the MLL gene.
Nonsupervised learning tools for hierarchical clustering of gene expression data and other clustering approaches are most useful for the discovery of intrinsic biology in patient cohorts and discovery of coincident patterns of gene expression. However, most unsupervised hierarchical clustering algorithms are not powerful enough to resolve multiple clusters in very large datasets (> 12,000 genes in > 100 cases) without the investigator first selecting a more limited subset of expressed genes on which to actually perform clustering (usually < 100), which may introduce significant bias and limit the analysis. In the retrospective infant leukemia cohort, nearly 7000 of the 12,625 genes and ESTs on the Affymetrix U95A.v2 chip were expressed at significant levels in at least 1 of the 127 infant leukemia cases. To attempt to avoid bias by limiting gene selection and to use higher dimensional methods for discovery of inherent clusters of patients based on common gene expression patterns, we turned to 2 methods: (1) principal component analysis (PCA: see Bioinformatics Core), and (2) VxInsight (http://www.cs.sandia.gov/projects/VxInsight.html), a new and very powerful tool for nonsupervised clustering and visualization of genomic data developed by our collaborators at Sandia National Laboratory. VxInsight has the capacity to cluster patients or genes, using all of the gene expression data without having to select smaller subsets of genes for actual clustering, in a novel and intuitive way.
When VxInsight or PCA was applied to the infant leukemia dataset, we discovered that there were 3 statistically significant, intrinsic biologic groups of infant leukemia and that these intrinsic biologic groups could not simply be predicted by ALL versus AML labels or by the presence or absence of cytogenetic abnormalities involving MLL as these labels were distributed among each of the intrinsic biologic clusters (Figure 8; see Appendix, page 612) (MP Mosquera-Caro et al, unpublished data). Importantly, when an alternative multidimensional clustering method (PCA) was used on this dataset (data not shown), we also identified 3 distinct clusters. And as the membership between the clusters defined by VxInsight and PCA was 99% correlated, it was satisfying that 2 highly different multidimensional clustering algorithms yielded a highly similar result. In each cluster, an individual patient is represented by a pyramid (highly similar patients in each cluster will be overlapping in this “high level” view). By querying VxInsight in “real-time,” we could determine which cases had been assigned an ALL versus an AML label (Figure 8B and C) or which cases had rearrangements of the MLL gene (Figure 8B). In addition, we performed the ANOVA (analysis of variance) function in VxInsight to provide the most statistically significant genes that distinguish each cluster (some of the most significant of these genes are listed in the boxes adjacent to each cluster in Figure 8C). The top cluster of cases (Figure 8A, labeled white), which we refer to as cluster A, contained 21 infant leukemia cases, 16 of which had been labeled ALL and 5 AML. The shared gene expression profile of these cases is unique when compared with the other clusters and is highly similar to recent reports of gene expression profiles in very primitive hematopoietic stem cells (HSC).25 The gene expression profile shared by cases in cluster A reflect the earliest hematopoietic antigens (high EPOR, AML1, KIT, CD34, FLK1, and HOX family members) as well as a number of genes associated with the development of endothelial cells, leading us to speculate that this distinct group of infant leukemias may have arisen by transformation of very primitive HSCs or even the HSC-endothelial cell precursor, the hemangioblast. Perturbations of the TGF-β/bone morphogenetic protein/SKI oncogene pathway involved in early mesoderm development are unique to this cluster and may provide new insights into novel therapeutic approaches. Interestingly, the majority of MLL-containing cases in this group were t(4;11) variants. The leftmost cluster (Figure 8A, labeled green) contains 52 cases, 51 of which were ALL. Many of these cases contained a t(4;11) or other MLL variant, but the gene expression profile of the t(4;11)-containing cases in this cluster were quite distinct from those in cluster A and are quite similar to the gene expression profiles obtained by Armstrong and colleagues.26 The cases in this relatively homogeneous cluster, which we refer to as cluster B, share a gene expression profile reflective of a committed B lymphocyte precursor, more differentiated than the cases in cluster A. Finally, the third distinct cluster of infant cases (Figure 8A, blue, bottom right) is quite heterogeneous, containing 54 cases, 42 with AML, and 12 with ALL morphology. The MLL variants seen in this group were more frequently t(9;11) and other MLL rearrangements. The shared gene expression profiles distinguishing these interesting cases include expression of many members of the RAS family and genes that impinge upon, regulate, or are regulated by RAS. In addition to RAS signaling pathways, cases in this cluster are also characterized by expression of several DNA repair and GST genes, leading us to speculate that it is this group of infant leukemia cases that might uniquely result from environmental exposures. Clearly the 3 intrinsic biologic groups of infant leukemia that we have identified through gene expression profiling are not predicted by ALL or AML labels or MLL-containing cytogenetic abnormalities. If validated, the distinct sets of genes that can be used to identify each biologic group (Figure 8C) represent potentially important diagnostic and therapeutic targets. Using supervised learning techniques, we have also identified genes that are predictive of outcome at initial diagnosis in this infant cohort. While we could not statistically model outcome using all of the cases combined (ALL and AML) or when cases were divided into morphologically-defined groups (AML versus ALL), we could best model genes predictive of outcome when cases were grouped (conditioned) on which VxInsight (or PCA) cluster that they were assigned to (P = .01), providing further evidence that the VxInsight cluster assignment has biologic and clinical validity.
Gene Expression Studies in Pediatic ALL: Novel Biologic Clusters and New Genes Strongly Predictive of Outcome at Initial Diagnosis
To obtain gene expression profiles associated with outcome in a statistically significant fashion, we developed a care control cohort design that could compare and contrast gene expression profiles in distinct cytogenetic subgroups of ALL patients who either did or did not achieve a long-term remission (for example, comparing children with t(4;11) who failed versus those who achieved long-term remission. The design developed by Dr. Jonathan Shuster was constructed to look at a number of small independent case-control studies within B precursor ALL. These included t(4;11), t(9;22), t(1;19), monosomy 7, monosomy 21, female, male, African American, Hispanic, and POG AlinC15 arm A. Cases were selected from several completed POG clinical treatment trials, but the majority of cases came from the POG 9000 series. As standard cytogenetic analysis of the samples from patients registered to these older trials would not have usually detected the t(12;21), we performed RT-PCR studies on a large cohort of these cases to select ALL cases with t(12;21) who either failed therapy (n = 8) or achieved long-term remissions (n = 22). Cases who “failed” had failed within 4 years while “controls” had achieved a complete continuous remission of 4 or more years. A case-control study of induction failures (cases) versus complete remissions (CRs; controls) was also included in this cohort design as was a T-cell cohort. It is very important to recognize that the study was designed for efficiency and maximum overlap, without adversely affecting the random sampling assumptions for the individual case-control studies. As for the infant leukemia cases, gene expression arrays were completed using 2.5 micrograms of RNA per case (all samples had > 90% blasts) with double linear amplification. All amplified RNAs were hybridized to Affymetrix U95A.v2 chips.
Class Discovery: Unsupervised Learning in ALL
Excellent studies previously published by Yeoh et al27 and Ross et al28 have found that the gene expression profiles of pediatric ALL cases cluster according to the recurrent cytogenetic abnormalities associated with this disease, and thus, that cytogenetics essentially define the intrinsic biologic groups of disease. However, Yeoh et al first used supervised learning algorithms (primarily support vector machines) to identify expressed genes that were associated with each recurrent cytogenetic abnormality in ALL.27 Using a highly selected set of 271 genes that resulted from this supervised learning approach, hierarchical clustering or PCA was performed. As would be expected from this approach, distinct ALL clusters could be defined based on shared gene expression profiles and each cluster was associated with a specific cytogenetic abnormality. Similar to this approach, we also first used supervised learning methods for class prediction (Bayesian networks, support vector machines) to identify a set of 147 genes from the POG ALL case control study that could predict for the presence of the most frequent cytogenetic abnormalities seen in ALL; clustering with this limited set of genes did indeed yield clusters that correlated relatively well with specific karyotypes. However, when we performed a full unsupervised learning approach (VxInsight, principal component analysis), we discovered 9 novel biologic clusters of ALL (2 distinct T ALL clusters and 7 distinct B precursor ALL clusters) each with distinguishing gene expression profiles (Figure 9; see Appendix, page 612).29 Two distinct clusters of T lineage ALL (Figure 9, S1 and S2, and 7 distinct B precursor ALL clusters (A, B, C, X, Y, Z)) were identified. Using ANOVA, we identified over 100 statistically significant genes uniquely distinguishing each of these cohorts; review of these lists of genes reveals many interesting signaling molecules and transcription factors. While there were some trends, no cytogenetic abnormality precisely defined any specific cluster. Cases with a t(12;21) or hyperdiploidy, both conferring low risk and good outcomes, tend to cluster together and were seen primarily in clusters C and Z as well as the top component of the X cluster. On the terrain map from VxInsight (Figure 9, top), these 3 cluster regions (C, Z, and X) are actually fairly closely approximated indicating they are more related than for example cluster C to cluster S2. Similarly, the t(1;19) cases clustered in Y had a poorer outcome than those in clusters A and B. Finally, it is of interest that ALL cases with t(9;22) simply did not cluster; they appeared to be distributed among virtually all B precursor clusters. Results similar to our own were recently reported by Fine et al.30 While we do not understand the significance of this result, it suggests that the t(9;22) is a preleukemic or initiating genetic lesion that may not be sufficient for leukemogenesis, or alternatively, that clones with a t(9;22) are quite genetically unstable and transformation and genetic progression may occur along many pathways.
These studies suggest that cytogenetic-based classification schemes alone may not be sufficient for precise diagnosis and risk classification in ALL and that gene expression profiling may provide a more comprehensive view of the biologic potential and functional activation state of leukemic cells. Thus, in summary, unsupervised learning approaches reveal tremendous complexity and novel biologic clusters of ALL that are not precisely correlated with the frequent cytogenetic abnormalities seen in this disease. These results may imply that these genetic lesions are not fully sufficient for transformation or are initiating genetic lesions. Gene expression profiling may therefore provide a more comprehensive picture of the genetic progression in this heterogeneous disease and may be more capable of grouping cases with similar functional activation or inactivation of specific biologic pathways. As we further develop insight into the functional pathways that are perturbed in each of these clusters, we may develop new insights for improved molecular classification and therapeutic targeting.
Class Prediction: Identification of Novel Genes Highly Predictive of ALL Outcome
A second major goal of our recent studies was to identify sets of genes that could be used to predict outcome as well as therapeutic resistance and treatment failure. This is a more complex task, as the biologic or clinical labels for continuous complete remission (CCR) or long-term remission and “failure/relapse” are not pure or precisely defined, and are dependent on time and on numerous host and tumor factors. In addition, there is a tendency to “overfit” supervised learning algorithms and computational approaches to find predictive profiles and genes. Frequently, genes discovered by these approaches cannot be independently validated in test sets. Thus, these methods require carefully designed cohorts, cross-validation, and statistical analyses. It is best to confirm preliminary results on totally independently derived datasets.
To identify genes strongly predictive of outcome in pediatric ALL, we divided the retrospective POG ALL case control cohort (n = 254) into training (2 of 3) and test (1 of 3) sets and applied supervised learning methods (support vector machines, Bayesian networks, VxInsight) and performed statistical analyses. Through this approach, we identified a limited set of novel genes that were predictive of outcome in pediatric ALL.29 A particularly strong set of candidate genes (G0, G1, G2) predictive of outcome was identified through a Bayesian network approach; further attesting to their potential validity, these genes were also identified using other supervised learning methods (SVM, VxInsight) (Table 2 ).
Our analyses have shown that pediatric ALL patients whose leukemic cells contain relatively high levels of expression of each of these genes have an extremely good outcome while low levels of expression of these genes is associated with treatment failure. G0, a novel gene identified only as an EST and a hypothetical protein on the Affymetrix U95A chip, which we have now fully cloned and named OPAL1 (for Outcome Prediction in Acute Leukemia number 1) conferred the strongest predictive power and was at the top of predictive lists in all supervised learning methods employed. By assessing the level of OPAL1 alone, ALL cases could be split into those with good outcomes (OPAL1 high: 87% long-term remissions) versus those with poor outcomes (OPAL1 low: 32% long-term remissions, 68% treatment failure). Detailed statistical analyses of the significance of OPAL1 expression in the retrospective cohort revealed that low OPAL1 expression was associated with induction failure while high OPAL1 expression was associated with long-term event-free survival, particularly in males. Higher levels of OPAL1 were also associated with certain cytogenetic abnormalities (such as t(12;21) and normal cytogenetics). Although the number of cases were limited in our initial retrospective cohort, low levels of OPAL1 appeared to define those patients with low-risk ALL who failed to achieve long-term remission, suggesting that OPAL1 may be useful in prospectively identifying children with low or standard risk disease who would benefit from further intensification. As a very significant validation of the importance of this novel gene in outcome prediction, we validated its predictive ability in an independent array cohort of cases developed by St Jude and reported by Yeoh et al.27 OPAL1 was also highly predictive of outcome in this set of cases, a critical validation step. We also found a trend between high OPAL1 and improved outcome in our retrospective cohort of infant ALL cases. OPAL1 was also highly predictive of outcome in T ALL (P = .02), as well as B precursor ALL. In summary, supervised learning and computer learning algorithms are identifying new genes that may significantly contribute to the refinement of risk classification in acute leukemia and which may be further developed as diagnostic and therapeutic targets.
V. Apoptosis-Targeted Therapies for Hematological Malignancies
John C. Reed, MD, PhD*
The Burnham Institute, 10901 N. Torrey Pines Road, La Jolla CA 92037
Defects in programmed cell death (apoptosis) mechanisms play important roles in the pathogenesis and progression of hematological malignancies, allowing neoplastic cells to survive beyond their normally intended life-spans and subverting the need for exogenous survival factors. Apoptosis defects also serve as an important complement to proto-oncogene activation, as many deregulated oncoproteins that drive cell division also trigger apoptosis (e.g., Myc; E1a; Cyclin-D1).1 Similarly, errors in DNA repair and chromosome segregation normally trigger cell suicide as a defense mechanism for eradicating genetically unstable cells, and thus apoptosis defects permit survival of the genetically unstable cells, providing opportunities for selection of progressively aggressive clones.2 Lesions in apoptosis pathways also promote resistance to the immune system, inasmuch as many of the weapons cytolytic T-cells (CTLs) and natural killer (NK) cells use for attacking tumors depend on integrity of the apoptosis machinery.3 Finally, cancer-associated defects in apoptosis play a role in chemoresistance and radioresistance, increasing the threshold for cell death, and thereby requiring higher doses for tumor killing.4
Apoptosis Pathways
Apoptosis is caused by proteases known as “Caspases” for Cysteine Aspartyl-specific Proteases.5,6 Caspases constitute a family of intracellular cysteine proteases (n = 11 in humans), which collaborate in proteolytic cascades where Caspases activate themselves and each other. Within these proteolytic cascades, Caspases can be positioned as either upstream “initiators” or downstream “effectors” of apoptosis.
Several pathways for activating Caspases exist (Figure 10; see Appendix, page 613). First, mitochondria induce apoptosis by releasing Cytochrome-c (cyt-c) into the cytosol, which then causes assembly of a multiprotein Caspase-activating complex, referred to as the “apoptosome.”7 The central component of the apoptosome is Apaf1, a Caspase-activating protein that oligomerizes upon binding cyt-c, and then binds pro-Caspase-9 via interaction with its Caspase recruitment domain (CARD) domain. The mitochondrial pathway for apoptosis, also known as the “intrinsic pathway,” is activated by myriad stimuli, including growth factor deprivation, oxidants, Ca2+ over-load, and DNA-damaging agents. In addition to cyt-c, mitochondria also release several other proteins, including endonuclease G, AIF (a death-modulating flavoprotein), and IAP-antagonists SMAC (DIABLO) and OMI (HtrA2) (see below), some of which may promote Caspase-independent (non-apoptotic) cell death.8 A second pathway for apoptosis induction is specific to CTL and NK cells, which spray apoptosis-inducing protease, granzyme B (GraB), onto target cells. GraB then piggy-backs into cells via mannose-6-phosphate receptors (IGFR2) and enters effective cellular compartments via perforin channels.9 GraB is a serine protease, but similar to the Caspases, it cleaves substrates at Asp residues, including several Caspases and some Caspase substrates. The third pathway involves receptors of the TNF family. Of the ~30 members of the TNF family receptors, 8 contain a so-called death domain (DD) in their cytosolic tails.10 Several of these DD-containing TNF-family receptors use Caspase-activation as a signaling mechanism, including TNFR1/CD120a; Fas/APO1/CD95; DR3/Apo2/Weasle; DR4/TrailR1; DR5/TrailR2; and DR6. Ligation of these receptors at the cell surface results in the recruitment of several intracellular proteins, including certain pro-Caspases, to the cytosolic domains of these receptors, forming a “death-inducing signaling complex” (DISC) that triggers Caspase activation, constituting the so-called “extrinsic” pathway for apoptosis.11 The specific Caspases summoned to the DISC are Caspase-8 and, in some cases, Caspase-10. These Caspases contain so-called death effector domains (DEDs) in their N-terminal prodomains that bind to a corresponding DED in the adapter protein, FADD, thus linking them to the TNF-family death receptor complexes. A nuclear pathway for apoptosis regulation, a fourth pathway, may exist which centers on discrete nuclear organelles, called PML oncogenic domains (PODs) or nuclear bodies (NBs). PML is the protein encoded by the fusion partner of the retinoic acid receptor-α (RARα) involved in t(15;17) translocations seen commonly in acute promyelomonocytic leukemia.12 Targeted ablation of the pml gene in mice results in general resistance to apoptosis through unknown mechanisms.13 Several proteins that can promote apoptosis have been localized to PODs, including Daax, Zip Kinase, and Par4, and defects in assembly of these nuclear structures are documented in cancers.12 It remains to be determined how PODs are linked to Caspase activation pathways. Finally, a Caspase activation pathway linked to endoplasmic reticulum (ER)/Golgi stress has been proposed,6 though many details are lacking.
Apoptosis Suppressors
Multiple endogenous antagonists of the Caspase-activation pathways have been discovered, and examples of dysregulation of their expression or function in lymphoma, leukemia, and solid tumors have been obtained. These apoptosis-suppressors have become targets for drug discovery, with the idea of nullifying their cytoprotective functions and restoring apoptosis sensitivity to malignant cells. Alternatively, one can envision directly activating agonists of apoptosis in cancer cells. A summary of some of the more promising strategies for modulating the activity of apoptosis genes and proteins for improved therapy of hematological malignancies is provided below, according to the apoptosis pathways they affect.
Intrinsic pathway
Bcl-2 family proteins (n = 24 in humans) are central regulators of the intrinsic pathway, which either suppress or promote changes in mitochondrial membrane permeability required for release of cyt-c and other apoptogenic proteins.7,14 Overexpression of anti-apoptotic Bcl-2 or Bcl-XL probably occurs in more than half of all hematological malignancies, rendering neoplastic cells resistant to many types of apoptotic stimuli, including most cytotoxic anticancer drugs. Attempts to overcome the cytoprotective effects of Bcl-2 and Bcl-XL in cancer include three strategies: (1) directly attacking the proteins with small-molecule drugs; (2) inducing mRNA degradation with antisense oligonucleotides; and (3) shutting off gene transcription.
First, strategies for attacking the Bcl-2 and Bcl-XL proteins using small-molecule drugs have emerged from an understanding of how our own cells keep these anti-apoptotic proteins under control. In this regard, a large family of endogenous antagonists of Bcl-2 and Bcl-XL has been revealed (so-called “BH3-only proteins”), possessing a conserved amphipathic α-helical domain (BH3) that binds a hydrophobic crevice on the surface of Bcl-2 and Bcl-XL.15 Proof-of-concept experiments using synthetic BH3 peptides have confirmed the validity of strategies based on generating small-molecule drugs that occupy the BH3-binding site on Bcl-2 or Bcl-XL, abrogating their cytoprotective functions. Indeed, several groups have presented pre-clinical data regarding BH3-mimicking compounds. The optimal structure-activity relation (SAR) profile of these compounds remains to be determined, with respect to the issue of selective versus broad-spectrum activity against the various anti-apoptotic members of the Bcl-2-family (Bcl-2; Bcl-XL; Mcl-1, Bcl-W, Bfl-1, Bcl-B), in terms of balancing anti-tumor efficacy against side effects.
Second, nuclease-resistant (phosphorothioate) antisense oligonucleotides targeting the BCL-2 mRNA have advanced to Phase III clinical trials for melanoma, myeloma, CLL, and AML. Phase II trials are also underway for a variety of solid tumors. Because BCL-2 antisense enhances sensitivity to cytotoxic anticancer drugs in vitro and in xenograft models, most clinical trials combine the antisense agents with conventional chemotherapy.16 Uncontrolled Phase II data suggest a benefit to adding BCL-2 antisense to conventional therapy, but definitive proof awaits the Phase III results. Also at issue is whether phosphorothioate-based oligonucleotides work entirely through antisense mechanisms.
Third, with regards to transcriptional silencing, among the “drugable” modulators of BCL-2 and BCL-XL gene transcription are members of the steroid/retinoid super-family of ligand-activated transcription factors (SRTFs). Depending on the tissue of origin of the malignancy, expression of Bcl-2 or Bcl-XL can be down-regulated in specific types of cancer and leukemia cells by small-molecule drugs that modulate the activity of retinoic acid receptors (RAR), retinoid X receptors (RXR), PPARγ, vitamin D receptors (VDR), and certain other members of the SRTF superfamily. RAR and RXR-ligands are already approved for treatment of some types of leukemia and lymphoma, and are in advanced clinical testing for solid tumors. PPARγ modulators have demonstrated antitumor activity in xenograft models of breast and prostate cancer,17 sometimes displaying synergy with retinoids, probably due in part to the fact that PPARγ binds DNA as a heterodimer with RXR. Chemical compounds that inhibit histone deactylases (HDACs), transcriptional repressors that interact with retinoid receptors and other transcription factors, can also favorably modulate expression of Bcl-2 or Bcl-XL in some tumor lines.18 These observations portend opportunities for exploiting endogenous transcriptional pathways for suppressing expression of anti-apoptotic BCL-2-family genes in cancer, with the idea of employing them as chemo- or radio-sensitizers, rather than relying on their antitumor activity as single agents. However, much remains to be learned about the genetic characteristics of malignant cells that dictate responsiveness versus resistance to SRTF-ligands, as well as the complex pharmacological interplay between this class of agents and convention cytotoxic drugs.
Extrinsic pathway
The extrinsic pathway is activated in vivo by TNF-family ligands that engage DD-containing receptors, resulting in activation of DED-containing Caspases. The “death ligands” are expressed on CTLs, NK cells, and other types of immune-relevant cells (activated monocytes/macrophages and dendritic cells) and are used as weapons for eradication of transformed cells.10 Various studies using mice with genetic alterations in genes encoding death ligands or their receptors, as well as use of neutralizing antibodies and Fc-fusion proteins, have provided evidence of important roles in tumor suppression by cellular immune mechanisms. For example, Fas-Ligand (FasL) is important for CTL-mediated killing of some tumor targets, and TRAIL (Apo2-Ligand) is critical for NK-mediated tumor suppression.
Since chemorefractory cells tend to have defects in their intrinsic pathway, given the predominant reliance of cytotoxic drug and x-irradiation on the mitochondrial pathway for cell death,7 interest has emerged in developing therapeutics that kill cancer cells via the extrinsic pathway. In this regard, hope of applying TNF as a biological agent for cancer treatment were dashed by the proinflammatory effects of this cytokine, due to its simultaneous induction of both Caspase-activation pathways and NF-κB. In contrast, the TNF-family cytokines FasL and TRAIL trigger activation of the Caspase pathway, without concomitant induction of NF-κB—raising hopes they might be successfully exploited for cancer therapy, where TNF failed due to toxicity. While agonistic antibodies that trigger the receptor, Fas (CD95), are unfortunately highly toxic to liver,19 TRAIL and agonistic antibodies that bind TRAIL-receptors appear to be well tolerated in vivo. Indeed, a Phase I trial in humans was recently completed using an agonistic antibody directed against TRAIL-Receptor-1 (TRAIL-R1) (also known as Death Receptor-4 [DR4]). In mouse xenograft models using selected human tumor cell lines, TRAIL and agonistic antibodies directed against TRAIL-receptors have been demonstrated to possess potent antitumor activity,20 raising hopes of using these biological agents as a novel approach to cancer treatment and thereby mimicking some of the effector mechanisms normally employed by the immune system in its defense against transformed cells, and potentially bypassing the roadblocks to apoptosis within the intrinsic (mitochondrial) pathway.
Unfortunately, many tumor cell lines display intrinsic resistance to TRAIL, despite expressing the necessary cell-surface receptors, particularly hematological malignancies. This observation implies that during evolution of tumors in vivo, selection occurs for malignant clones capable of withstanding immune attack, and thus successful biological therapy depends on restoring competency of the extrinsic pathway. A possible mechanism for accomplishing this goal has recently been revealed by studies of synthetic triterpenoids, such as CDDO and CDDOm, that trigger a poorly defined pathway for ubiquitination and degradation of FLIP,21,22 an apoptosis-modulating protein that binds pro-Caspase-8 and -10.3 At least in vitro, these compounds sensitize solid tumor cell lines to TRAIL, and induce apoptosis as single agents in leukemia cells, through a Caspase-8-dependent mechanism that remains operative even in chemorefractory cells.21– 23 It remains to be determined whether these promising observations will hold-up in vivo.
Other routes to engaging the extrinsic pathway have recently been revealed that may apply at least to leukemia. For example, the protein kinase C (PKC) modulator Byrostatin induces myeloid leukemia cell lines to produce TNF, resulting in autocrine engagement of TNF-receptors and apoptosis induction through a mechanism that is suppressible by TNFR-Fc fusion protein and Caspase-8 dominant-negative.24 Similarly, all-trans-retinoic acid (ATRA) induces TRAIL production and triggers autocrine induction of apoptosis in acute promyelomonocytic leukemia (APML) cells that harbor the RARα-PML fusion protein resulting from t(15;17) chromosomal translocations.25
Convergence pathway
The intrinsic and extrinsic pathways for Caspase activation converge on downstream effector Caspases (Figure 10; see Appendix, page 613). In this regard, the inhibitor of apoptosis proteins (IAPs) constitute a family of evolutionarily conserved apoptosis suppressors (n = 8 in humans), many of which function as endogenous inhibitors of Caspases.26 All members of this family, by definition, contain at least one copy of a so-called BIR (baculovirus iap repeat) domain, a zinc-binding fold, which is important for their anti-apoptotic activity, present in 1-3 copies. Among the Caspases directly inhibited by human IAP family members XIAP, cIAP1, and cIAP2 are effector Caspases-3 and -7, as well as initiator Caspase-9 (intrinsic pathway). For XIAP, the second BIR domain and the linker region between BIR1 and BIR2 are required for binding and suppressing Caspases-3 and -7, while the third BIR domain binds Caspase-9. Thus, different domains in the multi-BIR containing IAPs are responsible for suppression of different Caspases. IAP family member Livin (ML-IAP), contains a single BIR and inhibits Caspase-9 but not Caspase-3 and -7. Survivin also contains a single BIR and associates with the pro-form of Caspase-9, with assistance from the protein HBXIP, suppressing Apaf1-mediated activation of this important protease in the cytochrome c pathway for apoptosis.27
Pathological overexpression of IAPs occurs in many cancers and leukemias,28 though more detailed information is needed about which of the 8 members of this gene family is deregulated in specific types of malignancies. Antisense-mediated reductions in IAP-family members XIAP, cIAP1, Survivin, or Apollon can induce apoptosis of tumor cell lines in culture, or sensitize cells to cytotoxic anticancer drugs, thus providing proof of concept evidence that the pathological elevations of IAPs found in cancers are important for maintaining cell survival and resistance to chemotherapy.
Endogenous antagonists of IAPs help to keep these apoptosis-suppressors in check, promoting apoptosis. Two of these naturally occurring IAP antagonists, SMAC (Diablo) and HtrA2 (Omi), are sequestered inside mitochondria, becoming released into the cytosol during apoptosis.18 SMAC and HtrA2 have N-terminal leader sequences that are removed by proteolysis upon import into mitochondria, exposing a novel tetra-peptide motif that binds the BIR domains of IAPs. These IAP antagonists compete with Caspases for binding to IAPs, thus freeing Caspases from the grip of the IAPs and promoting apoptosis. Synthetic peptides that mimic SMAC and HtrA2 trigger apoptosis or sensitize tumor cell lines to apoptosis induced by cytotoxic anticancer drugs or TRAIL in vitro and even in some tumor xenograft models.29 Structural information derived from high-field NMR and x-ray crystallography indicates that a tetra-peptide motif is sufficient to bind the BIRs of IAPs, suggesting a path to drug discovery where small-molecule chemical compounds that occupy the same tetra-peptide-binding motif on BIRs can be envisioned as new apoptosis-sensitizing agents for cancer.30 However, many issues remain unresolved, including the questions of which IAP-family members, and which BIR domains within those IAPs, should be targeted by drugs.
Signal Transduction and Apoptosis Regulation
Certain signaling proteins have emerged as potential drug targets for indirectly modulating apoptosis pathways, based on their ability to influence the expression or function of other proteins. Some of these apoptosis-modulating proteins may impact several apoptosis pathways simultaneously, possibly at multiple points. For example, NFkB family transcription factors can induce expression of anti-apoptotic proteins that oppose the intrinsic (Bcl-XL; Bfl-1), extrinsic (FLIP), and convergence (cIAP2) pathways, as well as suppressing expression of death-inducer Bax (intrinsic pathway) in some types of tumor cells.31 Further, hyper-activity of NF-κB is now well documented in many types of lymphoma and leukemia.32 Consequently, agents that inhibit NF-κB activation are desired, and are currently being pursue, based on strategies that seek to maintain levels of endogenous NF-κB-inhibiting IκB-family proteins, by either suppressing the IκB kinases (IKKs) responsible for phosphorylation of IκBα (which induces its polyubiquitination) or blocking the proteolytic activity of the 26s proteasome responsible for degrading IκB following ubiquitination.
Similarly, Akt (PKB) directly phosphorylates multiple protein targets of relevance to apoptosis, suppressing cell death clearly within the intrinsic pathway (e.g., BAD inactivation; human Caspase-9 suppression) and possibly also the extrinsic pathway (e.g., FasL expression) in some types of cells. Pathological elevations in Akt activity are a common occurrence in tumors, due to loss of tumor suppressor PTEN, hyper-activity of PI3K-activating protein tyrosine kinase growth factor receptors and oncoproteins such as the product of the BCR-ABL fusion gene of CML, as well as other mechanisms.33 Small-molecule drugs that occupy the ATP-binding pocket of the catalytic domain of Akt thus represent a highly attractive approach to restoration of apoptosis sensitivity in cancers, and would also be expected to have additional benefits on cell proliferation and cell growth. While the PH domain of Akt that binds second-messenger phospholipids involved in Akt activation could theoretically be targeted by drugs, the hydrophilic interactions associated with coordinating polyphosphorylated lipids do not lend themselves well to membrane permeable compounds.
The transcription factor p53 reportedly regulates the expression of multiple apoptosis-regulating genes that affect either the intrinsic or extrinsic pathways, though the specific targets of p53 that are most relevant probably differ widely among tissues and tumor-types.34 While clinical trials are underway using gene therapy-based approaches for restoring p53 activity in deficient cancers, which might permit better locoregional control of cancer in combination with radiotherapy, this strategy is less attractive for hematological malignancies.
In addition to NF-κB, Akt, and p53, multiple cancer-relevant proteins that operate upstream of these factors could to some extent be viewed as apoptosis-modulators, though acting rather indirectly on the core cell death machinery. Therapies targeting these upstream inducers include many biological agents, such as: (a) monoclonal antibodies recognizing cell surface receptors (e.g., anti-HER2 [Herceptin]; anti-EGFR [Erbitux]), or small molecule drugs that target their kinase domains, which presumably suppress PI3K/Akt activation; and (b) anti-CD40L antibodies and CD40L adenovirus gene therapy, which modulate NF-κB activity. Monoclonal antibodies targeting the tetra-span membrane protein CD20 (Rituximab) also can downregulate expression of Bcl-2-family member Mcl-1 (intrinsic pathway) and XIAP (convergence pathway) in certain leukemias, providing yet another example of unexpected links to apoptosis pathways through receptor-mediated signaling, in a context relevant to cancer treatment.35 Thus, abundant opportunities may exist for exploiting receptor targeting agents for indirectly restoring apoptosis sensitivity in cancer, representing an area worthy of further in-depth exploration.
Summary
Essentially all cytotoxic anticancer drugs currently in clinical use, when they work, induce apoptosis of malignant cells. While microtubule-binding drugs, DNA-damaging agents, and nucleosides are important weapons in the treatment of cancer, a new class of targeted therapeutics may soon be forthcoming based on strategies that have emerged from a deeper understanding of the molecular mechanisms that underlie the phenomenon of apoptosis. From an improved understanding of apoptosis mechanisms, multiple novel strategies have emerged for restoring apoptosis sensitivity in cancer, and clinical testing of several of these approaches is underway. Much remains to be learned about how best to exploit these new potential therapies, matching genetic lesions in malignant cells to the optimal agent. It remains to be determined to what extent toxicity to normal tissues will limit application of apoptosis-based therapies for cancer treatment. Malignant cells, however, may be relatively more dependent on apoptosis suppression than normal cells, due to aberrations in proto-oncogene activity and cell cycle checkpoint control, and other factors. Exploiting this differential dependence on apoptosis-suppressing mechanisms offers hope that improved clinical outcomes may not be far from realization.
Recurrent genetic subtypes of B- and T-cell acute lymphoblastic leukemia (ALL).
| Subtype . | Associated Genetic Abnormalities . | Frequency in Children . | Risk Category . |
|---|---|---|---|
| B-precursor ALL | Hyperdiploid DNA content; trisomies of chromosomes 4, 10, 17 | 25% of B precursor cases | Low |
| t(12;21)(p13;q22): | 28% of B precursor cases | Low | |
| TEL/AML1 11q23/ rearrangements; | 4% of B precursor cases; | High | |
| particularly t(4;11)(q21;q23) | < 80% of infant ALL | ||
| t(1;19)9q23;p13) – E2A/PBX1 | 6% of B precursor cases | High | |
| t(9;22)(q34;q11): BCR/ABL | 2% of B precursor cases | Very high | |
| Hypodiploidy | Relatively rare | Very high | |
| B-ALL | t(8;14)(q24;q32) – IgH/MYC | 5% of all B lineage ALL cases | High |
| T-ALL | Numerous translocations involving the TCR αβ (7q35) or TCR γδ (14q11) loci | 7% of ALL cases | Not clearly defined |
| Subtype . | Associated Genetic Abnormalities . | Frequency in Children . | Risk Category . |
|---|---|---|---|
| B-precursor ALL | Hyperdiploid DNA content; trisomies of chromosomes 4, 10, 17 | 25% of B precursor cases | Low |
| t(12;21)(p13;q22): | 28% of B precursor cases | Low | |
| TEL/AML1 11q23/ rearrangements; | 4% of B precursor cases; | High | |
| particularly t(4;11)(q21;q23) | < 80% of infant ALL | ||
| t(1;19)9q23;p13) – E2A/PBX1 | 6% of B precursor cases | High | |
| t(9;22)(q34;q11): BCR/ABL | 2% of B precursor cases | Very high | |
| Hypodiploidy | Relatively rare | Very high | |
| B-ALL | t(8;14)(q24;q32) – IgH/MYC | 5% of all B lineage ALL cases | High |
| T-ALL | Numerous translocations involving the TCR αβ (7q35) or TCR γδ (14q11) loci | 7% of ALL cases | Not clearly defined |
Genes strongly predictive of outcome at initial diagnosis in pediatric acute lymphoblastic leukemia (ALL).
| Gene Identifier: Bayesian Network . | Gene/Protein Name . | Previously Known Function / Comment . |
|---|---|---|
| Abbreviations: IL, interleukin; UNM, University of New Mexico. | ||
| G0 | OPAL1 (Assigned by UNM) | Unknown human EST, not previously fully cloned |
| G1 | GNB2L1: G protein β2, related sequence 1 | Signal transduction; activator of protein kinase C |
| G2 | IL-10 receptor alpha | IL-10 receptor alpha |
| Gene Identifier: Bayesian Network . | Gene/Protein Name . | Previously Known Function / Comment . |
|---|---|---|
| Abbreviations: IL, interleukin; UNM, University of New Mexico. | ||
| G0 | OPAL1 (Assigned by UNM) | Unknown human EST, not previously fully cloned |
| G1 | GNB2L1: G protein β2, related sequence 1 | Signal transduction; activator of protein kinase C |
| G2 | IL-10 receptor alpha | IL-10 receptor alpha |
Event free survival curves showing outcomes of infants and older children with blasts that harbor 11q23 translocations.
Outcomes are shown for the last series (1800s/1922) of CCG ALL trials (historical controls) compared to the most recent series (1950/1960s). Outcomes are superior for children ≥ one year of age in both the historical control group (P = .01), and in the 1950/1960s series (P = .06).
Abbreviations: HC, historical controls
Event free survival curves showing outcomes of infants and older children with blasts that harbor 11q23 translocations.
Outcomes are shown for the last series (1800s/1922) of CCG ALL trials (historical controls) compared to the most recent series (1950/1960s). Outcomes are superior for children ≥ one year of age in both the historical control group (P = .01), and in the 1950/1960s series (P = .06).
Abbreviations: HC, historical controls
Event-free survival of patients with M1, M2, and M3 bone marrows at day 7 of induction therapy.
Outcomes are shown for the last series of CCG ALL trials (1800s/1922) compared to the most recent series (1950/1960s).
Abbreviations: HC, historical controls
Event-free survival of patients with M1, M2, and M3 bone marrows at day 7 of induction therapy.
Outcomes are shown for the last series of CCG ALL trials (1800s/1922) compared to the most recent series (1950/1960s).
Abbreviations: HC, historical controls
Diagram of the mitochondrial and death receptor pathways of cell death. See text for description.
Diagram of the mitochondrial and death receptor pathways of cell death. See text for description.
Probability of complete remission vs thiopurine methyltransferase (TPMT) genotype on St Jude Children’s Research Hospital Protocol Total XII (SJCRH Total XII).
Probability of complete remission vs thiopurine methyltransferase (TPMT) genotype on St Jude Children’s Research Hospital Protocol Total XII (SJCRH Total XII).
Genes (italicized) whose products interact with methotrexate include DHFR (dihydrofolate reductase), GGH (gamma glutamyl hydrolase), MTHFR (methylenetetra-hydrofolate reductase), and FPGS (folylpoly-glutamate synthetase). All are subject to common genetic polymorphisms.
Genes (italicized) whose products interact with methotrexate include DHFR (dihydrofolate reductase), GGH (gamma glutamyl hydrolase), MTHFR (methylenetetra-hydrofolate reductase), and FPGS (folylpoly-glutamate synthetase). All are subject to common genetic polymorphisms.