Abstract
It is increasingly evident that molecular diagnostics, that is, the use of diagnostic testing to understand the molecular mechanisms of an individual patient’s disease, will be pivotal in the delivery of safe and effective therapy for many diseases in the future. A huge body of new information on the genetic, genomic and proteomic profiles of different hematopoietic diseases is accumulating. This chapter focuses on new technologies and advancements in understanding the molecular basis of hematologic disorders, providing an overview of new information and its significance to patient care.
In Section I, Dr. Braziel discusses the impact of new genetic information and research technologies on the actual practice of diagnostic molecular hematopathology. Recent and projected changes in methodologies and analytical strategies used by clinical molecular diagnostics laboratories for the evaluation of hematologic disorders will be discussed, and some of the challenges to clinical implementation of new molecular information and techniques will be highlighted.
In Section II, Dr. Shipp provides an update on current scientific knowledge in the genomic profiling of malignant lymphomas, and describes some of the technical aspects of gene expression profiling. Analysis methods and the actual and potential clinical and therapeutic applications of information obtained from genomic profiling of malignant lymphomas are discussed.
In Section III, Dr. Liotta presents an update on proteomic analysis, a new and very active area of research in hematopoietic malignancies. He describes new technologies for rapid identification of different important proteins and protein networks, and the potential therapeutic and prognostic value of the elucidation of these proteins and protein pathways in the clinical care of patients with malignant lymphomas.
I. Molecular Diagnosis of Hematopoietic Disorders
Rita M. Braziel, MD*
Oregon Health and Sciences University, Department of Pathology, L471, 3191 SW Sam Jackson Park Rd, Portland OR 97239
The monumental research advances in genomic and protein research over the past several years have made it possible to envision, in the not too distant future, the development of medical care that is truly tailored to each individual patient. Clinicians are anxious to incorporate this new knowledge into the selection of more specifically targeted therapies, and there is general agreement that new research insights need to be translated into useful clinical tests. Ideally, information from all available studies, including traditional morphologic and immunophenotypic findings as well as data from new genomic, proteomic, and pharmacogenomic studies would be applied to define a patient’s disease profile. This profile could be used not only to confirm and refine primary diagnoses, but potentially might also be used to determine the risk for development of cancer; to detect cancer at an early, more curable stage; to predict drug efficacy against a cancer and also drug toxicity; to improve disease staging for determination of risk of distant spread and subsequent relapse; and to assess response to therapy by monitoring for residual disease.
How close are we in 2003 to providing our patients with this type of individualized medical approach? Unfortunately, it must be acknowledged that we are still far from this ideal. Thus far, few of the exciting scientific advances of the recent past have been translated into improved patient care or better technology for molecular diagnostics. Nonetheless, the field of molecular diagnostics is growing, adapting, changing constantly, and beginning to permeate virtually every area of medicine. Table 1 provides a listing of some of the current and anticipated techniques used in molecular diagnostics today. These techniques are discussed below.
Tests for Genome-Wide Screening for Chromosomal Abnormalities
Routine cytogenetics is the traditional method for a survey of genome-wide chromosomal abnormalities, but standard karyotyping studies, even with chromosome banding, miss many subtle chromosomal abnormalities. Other methodologies for genomic profiling of chromosomal abnormalities have been developed, which have considerably augmented our knowledge of the genetic features of various hematopoietic malignancies. These assays for genomic profiling are based on screening of chromosomes or DNA for loss or gain of chromosomes or genes, in contrast to gene-expression profiling performed on RNA. These assays detect changes in chromosome/gene location and number, not gene expression and function; for all of these studies, at least for disomic loci, the normal reference copy number is 2. Spectral karyotyping (SKY) and comparative genomic hybridization (CGH) are complementary fluorescent molecular genetic techniques for detection of whole genome chromosomal abnormalities. With SKY, 24 differentially labeled painting probes representing all chromosomes are cohybridized, Fourier spectroscopy is used to distinguish the different spectrally overlapping probes, and special imaging software is used for analysis. This technique has been found to greatly facilitate the detection of many previously cryptic chromosomal translocations and rearrangements, and is already available for clinical purposes in many institutions.1–,3 CGH uses the hybridization of differentially labeled tumor DNA and reference DNA to produce a map of the DNA copy number changes in the tumor genome.3– 4 CGH assays are not yet available for routine clinical use, but technical permutations of this research methodology are reputedly in the pipeline for clinical laboratories. A variant of CGH, called matrix CGH, uses genomic cDNA fragments instead of the chromosome targets used in standard CGH, and even more powerful is the use of arrayed cDNA sequences with CGH. These latter techniques allow detection of unknown amplified genes, not just gene regions, and provide even higher resolution for identification of genomic imbalances.
Tests Targeting Specific Chromosomal Abnormalities
Multiple methods can be used for the detection of specific chromosomal abnormalities, including various permutations of the polymerase chain reaction (PCR), Southern blotting, and fluorescence in situ hybridization (FISH) with molecular probes. PCR-based methods, often multiplexed, have been the screening test of choice for most molecular laboratories if the chromosomal abnormality of interest was amenable to PCR analysis. However, as the number of genes important in diagnosis and prediction of prognosis has increased almost exponentially over the past few years, a different molecular testing algorithm has evolved for hematopoietic malignancies. The development, validation, and maintenance of numerous PCR analyses for detection of the ever-increasing important chromosomal abnormalities in hematopoietic malignancies is simply not practical for most laboratories. Fortunately, the development of molecular probes for use in FISH assays has provided a valuable alternative method to standard PCR analyses. FISH assays are not as sensitive as PCR assays, but FISH analyses are used predominantly at diagnosis and relapse, a time when only a low level of analytical sensitivity is needed since there are usually high levels of abnormal cells. The use of FISH assays for molecular evaluation of malignant lymphomas and leukemias has increased remarkably over the past 1–2 years, and has blurred the lines between classical cytogenetics and molecular pathology.
FISH is a very useful technique for detection of targeted chromosomal abnormalities. It can be done on blood, bone marrow, tissue touch preparations, body fluids, and even paraffin-embedded fixed tissue, so it is applicable to many specimen types. FISH overcomes one of the biggest problems with routine cytogenetic analysis of many lymphoma and chronic leukemia samples (i.e. the need for metaphases), as FISH can be done with either metaphase or interphase preparations. In FISH assays, the target is usually nuclear DNA of interphase or metaphase cells attached to glass microscope slides. Most FISH assays are based on the ability of single stranded DNA to bind (hybridize) to complementary DNA, although some RNA FISH assays are available.5 The molecular test probes (DNA) can be labeled with biotin or digoxigenin-labeled nucleotides and detected with fluorophor-conjugated antibodies, or may be directly fluorophor-labeled. With the use of dual or triple pass filters, multicolor FISH can be done.6,7
There are several different strategies for the design of FISH assays. Single fusion-dual color FISH assays for translocations utilize 2 probe hybridization targets located on 1 side of each of the 2 genetic breakpoints; the usual level of false positive background cells from incidental overlap of signal in this type of assay is 5%–10%. Dual fusion-dual color FISH assays for translocation utilize large probes that span 2 breakpoints on the different chromosomes. Dual fusion-dual color FISH is optimal for detection of low levels of nuclei possessing a simple balanced translocation, as it greatly reduces the number of normal background nuclei with an abnormal signal pattern. FISH using dual color-break apart probes is very useful in the evaluation of genes known to have multiple translocation partners; the differently colored probes hybridize to targets on opposite sides of the breakpoint in the known gene. Multicolor FISH using 3 to 4 differently colored probes can be done in selected cases to determine the overlap of different genetic abnormalities in different cell populations. FISH with centromeric probes is useful for detection of changes in chromosome number (i.e., monosomy, diploidy, trisomy).6– 8
Genomic probes for the genetic abnormalities of many leukemias, lymphomas, and even myeloproliferative and myelodysplastic disorders are now readily available from commercial sources. The most notable expansion in the library of molecular FISH probes is for the B-cell malignancies; some of the B-cell lymphoma-associated chromosomal abnormalities for which FISH analysis can be performed are listed in Table 2 . FISH assays are particularly useful in detection of chromosomal translocations that are not amenable to PCR detection due to widely distributed breakpoints, because FISH probes are much larger than the probes and primers used in PCR analysis. Like SKY, FISH assays will detect some genetic abnormalities that are karyotypically silent.
It should be remembered that FISH assays are useful mainly around the time of initial diagnosis or at relapse, when there is a relatively high level of abnormal cells. FISH is not useful for detection of low level minimal residual disease (MRD) following therapy, as the sensitivity of even the best dual fusion-dual color FISH assay is only approximately 1 positive cell in 100 normal cells, not sufficient for detection of MRD. It is also important to remember that, despite the glamor of some of the newer techniques, standard cytogenetics continues to be extremely important in the initial diagnosis and follow-up of patients with hematopoietic malignancies. Focusing only on tests that target specific genetic abnormalities, like FISH and PCR, can result in the failure to detect the additional important cytogenetic abnormalities that may be present initially or that may occur following therapy. For example, the need for intermittent cytogenetic analysis is very clear in chronic myelogenous leukemia (CML) patients. A number of these CML patients have developed clonal karyotypic abnormalities in Philadelphia chromosome–negative cells while on therapy with imatinib mesylate; these abnormalities would not have been detected by FISH or PCR analyses for BCR/ABL.9
Genotyping for single nucleotide polymorphisms (SNPs) is relevant to both research and routine molecular diagnosis. Allele-specific PCR amplification techniques using sequence-specific primers (PCR-SSP) are widely employed for detection of SNPs in the genes that encode immunogenic proteins such as alloantigens. The status of certain alloantigens (i.e., human leukocyte antigens, blood group antigens, human platelet alloantigens) is frequently investigated by this methodology before organ transplantation. Technical advances in genotyping of SNPs have improved the ability to perform testing for HLA and blood group antigens on small samples of DNA, such as those obtained from patients with low leukocyte counts.10,11
Gene Expression Profiling
Despite glowing predictions of future clinical utility and multiple published reports on the use of microarrays for gene expression profiling (GEP) in lymphomas and leukemias,12– 18 no microarrays are available yet for molecular diagnostics. However, focused arrays utilizing fewer, but highly significant genes, are currently available for research studies and are in the development pipeline for B-cell non-Hodgkin lymphomas and acute leukemias. The diffuse large B-cell lymphomas are likely to be the lymphoma subtype for which focused microarrays will first be used for routine clinical purposes. Needless to say, molecular laboratories are anxiously awaiting the shift of this technology into the clinical arena.
Although microarrays for GEP have not yet made it into the clinical molecular laboratory, information gained from GEP data is impacting clinical laboratory testing. One example is flow cytometric analyses of ZAP-70 expression in chronic lymphocytic leukemias/small lymphocytic lymphomas (CLL/SLL). Pilot GEP studies in patients with CLL/SLL identified genes that were differentially expressed between leukemic clones that did not have mutated IgHV regions and those that did. The best discriminator was a gene called ZAP-70; a high level of ZAP-70 expression correctly predicted unmutated IgHv gene status in most patients.19,20 This is clinically relevant because the absence of somatic mutations in the variable regions of the IgH gene has been determined to have adverse prognostic significance in CLL/SLL patients; those with unmutated IgHv regions often have progressive disease while those with mutated IgHv regions often pursue a more indolent course. Since molecular testing for IgHv mutation status involves multiple complex PCR reactions and sequencing procedures, the analysis is impractical for clinical testing. Fortunately, the ZAP-70 protein is readily detected by either flow cytometric analysis or immunohistochemical staining,20 and these procedures are currently being set up in many clinical laboratories in lieu of molecular analysis for somatic mutation of the IgHV regions or microarray GEP.
There are some caveats about GEP data; these may be part of the reason for delay in implementation of this technique in clinical practice. Numerous GEP databases are now available in the public domain for different lymphomas, acute leukemias, and myelomas, and multiple publications have resulted from independent analyses of these databases. It turns out that different investigators do not always find the same results and draw the same conclusions from analyses of the same databases. This has made it apparent that, because of the extreme complexity, there are potential problems with analyses of GEP microarrays and databases that can produce erroneous GEP results.21,22 A few of the problems that have been described include sampling variability of tumors, chip differences and defects, differences and biases in analysis of GEP data, and sources of systematic error in microarray analysis. Clearly, sifting the real GEP changes from artifacts and noise in microarray experiments is often difficult, but rapid identification and neutralization of spurious results is essential to prevent them from becoming accepted facts.
Another potential problem with GEP is the possibility of missing relevant cell populations present at a low level in the tumor specimen. Since GEP provides an average expression profile for an entire cellular population, small subpopulations of important cells are unlikely to be recognized. The application of new methods for microdissection, followed by RNA amplification, would allow targeting of specific populations of interest that were previously missed by GEP. In conclusion, there are enough problems with GEP microarrays and interpretations that it is important to have validation of significant GEP changes from more than one laboratory/database before important clinical decisions are based on this data.
Molecular Tests for Minimal Residual Disease Detection
Although many patients with hematologic malignancies achieve a complete clinical remission (CR) and even a complete pathologic remission by standard morphologic and immunologic criteria, a relatively high proportion of them will ultimately relapse. The source of this relapse is clearly from a persistent malignant cellular population that is present at a low level, below the limit of detection by standard techniques. For this reason, considerable effort has been devoted by molecular laboratories in the past 5 to 10 years to develop new molecular techniques to increase the sensitivity of detection of neoplastic cells. The application of these techniques has demonstrated the presence of residual neoplastic cells in many patients in CR. This reservoir of neoplastic cells, detected only by sensitive molecular methods, is commonly referred to as minimal residual disease (MRD). The detection of MRD in a variety of hematologic malignancies suggests that obtaining a molecular remission should be a goal of therapy, and the results of most studies of MRD detection support this concept. However, it has still not been clearly established for many hematologic malignancies that patients with only a few residual malignant cells, detected only by very sensitive techniques, will benefit from additional therapy.
If achieving a molecular remission is confirmed to be an important goal following therapy for most hematologic malignancies, as seems likely, then MRD testing will become a much larger component of testing in molecular diagnostics laboratories. Ideally, techniques used for MRD detection should have a sensitivity level in the 105 to 106 range, be applicable to almost all patients with the disease, provide some quantification of the target, and be rapid, inexpensive, readily standardized, and disease-specific. Also of critical importance for the clinical utility of tests for MRD detection is good interlaboratory reproducibility and standardization of reporting. In reality, most commonly used molecular analyses for MRD detection do not meet many of these criteria. A particular problem for clinicians is the lack of standardization of testing techniques and primers between laboratories, which essentially mandates follow-up testing for MRD be performed in the laboratory that did the previous testing to allow comparison of results. With frequent shifts in patient locations and changing insurance carrier requirements, sending follow-up specimens to the same laboratory may be impossible.
Only a few commonly used techniques are sensitive enough for detection of MRD in leukemias and lymphomas. Nested PCR and quantitative real-time PCR can be used for disease-associated translocations, without the need for patient-specific primers. If the malignant clone does not carry a good translocation target for PCR analysis, patient-specific gene rearrangements may be targeted, using either nested or quantitative real-time PCR. Nested PCR analyses can detect up to 1 malignant cell in 106 normal cells. Quantitative real-time PCR assays, with a sensitivity of 1 in 104–105, are almost as sensitive as the nested PCR. A substantive number of studies of MRD detection have been performed in only a few hematopoietic malignancies, specifically chronic myelogenous leukemia, follicular lymphoma, and childhood acute lymphoblastic leukemia. The different methods used for detection of MRD in these 3 different hematopoietic malignancies are discussed below.
Chronic myelogenous leukemias
With imatinib mesylate therapy, a complete cytogenetic response (CCR) can be achieved for most patients with newly diagnosed chronic myelogenous leukemia (CML).23 Quantitative real-time RT-PCR analysis (Q-RT-PCR) is most often used to monitor for MRD in patients who have achieved a CCR by bone marrow cytogenetics and/or FISH. Interestingly, Q-RT-PCR monitoring for BCR/ABL can be performed on either peripheral blood or bone marrow; comparable results have been found on analysis of simultaneous blood and marrow specimens24 (R Braziel, unpublished data). This facilitates follow-up of imatinib-treated CML patients.
Real-time PCR is a relatively new molecular technique that allows simultaneous PCR amplification and detection of target DNA or cDNA sequences. The specimen is normalized against an internal control, typically a single copy gene; for CML MRD testing, ABL or G6PDH is typically used as the internal control. A standard curve is made from a dilution series of a BCR/ABL-positive cell line, and the amount of residual leukemia cells is calculated by using this standard curve (Figure 1; see Appendix, page 600). Advantages of real-time PCR over standard nested PCR for BCR/ABL include a decreased turnaround time, decreased chance for post-PCR contamination, decreased variability of results because the data collection occurs in the exponential phase of the PCR reaction, high throughput, and the possibility of obtaining quantitative results. Real-time PCR procedures are much more amenable to interlaboratory standardization than nested PCR analyses. The major disadvantage of real-time PCR testing is the inability to compare the size of any detected rearrangements to that of the original malignant clone without additional testing. However, this is not a problem with MRD testing in CML, as there is not a background population of normal cells carrying the BCR/ABL translocation. Quantitative real-time PCR technology can be used with many translocation targets, and can also be used for antigen receptor gene rearrangement analysis. The determination of the trend in the quantitative numbers of residual BCR/ABL-positive cells over a period of time is thought to provide important therapeutic information in the follow-up of CML patients.25,26 Optimal methods for quantitative real-time PCR detection of MRD in CML patients have not yet been established, and this testing is currently performed mainly in a few reference laboratories.
Once again, the importance of remembering the limitations of very targeted molecular testing in CML must be stressed. The clinician must be alert for development of clonal abnormalities in BCR/ABL negative cells9 or the development of resistance to imatinib mesylate. The presence of mutations or amplification of BCR/ABL is known to be associated with resistance to imatinib mesylate in CML patients,27 and testing for these abnormalities is likely to become a standard part of the evaluation of CML patients in the future, at least for those who fail to achieve and maintain a CCR.
Follicular lymphomas
The recent use of therapeutic modalities such as autologous bone marrow transplant following ex vivo purging of bone marrow B cells, monoclonal antibody therapy, and vaccine therapy has resulted in improved clinical outcomes of patients with follicular lymphomas (FL). PCR analysis for MRD performed on serial bone marrow samples in treated FL patients in complete remission has shown that some patients do achieve a molecular remission, and that the failure to achieve or maintain a molecular remission is predictive of relapse.28– 34 Although the optimal methodology and timing for detection of MRD has yet to be determined, the t(14;18)(q32;q21)–IgH/BCL2 translocation, seen in 80%–90% of FL, is a good target for MRD detection. Unlike MRD testing in CML, in patients with FL, bone marrow analysis is clearly more sensitive for detection of MRD than peripheral blood. Many FL patients clear FL cells from the blood, while they still have persistent marrow involvement.
Nested PCR assays have been used historically and remain the most sensitive methodology for FL MRD detection; nested PCR can detect one translocation-carrying cell in 106 normal cells and is still used in many laboratories for detection of this translocation. However, other molecular laboratories have switched from nested PCR to quantitative real time PCR. Real-time PCR for IgH/BCL2 is less labor-intensive and lacks the risk of contamination of standard nested PCR, but does have an analytical sensitivity that is usually at least 1 log less than that of nested PCR for IgH/BCL2. An additional and under-recognized problem in interpretation of real-time PCR analyses for MRD in FL is the potential for false positive results from occasional benign IgH/BCL2 translocation-carrying cells. These are present in 30–40% of normal individuals, and with a highly sensitive test, a false positive result could occur if a comparison to the original FL clone is not made.35– 37 This comparison is readily performed with nested PCR (Figure 2 ), but would require substantial additional molecular analysis with real-time PCR. Using a quantitative real-time PCR method on serial bone marrows for MRD detection in FL, determining a trend over time, may obviate the necessity for this additional testing. Additional studies are needed to evaluate the clinical efficacy of MRD detection in FL patients in general, and to compare the relative clinical value of the nested and quantitative real-time PCR MRD detection methods.
Precursor B-cell lymphoblastic leukemias
Multiple large prospective studies have clearly demonstrated the high prognostic value of MRD monitoring in children with precursor B-cell lymphoblastic leukemias (pre-B-ALL).38– 43 In childhood pre-B-ALL, studies of MRD have generally targeted patient-specific IgH antigen receptor gene rearrangements. This method takes advantage of the fingerprint-like sequences of the junctional regions of rearranged IgH genes, which differ in length and composition for each lymphocyte clone. To obtain these sequences, standard IgH PCR analysis is performed at diagnosis and/or relapse and the PCR products are Southern blotted, followed by sequencing of junctional regions of the clonal IgH rearrangements. The different IgH rearrangements are then used for design of patient-specific oligonucleotide primers that are subsequently used in real-time PCR assays to follow the patient. Patient-specific IgH primers increase PCR sensitivity up to 1000-fold compared to standard consensus primers for IgVH gene rearrangements; reactive background rearranged B cells do not obscure the clonal PCR products. At the present time, patient/clone-specific IgH PCR is not practical outside of a funded clinical trial setting, but this technique clearly offers the best potential for a sensitive, specific, and rapid analysis method that could be used over the course of therapy in most patients with pre-B-ALL.
Indeed, this same type of patient/clone-specific IgH PCR technology could be used for MRD detection in most other B-cell lymphomas (BCL) also, in which either no translocation-associated molecular event is available for MRD testing or the recurrent translocations occur in too low a proportion of the BCL subtype to be clinically useful. Standard IgH gene PCR with consensus rather than patient-specific IgH primers can usually detect only 1 malignant cell in 102–3 normal cells, so the only sufficiently sensitive and specific method of testing for MRD detection in most BCL is therefore the use of patient/clone-specific Ig gene rearrangements. Quantitative real-time PCR techniques using standard IgH primers may provide some early information about the trend of the disease course over time, but will become negative when the patient could still have substantial residual disease. The combination of patient-specific IgH primers and quantitative real-time PCR could make a major contribution to the achievement of standardized MRD detection in BCL.
Conclusion
Whether offering or ordering a molecular test, the physician should know the circumstances in which the test should be ordered, the circumstances in which the test would not be useful, the advantages and the limitations of the test, and how to interpret the results. Many clinicians and pathologists are unfamiliar with molecular tests for hematologic malignancies, and misinterpret results of molecular testing. To avoid this, clinicians must be knowledgeable about the molecular test they are ordering and cautious about overinterpretation of results. Physicians ordering molecular tests must be prepared to offer counseling on them, either personally or by referral. Good patient consent forms for molecular testing are crucial, and should explain to patients the meaning of a positive test, a negative test, and an inconclusive test. The consent form should inform the patient that the test could uncover other clinically relevant information, things that were not even being looked for.
Clinical molecular laboratories today are faced with two daunting tasks. First and foremost is the necessity of expanding test menus to meet the increasing clinical demand for testing for new genetic markers in hematologic disorders. Expanding test menus to meet clinical needs will require technical advances, as the current technology in clinical molecular laboratories does not allow rapid screening for a broad panel of relevant genes at a reasonable cost. However, equipment development has not been aimed at clinical laboratories, which traditionally have low budgets for new equipment, but at large pharmaceutical companies with abundant cash for new purchases. This disconnect must clearly be addressed if clinical molecular testing is to be advanced, but even if the technological bottleneck preventing translation of new genetic knowledge to the clinical arena is alleviated, the lack of a reasonable level of reimbursement for molecular testing in general is still a major roadblock to successful implementation of new techniques for clinical molecular diagnostic testing. In most cases, the amount reimbursed for molecular diagnostic testing in hematopoietic malignancies in the United States today is inadequate to even cover the costs of test reagents. Little progress in the ideal model of “personalized” medicine will occur if this lack of funding persists.
II. Molecular Signatures of Lymphoid Malignancies: Identification of Novel Disease Subtypes and Rational Therapeutic Targets
Margaret A. Shipp, MD*
Dana-Farber Cancer Institute, 44 Binney Street, Room D940, Boston MA 02115-6084
Lymphoid malignancies are currently classified on the basis of morphology, immunophenotype, genetic features, clinical characteristics, and possible normal cells of origin.1 With the sequencing of the human genome and associated development of representative DNA microarrays, it is now possible to obtain broad-based transcriptional profiles of specific lymphoid malignancies and previously unidentified disease subtypes.
The most commonly used platforms for gene expression profiling are cDNA and oligonucleotide microarrays (Figure 3; see Appendix, page 600).2 With cDNA arrays, polymerase chain reaction (PCR) products of cDNA clones are spotted on filters or glass slides. A potential advantage of cDNA arrays is that they can be designed to address specific biologic questions. For example, the recently described “lymphochip” cDNA arrays are enriched for genes with documented importance in lymphocyte biology.2 Oligonucleotide microarrays include oligonucleotide probes deposited or synthesized directly on the surface of a silicon wafer. Oligonucleotide microarrays can potentially offer additional specificity by tailoring probes to reduce cross-hybridization and discern splice variants.2 A common oligonucleotide array platform also facilitates comparisons across datasets of different tumor types3 (Figure 3; see Appendix, page 600).
Two main approaches have been used to analyze gene expression datasets: unsupervised and supervised learning (Table 3 ).2 Unsupervised learning methods aggregate samples into groups based on the overall similarity of their gene expression profiles without a priori knowledge of specific relationships (Table 3 ). Commonly used unsupervised learning algorithms include self-organizing maps (SOMs), hierarchical clustering, and probabilistic clustering (Table 3 ). In contrast, supervised learning techniques group tumors based on known differences (i.e., cured versus fatal disease) and develop transcriptional profiles of the defined groups (Table 3 and Figure 4; see Appendix, page 601). Frequently used supervised learning algorithms include weighted voting, k-NN, support vector machine (SVM), and IBM SPLASH (Table 3 ).
One of the lymphoid malignancies in which gene expression profiling has been informative is diffuse large B-cell lymphoma (DLBCL). The most common lymphoid malignancy in adults, DLBCL comprises almost 40% of all lymphoid tumors. Although a subset of DLBCL patients can be cured with standard adriamycin-containing combination chemotherapy, the majority die of their disease. Robust clinical prognostic factor models such as the International Prognostic Index can be used to identify patients who are less likely to be cured with standard therapy.4 However, such models do not provide specific insights regarding more effective treatment strategies. For these reasons, additional insights into molecular bases for the observed clinical heterogeneity in DLBCL are critically needed. In addition, the multiple genetic abnormalities associated with subsets of DLBCL reflect additional molecular heterogeneity in this disease.5,6
Investigators have utilized gene expression profiling to elucidate molecular bases for observed differences in DLBCL, identifying possible normal cells of origin,7 tumors with different responses to standard combination chemotherapy,8,9 novel rational treatment targets,8 and related disease entities (Savage et al, unpublished material). For these reasons, the lessons from gene expression profiling in DLBCL are likely to be broadly applicable to other lymphoid malignancies.
In one of the earliest applications of gene expression profiling, cDNA microarrays (lymphochips) and unsupervised learning techniques (hierarchical clustering) were used to characterize the transcriptional profiles of DLBCL and normal lymphocytes, including germinal center (GC) B cells and in vitro activated peripheral blood B cells.7 In a pilot study, subsets of DLBCLs were found to share gene expression patterns with normal GC B cells or in vitro activated PB B cells.7 In an expanded analysis, a refined cell-of-origin signature (100 genes that distinguished GC-B-cell-like and activated-B-cell-like lymphomas at a significance level of P < .001) was used to identify tumors with features of above-mentioned normal B cells and a third unrelated subset.9
Additional investigators have utilized supervised learning methods to develop transcriptional profiles of cured versus fatal/refractory DLBCLs.8 Genes implicated in outcome signatures included ones that regulate B-cell receptor signaling, critical serine/threonine phosphorylation pathways, and apoptosis.8 Two of the genes and pathways identified in this supervised outcome analysis8 have already been credentialed as possible rational therapeutic targets in DLBCL. In additional analyses, a combination of unsupervised and supervised learning methods were used to develop a DLBCL outcome model that included the cell-of-origin distinction and additional parameters including HLA class II expression and indices of proliferation.9
These extremely powerful computational strategies provide new mechanisms for identifying discrete subsets of DLBCL and other lymphoid malignancies.10 The next challenges will be to link the molecular signatures of cell-of-origin and prognosis in lymphoid tumors with implicated biological pathways, specific pathogenetic mechanisms,11,12,13 and associated rational targets of therapy.14
III. Proteomic Analysis of Hematolymphoid Neoplasms: Diagnostic, Biologic, and Therapeutic Implications
Andrew L. Feldman, MD, Virginia Espina, MS, Mary Winters, BS, Elaine S. Jaffe, MD, Emanuel F. Petricoin III, PhD, and Lance A. Liotta, MD, PhD*
National Cancer Institute, National Institutes of Health, 10 Center Drive, MSC 1500, Bethesda MD 20892-1500
Hematolymphoid neoplasms are responsible for over 60,000 deaths annually in the United States, and are the most commonly occurring cancers in children.1 Despite these sobering statistics, it is within this field that molecular medicine has made its earliest and greatest strides, the promise of which is just beginning to be realized. The past few decades have seen the discovery of the t(9;22) BCR/ABL translocation in chronic myelogenous leukemia (CML),2 the characterization of the role in apoptosis of the BCL-2 family of proteins,3 the use of microarray analysis to delineate new subsets of diffuse large B-cell lymphoma (DLBCL),4 and the introduction of novel biologic agents such as rituximab5 and imatinib,6 which already have had far-ranging impact in reducing the burden of cancer in selected patients. The field of hematolymphoid neoplasms remains fertile ground for the application of technology in the molecular diagnosis, characterization, and treatment of human disease.
Overview of Proteomics
The functional effectors of cellular pathways and processes are proteins. While these proteins are encoded by the genome, only a subset of the possible protein products of the genetic code are produced, and the functional status of these proteins often depends heavily on posttranslational modifications that are not reflected in their genomic sequences. Thus, while significant advances have been made from the analysis of the genome and its transcribed complement of mRNA, the protein end products of these processes are the effector arm of cellular events and offer an in vivo, functionally relevant window into the workings of the cell. The study of this wide complement of proteins derived from the genome is known as proteomics, and the proteins collectively are called the proteome.
The analysis of proteins is used daily in the clinical diagnosis and treatment of hematolymphoid neoplasms. An example is BCL-2 protein, an antiapoptotic protein overexpressed as a result of the t(14;18) translocation in follicular lymphoma.7 Detection of BCL-2 protein by immunohistochemistry is used routinely in the diagnostic evaluation of B-cell lymphomas. The presence of BCL-2 protein can yield prognostic information as well, such as in DLBCL, where BCL-2 protein expression has been correlated with decreased survival.8 Biologically, the elucidation of the mechanism by which the BCL-2 protein acts has led to the discovery of a large family of proteins related to apoptosis, with relevance to a wide variety of human diseases in addition to follicular lymphoma.9 Finally, understanding the role of BCL-2 has led to molecular therapies such as antisense oligonucleotide approaches to lessen the antiapoptotic effects of this protein.10 Thus, clinicians may use a single protein to assist in diagnostics, prognostics, biologic understanding, and development of targeted therapies. However, analyzing proteins one-by-one is laborious, time consuming, and likely to miss critical biologic events due to the sheer number of existing proteins that could be assayed. New proteomic technology allows us to gain an overview of thousands of proteins simultaneously as a proteomic pattern, analyze the individual protein signaling pathways being utilized by neoplastic cells, characterize the neoplasm biologically, and select specific targeted treatment modalities, known as “personalized” molecular medicine.11
Diagnostics
The power of molecular diagnostics has been demonstrated for hematolymphoid neoplasms perhaps more than in any other field. The ability to assay for the presence of characteristic translocations and detect clonal immunoglobulin gene rearrangements not only is used for diagnosis on a routine basis, but has helped support the classification of distinct disease entities, such as anaplastic large cell lymphoma.12 A major advance has been the use of large-scale gene expression analysis to develop genomic patterns or “fingerprints” to aid in the classification of hematolymphoid neoplasms. Alizadeh et al4 demonstrated the ability of microarray data obtained from diffuse large B-cell lymphomas to delineate distinct patterns of gene expression, which not only had biologic correlates in terms of the phenotype of the neoplastic cells, but also conveyed prognostic information beyond that obtained using the International Prognostic Index (IPI) alone.
Proteomic technology has now advanced to the point where proteomic patterns can be analyzed as “fingerprints” in much the same way DNA array data have been analyzed.13 Importantly, while conventional protein assays query individual protein biomarkers, proteomic pattern analysis uses complex bioinformatic tools to interpret spectra representing thousands of proteins to distinguish between clinical entities, such as the presence or absence of cancer. Unlike DNA microarrays, where target sequences must be available to be printed on the array, proteomic pattern analysis does not mandate identification or isolation of each protein comprising the overall pattern.14 This is important since the proteome is significantly more complex than the genome, incorporating the results of multiple alternative splicing variants and posttranslational modifications. While the genome is now thought to encode approximately 30,000 genes, the number of protein species may exceed 300,000.15
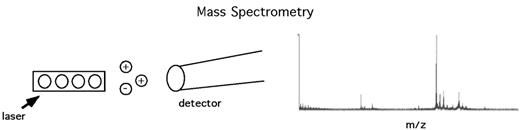
Proteomic pattern analysis has been greatly facilitated by advances in high-throughput mass spectrometry, especially surface-enhanced laser desorption ionization time-of-flight (SELDI-TOF) analysis (Figure 5 ). In SELDI-TOF, proteins from a patient sample (e.g., serum, urine, tissue lysate) are bound to a chip. After washing off unbound proteins and impurities, a matrix is applied that is subjected to photoactivation by laser energy. As proteins are desorbed by the laser they are launched as charged ions, and analysis of their time-of-flight (TOF) allows calculation of their mass-to-charge ratios (m/z). The spectrum of m/z ratios thus obtained is processed using self-learning bioinformatic pattern recognition software. Analysis of large cohorts of data using learning algorithms based on artificial intelligence approaches can allow the discrimination between two groups of samples (“supervised” learning, such as distinguishing the presence or absence of cancer), or to identify data clusters within a population set that may represent novel disease entities (“unsupervised” learning).
An example of the power of this approach is the recent report of the ability to characterize the proteomic spectra derived from sera of women with and without ovarian cancer.16 SELDI-TOF mass spectra were used as a training set to develop an artificial intelligence algorithm that, when applied to blinded samples, could accurately identify 100% of patients with ovarian cancer and 95% of controls. This approach was sensitive enough to accurately identify all cancer patients, even those with stage I disease; specificity was demonstrated by including women with benign ovarian conditions in the control group, as well as finding the algorithm incapable of detecting the presence of cancer in sera from patients with prostate cancer.17 A recent improvement in mass spectrometry allowed correct identification of sera from ovarian cancer patients and controls with 100% sensitivity and 100% specificity.14
The classification of hematolymphoid neoplasms has evolved from a system based on morphology alone to one based on cell of origin, the determination of which is aided by detection of lineage-specific protein markers using flow cytometry or immunohistochemistry.18 This classification also utilizes detection of additional protein markers which are not lineage-specific but rather relate to the pathogenetic mechanism of the disease (e.g., BCL-2) or other clinically relevant genetic events (e.g., p53 mutations19). In many cases, panels of antibodies are chosen in part due to limitations of the techniques employed, such as assaying cell surface molecules by flow cytometry, or ability of antibodies to detect antigens in paraffin-embedded tissue sections. The use of proteomic patterns representing tens of thousands of protein species may be an enormously powerful tool to complement the ongoing efforts to classify hematolymphoid neoplasms in ways that are biologically accurate and clinically relevant.
Additional ways that proteomic profiles might be used diagnostically include screening for minimal residual disease after treatment,20 screening for the development of neoplasia in high-risk populations (e.g., posttransplant),21 screening for transformation of a low-grade neoplasm into a high-grade one,22 and characterizing/predicting responses to therapeutic interventions.13 For example, lymphoma cells in vitro have been shown to demonstrate characteristic proteomic patterns after exposure to chemotherapeutic agents,23 and characterization of in vivo patterns may serve as an early predictor of response and/or toxicity.
Molecular Characterization and Treatment
As previously mentioned, proteomic profiles can have important diagnostic and prognostic implications without reference to the individual proteins which constitute these profiles.14 Even at the single protein level, the protein CD20 has been used widely as a marker of B cells and a target for anti-B-cell monoclonal antibody therapy,5 although these uses are not primarily based on its specific cellular function. Clearly, however, the widespread analysis of the proteome will yield extensive data regarding function and utilization of critical protein pathways, as was discussed for the BCL-2 protein, and is expected to have far-ranging implications for the identification of molecular targets for pathway-specific biologic therapy.
The ability to analyze the nature of the proteome in human tissues has been greatly facilitated by rapid developments in the field of protein microarrays.24 The power of this technique has been enhanced by the development of laser capture microdissection,25 in which subpopulations of cells, such as lymphoid follicles,26 can be isolated from tissue sections using a laser pulse. Protein lysates prepared from these samples then can be robotically applied in miniature dilution curves to a solid phase array with multiple other samples. Hundreds of replicate arrays can be generated and probed for the expression of a large complement of proteins using specific antibodies, including those that differentially recognize cleaved and/or phosphorylated forms of key signal transduction molecules.27 In this way, protein microarrays can identify the particular signaling pathways utilized by a population of neoplastic cells to tailor specific targeted therapy to modulate the function of these pathways.
One particularly attractive use of this technology is characterizing the status of apoptotic pathways in hematolymphoid neoplasms. Apoptosis is an essential element for the normal development and maintenance of the immune system.28 The development of a healthy immune repertoire is a highly stringent process, necessitating the elimination of most developing lymphocytes; conversely, the ability to avoid apoptosis is critical to the rapid expansion of quiescent lymphocytes in response to foreign antigen. Families of proteins related to apoptosis, such as the BCL-2 family, therefore include both proapoptotic and antiapoptotic members to aid in the homeostatic regulation of these immune processes. Perturbation of these homeostatic mechanisms is exemplified by the t(14;18) translocation in follicular lymphoma, leading to overexpression of BCL-2. Further studies have indicated that antiapoptotic signals from BCL-2 are important in nonfollicular neoplasms as well, in which complex interactions among multiple BCL-2 family members appear to be critical.29 A downstream level of regulation is the inhibitor of apoptosis protein (IAP) family, which exerts its effect by inhibiting effector caspases.30 Because multiple proteins and pathways modulate apoptosis, protein microarrays are ideally suited to assay these pathways and determine which are functionally active in a neoplastic cell population (Figure 6 ) in order to select molecular targets for biologic therapy. A similar approach could be used assaying cell cycle regulatory genes, tumor suppressor genes, transcription factors, etc.
Conclusion
Revolutionary technological advances in the field of proteomics have generated powerful new tools for the analysis of human neoplasia and cell signaling pathways. High-throughput mass spectrometry has generated proteomic patterns from patient samples that have had powerful implications in the diagnosis of several human malignancies, and this technology is ready to be applied to hematolymphoid neoplasms to aid diagnosis, monitor response, and refine disease classification. Coupled with a new generation of protein microarrays capable of characterizing the functional status of multiple signaling pathways in neoplastic cells, these advances are leading to “personalized” molecular medicine, in which proteomic analysis will be used to select therapeutic agents to specifically target critical pathways in each individual neoplasm to optimize therapeutic efficacy and minimize toxicity. Trials to validate the clinical use of these technologies are currently under way.
Summary of techniques for molecular analysis of hematopoietic disorders.
|
|
Non-random chromosomal abnormalities in B-cell lymphomas.
| Lymphoma Subtype . | Nonrandom Chromosomal Alterations . | Genes Involved . | Assay Used for Diagnosis / Prognosis† . |
|---|---|---|---|
| Abbreviations: CLL/SLL, chronic lymphocytic leukemia/small lymphocytic lymphoma; LPL, lymphoplasmacytic lymphoma; MZL, marginal zone lymphoma; FL, follicular lymphoma; MCL, mantle cell lymphoma; DLBCL, diffuse large B-cell lymphoma; BL/BLL, Burkitt/Burkitt-like lymphoma. | |||
| †The assays listed are the most commonly used today for clinical testing today, but many of these abnormalities can also be detected by other techniques. | |||
| CLL/SLL | Del 13q14 | Unknown | FISH |
| Trisomy 12 | Unknown | FISH | |
| Del 11q22-23 | ATM | FISH | |
| Del 17p1 | TP53 | FISH, karyotyping | |
| LPL | t(9;14)(p13;q32) | PAX5/IgH | FISH, karyotyping |
| MZL | t(11;18)(q21;q21) | API2/MALT1 | FISH, PCR |
| t(1;14)(p22;q32) | BCL10/IgH | FISH, PCR | |
| t(14;18)(q32;q21) | IgH/MALT1 | FISH | |
| Trisomy 3 | ?BCL6 | FISH | |
| FL | t(14;18)(q32;q21) | IgH/BCL2 | PCR, FISH |
| MCL | t(11;14)(q13;q32) | Cyclin D1/IgH | FISH, PCR |
| DLBCL | 3q27 | BCL6 | FISH |
| t(14;18)(q32;q21) | IgH/BCL2 | FISH, PCR | |
| BL / BLL | t(8;14)(q24;q32) | c-MYC/IgH | FISH for c-MYC |
| t(2;8)(p11;q24) | Igk/c-MYC | FISH for c-MYC | |
| t(8;22)(q24;q11) | c-MYC/Igl | FISH for c-MYC | |
| Lymphoma Subtype . | Nonrandom Chromosomal Alterations . | Genes Involved . | Assay Used for Diagnosis / Prognosis† . |
|---|---|---|---|
| Abbreviations: CLL/SLL, chronic lymphocytic leukemia/small lymphocytic lymphoma; LPL, lymphoplasmacytic lymphoma; MZL, marginal zone lymphoma; FL, follicular lymphoma; MCL, mantle cell lymphoma; DLBCL, diffuse large B-cell lymphoma; BL/BLL, Burkitt/Burkitt-like lymphoma. | |||
| †The assays listed are the most commonly used today for clinical testing today, but many of these abnormalities can also be detected by other techniques. | |||
| CLL/SLL | Del 13q14 | Unknown | FISH |
| Trisomy 12 | Unknown | FISH | |
| Del 11q22-23 | ATM | FISH | |
| Del 17p1 | TP53 | FISH, karyotyping | |
| LPL | t(9;14)(p13;q32) | PAX5/IgH | FISH, karyotyping |
| MZL | t(11;18)(q21;q21) | API2/MALT1 | FISH, PCR |
| t(1;14)(p22;q32) | BCL10/IgH | FISH, PCR | |
| t(14;18)(q32;q21) | IgH/MALT1 | FISH | |
| Trisomy 3 | ?BCL6 | FISH | |
| FL | t(14;18)(q32;q21) | IgH/BCL2 | PCR, FISH |
| MCL | t(11;14)(q13;q32) | Cyclin D1/IgH | FISH, PCR |
| DLBCL | 3q27 | BCL6 | FISH |
| t(14;18)(q32;q21) | IgH/BCL2 | FISH, PCR | |
| BL / BLL | t(8;14)(q24;q32) | c-MYC/IgH | FISH for c-MYC |
| t(2;8)(p11;q24) | Igk/c-MYC | FISH for c-MYC | |
| t(8;22)(q24;q11) | c-MYC/Igl | FISH for c-MYC | |
Analysis of gene expression datasets.
|
|
An example of nested polymerase chain reaction (PCR) analysis for detection of minimal residual disease (MRD) in marrows from 2 follicular lymphoma (FL) patients.
Each sample is subjected to PCR for β-globin housekeeping gene and IgH/BCL2; PCR products undergo gel electrophoresis and (+) bands are detected by ethidium bromide staining (center gels = β-globin and bottom gels = IgH/BCL2 MBR). A band is seen only if an appropriate product is detected; specificity of bands is confirmed by Southern blot using BCL2 probe (top autoradiographs).
(Panel A): Multiple controls are run in parallel with patient samples (see box insert). (+) controls are serial dilutions of an IgH/BCL2 (+) cell line (lanes 4–7). The 106 dilution is run twice; as shown, a (+) result is often detected in only 1of these samples at the lower limit of analytical test sensitivity.
(Panels B andC): Parallel PCR analyses of 5 sequential marrows from 2 different FL patients are shown. For each patient, sample A was at diagnosis, before any treatment, B was 1–2 months after completion of CHOP chemotherapy but before treatment with anti-CD20 monoclonal antibody, and marrows C, D, and E were obtained at 2 months, 6 months, and 12 months after CD20 therapy. PCR of all clinical samples is run in duplicate. The patient in Panel B has a (+) nested PCR for IgH/BCL2 MBR at diagnosis and after CHOP, but the marrow becomes PCR (–) by 2 months after CD20 therapy and remains (–) at 6 and 12 months. The patient in Panel C is also PCR (+) at diagnosis and after CHOP, but also has a (+) band in 1 lane at 2 months after CD20 therapy. The (+) band is confirmed to represent a benign IgH/BCL2-carrying cell, not MRD, as it is not present in the duplicate sample and is clearly a different size from the patient’s FL clone. Note that results are more easily interpreted on the BCL2-probed Southern blots than on the ethidium bromide–stained gels.
An example of nested polymerase chain reaction (PCR) analysis for detection of minimal residual disease (MRD) in marrows from 2 follicular lymphoma (FL) patients.
Each sample is subjected to PCR for β-globin housekeeping gene and IgH/BCL2; PCR products undergo gel electrophoresis and (+) bands are detected by ethidium bromide staining (center gels = β-globin and bottom gels = IgH/BCL2 MBR). A band is seen only if an appropriate product is detected; specificity of bands is confirmed by Southern blot using BCL2 probe (top autoradiographs).
(Panel A): Multiple controls are run in parallel with patient samples (see box insert). (+) controls are serial dilutions of an IgH/BCL2 (+) cell line (lanes 4–7). The 106 dilution is run twice; as shown, a (+) result is often detected in only 1of these samples at the lower limit of analytical test sensitivity.
(Panels B andC): Parallel PCR analyses of 5 sequential marrows from 2 different FL patients are shown. For each patient, sample A was at diagnosis, before any treatment, B was 1–2 months after completion of CHOP chemotherapy but before treatment with anti-CD20 monoclonal antibody, and marrows C, D, and E were obtained at 2 months, 6 months, and 12 months after CD20 therapy. PCR of all clinical samples is run in duplicate. The patient in Panel B has a (+) nested PCR for IgH/BCL2 MBR at diagnosis and after CHOP, but the marrow becomes PCR (–) by 2 months after CD20 therapy and remains (–) at 6 and 12 months. The patient in Panel C is also PCR (+) at diagnosis and after CHOP, but also has a (+) band in 1 lane at 2 months after CD20 therapy. The (+) band is confirmed to represent a benign IgH/BCL2-carrying cell, not MRD, as it is not present in the duplicate sample and is clearly a different size from the patient’s FL clone. Note that results are more easily interpreted on the BCL2-probed Southern blots than on the ethidium bromide–stained gels.
Overview of mass spectrometry.Patient proteins bound to a chip are irradiated by a laser, launching desorbed proteins as charged ions. Detection of the time-of-flight (TOF) of these ions yields a spectrum of mass-to-charge ratios (m/z). This spectrum, reflecting the protein constituents of the original sample, is then analyzed by self-learning pattern recognition software and compared to previously encountered spectra.
Overview of mass spectrometry.Patient proteins bound to a chip are irradiated by a laser, launching desorbed proteins as charged ions. Detection of the time-of-flight (TOF) of these ions yields a spectrum of mass-to-charge ratios (m/z). This spectrum, reflecting the protein constituents of the original sample, is then analyzed by self-learning pattern recognition software and compared to previously encountered spectra.
Application of protein arrays to apoptotic signaling pathways.Jurkat cells were treated with Fas ligand to induce apoptosis. Arrays were printed using cell lysates in serial dilutions and probed using an antibody specific to the cleaved (active) form of caspase 3. Increased cleaved caspase 3 is noted in the Fas ligand–treated cells (upper left). Hundreds of replicate arrays can be printed and probed with other antibodies; arrays then can be scanned, quantified, and compared.
Application of protein arrays to apoptotic signaling pathways.Jurkat cells were treated with Fas ligand to induce apoptosis. Arrays were printed using cell lysates in serial dilutions and probed using an antibody specific to the cleaved (active) form of caspase 3. Increased cleaved caspase 3 is noted in the Fas ligand–treated cells (upper left). Hundreds of replicate arrays can be printed and probed with other antibodies; arrays then can be scanned, quantified, and compared.