Abstract
The advent of new technologies has contributed to improvements in the diagnosis and classification of the non-Hodgkin lymphomas (NHL). Use of a more extensive test menu of paraffin active monoclonal antibodies for immunohistochemistry, molecular cytogenetic studies including standard cytogenetics, multi-color fluorescence in-situ hybridization (FISH), polymerase chain reaction and locus-specific FISH, as well as developments in high-resolution techniques including microarray gene expression profiling and array comparative genomic hybridization (CGH) allow more accurate diagnosis and precise definition of biomarkers of value in risk stratification. The identification of disease-specific gene lists resulting from expression profiling provides a number of potential protein targets that can be validated using immunohistochemistry. We will highlight how improvements in our understanding of lymphoma biology rapidly facilitate the development of new diagnostic reagents that could be used to alter clinical practice. These changing trends allow the development of new diagnostic strategies used to render accurate sub-classification of entities within the category of indolent B-cell lymphomas, including their distinction from related but more aggressive disorders, such as mantle cell lymphoma. A comprehensive understanding of the biology of these distinct lymphoid tumors will allow us to identify novel disease-related genes and should facilitate the development of improved diagnostics, outcome prediction, and personalized approaches to treatment.
The indolent B-cell lymphomas are listed in Table 1 . Grade 3A follicular lymphoma (FL) is included, as preliminary data suggest that it may represent one end of the spectrum of indolent grade 1 and 2 FLs.1 The other entities are considered clinically indolent, with disease evolution measured in years, in contrast to the aggressive B cell lymphomas. Collectively the indolent B-cell lymphomas account for approximately 40% of all non-Hodgkin lymphomas (NHLs), although the frequency of FL varies significantly throughout the world.2 The frequency of small lymphocytic lymphoma (SLL) in this study is artificially inflated by inclusion of cases with chronic lymphocytic leukemia (CLL).3 Each of these disorders is considered a distinct disease with a spectrum of clinical behavior largely dictated by the genetic alterations inherent within the tumor cells. Their relative frequency among the NHLs is listed in Table 1 and was used to determine the relative space devoted to each in this review. Thus the focus of discussion will include primarily FL, MALT and SLL/CLL.
Advances in genome science and the subsequent development of novel technologies to interrogate cancer genomes have led to a number of technology platforms that are beginning to work their way into the clinic. In particular, microarray gene expression profiling has been applied to a large number of clinical cases, providing some insight into the numbers of deregulated genes and the mechanisms that control gene expression in lymphomas. Significant gains in our understanding of the biology of NHL provide the framework for building outcome predictors for assigning individual patient risk. Standard cytogenetic approaches and the use of locus-specific FISH probes have been of value in understanding the complex genetic alterations found in MALT lymphomas. Some of these observations have clinical impact, as their presence may denote more aggressive clinical behavior and/or antibiotic resistance. Knowledge of the presence of these alterations will therefore have an effect on treatment planning and predicting prognosis. This review will focus on work published this year and will attempt to build on discussions in Hematology 2004.4
Follicular Lymphoma
FL represents the single most common NHL in North America, accounting for 32% of all cases, but only 22% of all NHLs from an international perspective.3 The frequency of FL varies markedly around the world, suggesting the possibility that epidemiological factors, host genetic factors or both influence its incidence. However, a review of published work indicates that the involvement of the BCL2 translocation in FL in different regions is similar.5 A comprehensive discussion of FL pathology, phenotype and molecular genetics was recently presented. Therefore, this review will focus on current diagnostic strategies, new insights gleaned from microarray gene expression studies, high-resolution cytogenetic data and how these novel strategies might be used to complement diagnosis, prognosis and biological understanding.
A number of diagnostic strategies are used routinely to distinguish FL from reactive hyperplasia and from other small B cell lymphomas. Diagnostic clues and potential pitfalls are listed in Table 2 . For the most part, diagnosis includes careful morphologic evaluation, immunohistochemical studies such as Bcl-2, CD10, Bcl-6, MIB-1 (paraffin Ki-67), flow cytometric assessment for light chain restriction, PCR for IGH and/or BCL2 clonality, FISH for BCL2 translocation and standard cytogenetic analyses.6
A number of cytogenetic studies in FL have confirmed the heterogeneity of clonal evolution that can be found at diagnosis.4 These differences are believed to contribute to clinical diversity and to provide the background on which additional genetic alterations occur over time, thereby contributing to histologic transformation and/or disease progression. Certain of these stochastic events appear to provide the neoplastic cells with a growth advantage and thus accelerate the clinical aggressiveness of the lymphoma. An understanding of the relative contribution of these two molecular features and how they contribute to the biology of FL is paramount to fully appreciating the spectrum of clinical behavior that is FL.
Work by the Lymphoma/Leukemia Molecular Profiling Project (LLMPP) provided compelling evidence that the non-neoplastic cells in the FL microenvironment contribute significantly to the biology and outcome of FL.7 This work established that non-neoplastic cells in the biopsy, in particular T cells responsible for immune response-1 (IR-1) and macrophages accounting for immune response-2 (IR-2) were the dominant gene expression signatures used to construct an outcome predictor in FL. These informative gene expression profiles were present in the diagnostic biopsies of patients with FL, suggesting that much of our ability to predict outcome in FL is present at the time of initial diagnosis. Although survival parameters were highly predictive, the relationship between gene signatures derived from non-neoplastic cells and transformation risk has not yet been analyzed. In contrast, Glas et al were unable to substantiate these data but rather found an 81-gene predictor that could predict immediate post-biopsy clinical behavior, either at the time of diagnosis or relapse, but not long-term survival or the risk of subsequent transformation.8 This finding led the authors to conclude that ongoing stochastic genetic events were contributing to transformation, but that these genetic alterations were not yet present in the neoplastic cells and thus could not be detected.9 A signature of genetic instability based on a survey of DNA repair genes was not significantly different between cases showing rapid transformation versus more stable disease. Instead, genes involved in the immune response, cytokine and chemokine signaling and antigen processing by follicular dendritic cells (FDCs) appeared to characterize cases at risk for transformation.10
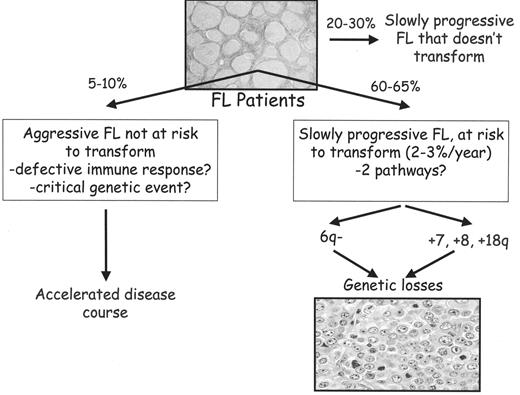
Recently, Farinha et al have shown that FL patients with a high content of macrophages in their biopsy samples show aggressive clinical behavior.11 To some extent, these findings parallel the results of gene expression profiling, suggesting a dominant role for immune response-2 (IR-2) as a negative outcome predictor.7 Given the significant contribution of transformation to survival in FL, it is of interest that none of these patients showed evidence of transformation. Histologic transformation is thought to be the dominant clinical event in the course of FL, variably reported to occur between 5% and 60% of patients at 10 years.12–16 These results are difficult to reconcile with the natural history and molecular alterations in FL, and suggest the possibility of two, or perhaps three distinct pathways of disease progression. In the largest subgroup, the disease may be cytogenetically unstable, allowing secondary cytogenetic alterations to occur, some of which result in transformation. Random molecular events that create a growth advantage would allow minor sub-clones to become dominant, altering both the clinical and morphological appearance of the tumor. If this risk was constant over time, then transformation events that produce these sub-clones would occur at a continuous rate. In contrast to these cytogenetically unstable cases, a small subgroup of FL may be characterized by clinically aggressive disease without transformation. This subgroup may be characterized by a unique cytogenetic alteration or some inheritable defect in the immune response that allows the FL to grow unabated. Lastly, there may be a third subgroup that does not transform. The possibility of at least two parallel pathways remains a hypothesis and is detailed in Figure 1 . A combination of gene expression profiling and array comparative genomic hybridization (CGH) will likely provide some insight into mechanisms that result in deregulated gene expression in FL. Knowledge of these genetic events may improve the ability of a molecular predictor to determine the subgroups of patients at risk for either aggressive FL or risk of transformation. Importantly, the identification of novel biomarkers could then be used in conjunction with the FL International Prognostic Index (FLIPI) to identify high-risk patients who might be candidates for alternative therapies. Such an approach would be beneficial for clinical trial design, allowing the selection of patients whose endpoints could be measured over a much shorter period of time. Correlating time to treatment failure to specific genes or signaling pathways might be important in identifying high priority targets. The study of sequential biopsy specimens in FL will similarly be important for an improved understanding of genetic events that underlie disease progression.
In addition to the FLIPI clinical index, a number of additional biomarkers exist. These include morphological features, immunophenotypic markers, cytogenetic alterations and more recently, a number of genes resulting from microarray gene expression profiling. These are listed in Table 3 , including their proposed mechanism of action.17–22
Preliminary array CGH data have been generated in FL.23 A number of recurrent alterations previously identified using chromosomal CGH were confirmed, lending credibility to the technique. However, the improved resolution of the array CGH strategy was highlighted by the discovery of a number of new genomic imbalances. As these data are matched to gene expression profiles, we should learn more regarding the relationship between DNA copy number changes and deregulated gene expression in FL.
Extranodal MALT Lymphomas
Low-grade, extranodal marginal zone B-cell lymphomas of MALT type account for approximately 8% of all NHLs.3,6 These tumors arise at a number of extranodal sites, including the stomach, lung, salivary gland, thyroid, skin, small bowel and a number of additional, less common sites. They demonstrate a unique morphology and phenotype reminiscent of normal MALT tissues (i.e., Peyer’s patches) that help to characterize this special arm of the immune system, features that distinguish them from the more common nodal lymphomas.24
The diagnosis of MALT lymphoma can be challenging, as extranodal sites of disease may be relatively difficult to access, resulting in small biopsy samples. The characteristic histologic findings must be sought, including sheets of centrocyte-like small B cells occupying the marginal zone and surrounding secondary lymphoid follicles. Movement of these cells into the follicles may be reminiscent of FL, a finding referred to as follicular colonization. Plasma cell differentiation may be present and may make it difficult to distinguish MALT lymphoma from lymphoplasmacytic lymphoma (LPL). Clinical information (serum protein electrophoresis, bone marrow status and morphology, presence of marrow mast cells) and specific histological findings (Russell and Dutcher bodies, lymphoepithelial lesions, etc.) are helpful in resolving this differential diagnosis. Transformation to a process resembling diffuse large B cell lymphoma (DLBCL) may occur and thus requires careful examination of the biopsy. MALT lymphomas are B cell tumors and thus express pan-B cell antigens including CD20 and CD79a. Demonstrating monotypic surface light chain expression has utility for diagnosis. Alternatively, cytoplasmic light chain restriction is seen in a variable percentage of cases with a plasma cell component and can be helpful. Unlike SLL and mantle cell lymphoma (MCL), MALT lymphomas are typically CD5-negative. In contrast to FL, the tumor cells in MALT lymphoma lack expression of CD10 and Bcl-6. CD43 is often expressed by MALT lymphoma cells and can be detected using paraffin section immunohistochemistry. CD11c is expressed in half of the cases but requires flow cytometry for detection. As noted above, small biopsy size can hamper a confident diagnosis in many cases. Thus, PCR strategies used to determine B cell clonality are very useful for diagnosis. Although clonality does not absolutely confirm malignancy, it is a helpful determinant in difficult, borderline lesions.
Collectively these lymphomas demonstrate a number of disparate cytogenetic alterations, the majority of which affect a common signaling pathway and thus share a common pathogenesis. The common karyotypic alterations that characterize MALT lymphomas include trisomies 3 and 18, translocations t(11;18)(q21;q21), t(1;14)(p22;q32), t(14;18)(q32;q21), t(3;14)(q27;q32) and the recently described t(3;14)(p14.1;q32).25,26 The apparent complexity of cytogenetic alterations that have now been implicated in the pathogenesis of extranodal MALT lymphoma serves as a paradigm for molecular cross talk in neoplastic disease. Recent data have shown that at least three of the very disparate translocations affect a common signaling mechanism, and thus unify all three under a common pathogenesis, resulting in the constitutive activation of the NF-κB pathway.27
All of these alterations can be detected using locus-specific FISH techniques that can be applied to routine, formalin-fixed and paraffin embedded material. As the presence of at least two of these karyotype abnormalities predicts for antibiotic resistance in gastric MALT lymphomas, such studies have an immediate impact on treatment decisions. Helicobacter pylori infection in the stomach has been clearly shown to be integral to the pathogenesis of gastric MALT lymphomas, but the role of other infectious agents in the etiology of MALT lymphomas at other anatomic sites remains speculative. A list of MALT sites, characteristic cytogenetic alterations, deregulated genes and suspected infectious organisms is provided in Table 4 .
Although murine models of gastric MALT lymphoma have been used to study gene expression profiling, there are essentially no gene expression profiling data or high-resolution cytogenetic studies currently available for human MALT lymphomas.28 However, a combination of adequate morphology, paraffin section immunohistochemistry and standard cytogenetics or locus-specific FISH techniques will allow an accurate diagnosis in most cases together with some useful biological data that might help to plan treatment. For example, the presence of either the t(11;18) or t(1;14) in a gastric MALT lymphoma not only helps substantiate the diagnosis but also defines patients for whom antibiotics alone would not be the sole therapeutic modality. As indicated in Table 4 , the diverse cytogenetic alterations found in MALT lymphoma are seen differentially based on anatomic site.25 For example, the t(11;18) frequently encountered in MALT lymphomas of the stomach or lung is rarely seen in the skin, ocular adnexa, salivary gland or thyroid. Indirectly this finding suggests the possibility that other infectious agents might be implicated at these sites. However, at the present time a cause and effect relationship remains speculative.
Small Lymphocytic Lymphoma
Small lymphocytic lymphoma (SLL) is an uncommon lymphoma, accounting for only 1%–2% of all NHLs.6 Its distinction from the more common CLL requires the lack of a peripheral blood lymphocytosis exceeding 5 × 109/L at diagnosis. For all intents and purposes, the two diseases are virtually identical. The histology, immunophenotype and cytogenetic alterations are indistinguishable. Clearly, poorly understood biological differences account for why some patients present with predominantly peripheral blood and bone marrow disease, while others remain more lymph node based. Differential expression of adhesion molecules may explain some of these differences. Cytogenetic abnormalities in small B cell NHLs are listed in Table 5 .
A number of biological prognostic factors have been recognized to be of value in SLL. Adverse factors include diffuse bone marrow involvement, atypical peripheral blood morphology with increased prolymphocytes, short lymphocyte doubling time (increased proliferation), poor-risk cytogenetics including 17p deletions, 11q deletions and complex karyotypes, mutated IGH genes and expression of CD38. The translocation, t(14;19) occurs infrequently in SLL but, when present, confers a more aggressive behavior.29 More recently, expression profiling has identified novel biomarkers that are of value in determining prognosis. In particular, ZAP-70 expression has been shown to be a useful surrogate for unmutated IGH status but is imperfectly correlated with CD38 expression.30 Its value lies in the ease with which it can be measured, including flow cytometric assessment and immunohistochemistry. More recently, additional candidate genes have been shown to correlate with mutational status in CLL, including lipoprotein lipase (LPL), disintegrin and metalloproteinase 29 (ADAM29), spartin (SPG20) and nuclear receptor-interacting protein 1 (NRIP1). Two of these genes, LPL and ADAM29, are overexpressed in unmutated and mutated CLL, respectively.31 The ratio of the two (LPL/ADAM29 expression ratio) was shown to outperform ZAP70 in a recent study of prognosis in CLL.32 The development of antibody reagents to several of these proteins or perhaps quantitative RT-PCR strategies using as few as 5 to 10 genes might improve risk stratification in CLL beyond current approaches.33
Splenic Marginal Zone Lymphoma
Splenic marginal zone lymphoma (SMZL) is an uncommon diagnosis, accounting for < 1% of all NHLs. Clinically patients are characterized by splenomegaly, often accompanied by peripheral blood and bone marrow involvement. Roughly 20%–30% of patients have a monoclonal serum protein, typically IgM type. Splenic histology is typically biphasic, with a unique pattern of involvement of the splenic white pulp. However, an uncommon red pulp pattern is poorly recognized and may be confused with benign, granulomatous diseases. Peripheral blood involvement is common, and SMZL likely accounts for a number of cases of chronic, CD5-negative leukemia. Some cases may show villous cytoplasmic projections, but the term splenic lymphoma with villous lymphocytes (SLVL) should be used with caution, as this entity as defined in the past may include a number of well-defined diseases. Bone marrow patterns are variable, but purely intrasinusoidal infiltration is classic and may be very difficult to appreciate on H&E stains alone. SMZL is a B cell lymphoma, expressing CD19, CD20, CD22 and CD79a. The neoplastic cells often express CD11c but are negative for both CD23 and CD43.6 Common cytogenetic alterations include del7q31–32, typically present as the sole abnormality.34 Trisomy 3 is also frequent. Recent gene expression profiling reveals that genes involved in B cell receptor signaling, AKT1 signaling, tumor necrosis factor (TNF) signaling and NF-κB activation are consistently deregulated in SMZL.33,35 Similarly, genes that map to 7q31 were consistently downregulated. Three genes were found to be very SMZL-specific, including ILF1, Senataxin and CD40. Inferior survival was associated with CD38 expression, unmutated IGH genes and the expression of NF-κB pathway genes including TRAF5, REL and PKC-α.
Nodal Marginal Zone Lymphoma
Nodal marginal zone lymphoma (NMZL) is a very uncommon diagnosis. It is often used as a diagnosis of exclusion, when other more common small B cell lymphomas are excluded, although this approach is not recommended. Local regional lymph node involvement in cases of MALT lymphoma is virtually indistinguishable from NMZL, requiring clinical input +/− cytogenetic data to diagnose it. NMZL is characterized by frequent trisomies, including +3, +7 and +18, but the characteristic translocations encountered in MALT lymphomas are never seen. There is evidence to suggest that NMZL as currently defined is not a homogeneous entity. Further analysis, including microarray gene expression profiling, will likely be required to determine whether NMZL represents more than a single entity.6
Lymphoplasmacytic Lymphoma
Lymphoplasmacytic lymphoma (LPL) is also an uncommon entity and represents a difficult diagnosis for most pathologists. It was recently found to be one of the least reproducible diagnoses during the International Lymphoma Classification Project used to validate the REAL classification.3 LPL accounts for only 1.2% of all NHLs. Involvement of lymph nodes and bone marrow are frequent. In contrast to CLL, the peripheral blood is infrequently involved. The disease represents a cytological spectrum from small B cell lymphocytes, plasmacytoid lymphocytes, through to mature plasma cells. Similarly, the immunophenotype represents a spectrum paralleling the morphology. The mature plasma cell component fails to express CD19, CD20 and CD5. Surface immunoglobulin (Ig) light chain restriction is not apparent, but cytoplasmic Ig is a requirement. Most cases of LPL are characterized clinically by a monoclonal serum and/or urine protein. Typically this is of the IgM class, but IgG and IgA are also encountered. The bone marrow is typically involved and infiltrates are often accompanied by frequent mast cells. Lymph node involvement is often interfollicular and perisinusoidal. Growth centers, characteristic of SLL/CLL, are not seen. Cytogenetic studies had previously implicated the t(9;14)(p13;q32) in LPL, but more recent studies question the accuracy of this association.36 No gene expression studies have been published. Clinical prognostic factors have been elucidated for LPL, but no large studies of relevant biomarkers have been published.
Indolent B-cell lymphomas.
| Lymphoma Subtype . | Frequency . |
|---|---|
| *The validation of the REAL classification almost certainly included cases of otherwise typical chronic lymphocytic leukemia (CLL).3 | |
| Follicular lymphoma, grade 1–3A | 22% |
| Extranodal marginal zone B-cell lymphoma, MALT-type | 8% |
| Small lymphocytic lymphoma/(CLL)* | 7% |
| Nodal marginal zone lymphoma | 2% |
| Lymphoplasmacytic lymphoma | 1.2% |
| Splenic marginal zone lymphoma | < 1% |
| Lymphoma Subtype . | Frequency . |
|---|---|
| *The validation of the REAL classification almost certainly included cases of otherwise typical chronic lymphocytic leukemia (CLL).3 | |
| Follicular lymphoma, grade 1–3A | 22% |
| Extranodal marginal zone B-cell lymphoma, MALT-type | 8% |
| Small lymphocytic lymphoma/(CLL)* | 7% |
| Nodal marginal zone lymphoma | 2% |
| Lymphoplasmacytic lymphoma | 1.2% |
| Splenic marginal zone lymphoma | < 1% |
Diagnostic clues and pitfalls in small B-cell lymphomas.
| Lymphoma Subtype . | Diagnostic Clues . | Pitfalls . |
|---|---|---|
| Abbreviations: LEL, lymphoepithelial lesion; SLL, small lymphocytic lymphoma; CLL, chronic lymphocytic leukemia; MZL, marginal zone lymphoma; PB, peripheral blood; BM, bone marrow; FL, follicular lymphoma; PCR, polymerase chain reaction; FISH, fluoresence in situ hybridization; RFH, reactive follicular hyperplasia; FDC, follicular dendritic cell | ||
| Follicular lymphoma | Follicle formation, admixed centroblasts, presence of tight FDC meshworks –CD10 +/− Bcl-6 expression between follicles –Low proliferative rate in contrast to RFH –PCR/FISH presence of t(14;18) | Not all FLs express Bcl-2 protein –BCL2 PCR can be negative (inherent high false- negative rate) –Discordance between morphology and proliferative rate in some cases |
| MALT lymphoma | Reactive follicles, LELs and centrocyte-like cells –Presence of light chain restriction –Absence of CD5/CD10 expression –FISH detection of characteristic translocations | Classic cytomorphology not always present –Small biopsies may produce false-positive PCR for IGH clonality –Distinction from LPL may be difficult |
| SLL | Presence of growth centers –Co-expression of CD5 and CD23 –Absence of t(11;14) | Growth centers may be ill-defined |
| Splenic MZL | Biphasic splenic morphology –Uncommon red pulp pattern –Frequent PB/BM involvement, may be intrasinusoidal in BM –Cells fail to express CD5, CD10, CD23 and CD43 | Occasional SMZL cases express CD5 –Some may weakly express CD23 –Diagnosis can be difficult without a splenectomy |
| Nodal MZL | Distinct marginal zone architecture –Rule out adjacent mucosal disease –B cells do not co-express CD5 or CD10 | Almost a diagnosis of exclusion –Absence of characteristic cytogenetic alterations found in MALT lymphomas |
| Lymphoplasmacytic lymphoma | Mature plasma cell component +/− Russell/Dutcher bodies –Usually not leukemic –Absence of growth centers and CD5 expression –Increased mast cells in BM –Presence of serum/urine monoclonal protein | Subtle inter-follicular pattern in some cases –Grey-zone cases with SLL/CLL occur –Clinical information often not available |
| Mantle cell lymphoma | Monomorphic appearance, absence of centroblasts; epithelioid histiocytes and mitoses –PB & BM often involved –CD5+, CD23− –Expression of cyclin D1 –Presence of t(11;14) | Blastoid variants may be pleomorphic Rare cases mimic MZL architecture –5% to 7% may be CD5− –Rare cases lack t(11;14) and cyclin D1 overexpression |
| Lymphoma Subtype . | Diagnostic Clues . | Pitfalls . |
|---|---|---|
| Abbreviations: LEL, lymphoepithelial lesion; SLL, small lymphocytic lymphoma; CLL, chronic lymphocytic leukemia; MZL, marginal zone lymphoma; PB, peripheral blood; BM, bone marrow; FL, follicular lymphoma; PCR, polymerase chain reaction; FISH, fluoresence in situ hybridization; RFH, reactive follicular hyperplasia; FDC, follicular dendritic cell | ||
| Follicular lymphoma | Follicle formation, admixed centroblasts, presence of tight FDC meshworks –CD10 +/− Bcl-6 expression between follicles –Low proliferative rate in contrast to RFH –PCR/FISH presence of t(14;18) | Not all FLs express Bcl-2 protein –BCL2 PCR can be negative (inherent high false- negative rate) –Discordance between morphology and proliferative rate in some cases |
| MALT lymphoma | Reactive follicles, LELs and centrocyte-like cells –Presence of light chain restriction –Absence of CD5/CD10 expression –FISH detection of characteristic translocations | Classic cytomorphology not always present –Small biopsies may produce false-positive PCR for IGH clonality –Distinction from LPL may be difficult |
| SLL | Presence of growth centers –Co-expression of CD5 and CD23 –Absence of t(11;14) | Growth centers may be ill-defined |
| Splenic MZL | Biphasic splenic morphology –Uncommon red pulp pattern –Frequent PB/BM involvement, may be intrasinusoidal in BM –Cells fail to express CD5, CD10, CD23 and CD43 | Occasional SMZL cases express CD5 –Some may weakly express CD23 –Diagnosis can be difficult without a splenectomy |
| Nodal MZL | Distinct marginal zone architecture –Rule out adjacent mucosal disease –B cells do not co-express CD5 or CD10 | Almost a diagnosis of exclusion –Absence of characteristic cytogenetic alterations found in MALT lymphomas |
| Lymphoplasmacytic lymphoma | Mature plasma cell component +/− Russell/Dutcher bodies –Usually not leukemic –Absence of growth centers and CD5 expression –Increased mast cells in BM –Presence of serum/urine monoclonal protein | Subtle inter-follicular pattern in some cases –Grey-zone cases with SLL/CLL occur –Clinical information often not available |
| Mantle cell lymphoma | Monomorphic appearance, absence of centroblasts; epithelioid histiocytes and mitoses –PB & BM often involved –CD5+, CD23− –Expression of cyclin D1 –Presence of t(11;14) | Blastoid variants may be pleomorphic Rare cases mimic MZL architecture –5% to 7% may be CD5− –Rare cases lack t(11;14) and cyclin D1 overexpression |
Prognostic factors in follicular lymphoma.
| Factor . | Effect on Outcome . | Mechanism . |
|---|---|---|
| * Dependent on treatment approach. Adriamycin-containing regimens suggest that grade 3 follicular lymphomas have improved survival parameters. | ||
| ** Initial studies indicated that marginal zone differentiation conferred an unfavorable outcome, but later studies refuted this finding. | ||
| # Initial studies suggest a favorable outcome associated with BCL6 translocations, but recent work suggests that these predispose to transformation. | ||
| ## Anti-tumor T cell response is associated with a favorable outcome in contrast to the macrophage signature that is associated with inferior survival. | ||
| ### Elevated cyclin B1 predicted favorable response to CHOP chemotherapy. | ||
| Increasing cytologic grade* | Unfavorable | Increased proliferation |
| Diffuse areas37 | Unfavorable | Early transformation |
| Marginal zone differentiation** | Equivocal | – |
| Intrafollicular proliferative rate | Unfavorable | Increased proliferation |
| Increased small vessel density19 | Favorable | ? |
| Bcl-2 expression | Unfavorable | Anti-apoptotic |
| Bcl-6 expression21 | Favorable | Germinal center phenotype |
| CD10 expression21 | Favorable | Germinal center phenotype |
| MDM2 expression38 | Unfavorable | Functional p53 loss |
| Bcl-XL expression17 | Unfavorable | Anti-apoptotic |
| Macrophage content11 | Unfavorable | Corrupt the microenvironment in favor of the tumor cells |
| Chromosomal gains18,20,39,40+7, +12q13–14, +18q | Unfavorable | Dominant oncogenes or dosage effect |
| Chromosomal losses18,41 del6q, −9p21, −17p13 | Unfavorable | Loss of tumor suppressor gene |
| BCL6 translocation42# | Variable | Marker of genomic instability? |
| Host immune response## | Variable | Increased anti-tumor T cell response (IR-1) or promote trophic microenvironment for tumor cells (IR-2) |
| 81-gene predictor | Variable | Indolent versus aggressive behavior |
| Cyclin B122### | Favorable | Cell cycle progression |
| Factor . | Effect on Outcome . | Mechanism . |
|---|---|---|
| * Dependent on treatment approach. Adriamycin-containing regimens suggest that grade 3 follicular lymphomas have improved survival parameters. | ||
| ** Initial studies indicated that marginal zone differentiation conferred an unfavorable outcome, but later studies refuted this finding. | ||
| # Initial studies suggest a favorable outcome associated with BCL6 translocations, but recent work suggests that these predispose to transformation. | ||
| ## Anti-tumor T cell response is associated with a favorable outcome in contrast to the macrophage signature that is associated with inferior survival. | ||
| ### Elevated cyclin B1 predicted favorable response to CHOP chemotherapy. | ||
| Increasing cytologic grade* | Unfavorable | Increased proliferation |
| Diffuse areas37 | Unfavorable | Early transformation |
| Marginal zone differentiation** | Equivocal | – |
| Intrafollicular proliferative rate | Unfavorable | Increased proliferation |
| Increased small vessel density19 | Favorable | ? |
| Bcl-2 expression | Unfavorable | Anti-apoptotic |
| Bcl-6 expression21 | Favorable | Germinal center phenotype |
| CD10 expression21 | Favorable | Germinal center phenotype |
| MDM2 expression38 | Unfavorable | Functional p53 loss |
| Bcl-XL expression17 | Unfavorable | Anti-apoptotic |
| Macrophage content11 | Unfavorable | Corrupt the microenvironment in favor of the tumor cells |
| Chromosomal gains18,20,39,40+7, +12q13–14, +18q | Unfavorable | Dominant oncogenes or dosage effect |
| Chromosomal losses18,41 del6q, −9p21, −17p13 | Unfavorable | Loss of tumor suppressor gene |
| BCL6 translocation42# | Variable | Marker of genomic instability? |
| Host immune response## | Variable | Increased anti-tumor T cell response (IR-1) or promote trophic microenvironment for tumor cells (IR-2) |
| 81-gene predictor | Variable | Indolent versus aggressive behavior |
| Cyclin B122### | Favorable | Cell cycle progression |
Anatomic localization of MALT lymphomas, cytogenetic alterations, deregulated genes and infectious etiologies.
| Anatomic Site . | Infectious Agent . | Translocation . | Gene . | Frequency . |
|---|---|---|---|---|
| * Frequency data based on references 25 & 26. | ||||
| ** Data supporting a definitive role for these organisms is lacking. | ||||
| Stomach | Helicobacter pylori | t(11;18)(q21;q21) t(1;14)(p22;q32) | API2-MALT1 fusion BCL10 | 22% 3% |
| Lung | ?? | t(11;18)(q21;q21) t(1;14)(p22;q32) | API2-MALT1 fusion BCL10 | 42% 7% |
| Intestine | Campylobacter jejuni** | t(11;18)(q21;q21) t(1;14)(p22;q32) | API2-MALT1 fusion BCL10 | 15% 10% |
| Ocular adnexa | Chlamydia psittaci** | t(3;14)(p14.1;q32) t(14;18)(q32;q21) | FOXP1 MALT1 | 20% 13% |
| Skin | Borrelia burgdorferi** | t(14;18)(q32;q21) t(3;14)(p14.1;q32) | MALT1 FOXP1 | 14% 10% |
| Salivary gland | Autoimmune? | t(14;18)(q32;p21) | MALT1 | 5% |
| Thyroid | Autoimmune? | t(3:14)(p14.1;q32) | FOXP1 | 50% |
| Anatomic Site . | Infectious Agent . | Translocation . | Gene . | Frequency . |
|---|---|---|---|---|
| * Frequency data based on references 25 & 26. | ||||
| ** Data supporting a definitive role for these organisms is lacking. | ||||
| Stomach | Helicobacter pylori | t(11;18)(q21;q21) t(1;14)(p22;q32) | API2-MALT1 fusion BCL10 | 22% 3% |
| Lung | ?? | t(11;18)(q21;q21) t(1;14)(p22;q32) | API2-MALT1 fusion BCL10 | 42% 7% |
| Intestine | Campylobacter jejuni** | t(11;18)(q21;q21) t(1;14)(p22;q32) | API2-MALT1 fusion BCL10 | 15% 10% |
| Ocular adnexa | Chlamydia psittaci** | t(3;14)(p14.1;q32) t(14;18)(q32;q21) | FOXP1 MALT1 | 20% 13% |
| Skin | Borrelia burgdorferi** | t(14;18)(q32;q21) t(3;14)(p14.1;q32) | MALT1 FOXP1 | 14% 10% |
| Salivary gland | Autoimmune? | t(14;18)(q32;p21) | MALT1 | 5% |
| Thyroid | Autoimmune? | t(3:14)(p14.1;q32) | FOXP1 | 50% |
Karyotypic abnormalities in small B-cell lymphomas.
| Disease Entity . | Cytogenetics . | Deregulated Genes . | Frequency (%) . |
|---|---|---|---|
| Abbreviations: SLL, small lymphocytic lymphoma; CLL, chronic lymphocytic leukemia; LPL, lymphoplasmacytic lymphoma; NMZL, nodal marginal zone lymphoma; SMZL, splenic marginal zone lymphoma; MCL, mantle cell lymphoma | |||
| SLL/CLL | +12, del11q, +13, −17p, rare t(14;19) | ?? BCL3 | 75 |
| LPL | Del6q | ?? | 30–70 |
| FL1-3A | t(14;18), t(3;14) or variants | BCL2, BCL6 | 95 |
| MALT | t(11;18), t(14;18), t(1;14), t(3;14), rare BCL6 translocations | API2-MALT1, MALT1, BCL10, FOXP1, BCL6 | Varies with anatomic site |
| NMZL | +3, +7, +18 | ?? | 50 |
| SMZL | Del7q31-32, del7q21 | ??, CDK6 | 70 |
| MCL | t(11;14) | Cyclin D1 | 96 |
| Disease Entity . | Cytogenetics . | Deregulated Genes . | Frequency (%) . |
|---|---|---|---|
| Abbreviations: SLL, small lymphocytic lymphoma; CLL, chronic lymphocytic leukemia; LPL, lymphoplasmacytic lymphoma; NMZL, nodal marginal zone lymphoma; SMZL, splenic marginal zone lymphoma; MCL, mantle cell lymphoma | |||
| SLL/CLL | +12, del11q, +13, −17p, rare t(14;19) | ?? BCL3 | 75 |
| LPL | Del6q | ?? | 30–70 |
| FL1-3A | t(14;18), t(3;14) or variants | BCL2, BCL6 | 95 |
| MALT | t(11;18), t(14;18), t(1;14), t(3;14), rare BCL6 translocations | API2-MALT1, MALT1, BCL10, FOXP1, BCL6 | Varies with anatomic site |
| NMZL | +3, +7, +18 | ?? | 50 |
| SMZL | Del7q31-32, del7q21 | ??, CDK6 | 70 |
| MCL | t(11;14) | Cyclin D1 | 96 |
Schematic of hypothetical disease progression in follicular lymphoma (FL). The left side would denote those cases with high macrophage content. The right side represents both the cytogenetically unstable cases that are at risk to transform and a subgroup of patients that may not be at risk to transform. Several pathways of clonal evolution are recognized as previously described.43
Schematic of hypothetical disease progression in follicular lymphoma (FL). The left side would denote those cases with high macrophage content. The right side represents both the cytogenetically unstable cases that are at risk to transform and a subgroup of patients that may not be at risk to transform. Several pathways of clonal evolution are recognized as previously described.43