Abstract
The genetic basis of a differential response to drugs has been understood for a limited number of agents for over 30 years. This knowledge has generated hope that the individual basis for response to a wide range of drugs would be quickly known, and individualized drug selection and dosing would be possible for many or all disorders. Understanding the variable response to drugs seems particularly pressing in the field of oncology, in which the stakes are high (failure to cure cancer usually leads to death), drugs commonly have a narrow therapeutic index, and toxicities can be severe (a significant frequency of toxic death is a feature of most acute myeloid leukemia protocols, for example). However, in common with many new technologies, the generalizability and clinical application of pharmacogenetics has proved more challenging than expected. Difficulties include, in many examples, a modest clinical effect relative to genotype, therapy-specific, not broad, applicability and the very major challenge of unraveling the complexity of gene-gene interactions. In addition, ethical and economic challenges to the application of pharmacogenetics have moved to the fore in recent years, particularly in the context of racial differences in outcome of therapy. Genomic, rather than candidate gene approaches to identification of relevant loci are increasingly being explored, and significant progress is being made. However, greater understanding of the complexities of multiple gene modifiers of outcome, and the statistical challenge of understanding such data, will be needed before individualized therapy can be applied on a routine basis.
Where We Began: Identification of Common Genetic Variants That Influence Response to Drugs
Differential response to the same drug in different patients is a common clinical experience. Many different factors may contribute to differential response including variable age and body size, diet, gastro-intestinal absorption, compliance with therapy and characteristics of the drug target, e.g., bacterial resistance to a specific antibiotic or mutation in the EGFR receptor in lung cancer treated with gefitinib.1–3 A heritable component to variable drug response was recognized in the middle of the twentieth century with demonstration of fast and slow metabolizers of drugs such as debrisoquine and isoniazid, and of a variable response to the anti-inflammatory phenylbutazone.4,5 A significant genetic component in the metabolism of phenylbutazone was reported in a study published in Science in 1968.5 This gloriously simple study used pairs of volunteer fraternal or monogenic twins, administered a dose of phenylbutazone and monitored clearance. The data showed close concordance of pharmacokinetic profiles between monogenic twins and significantly more variation between fraternal twins.
Understanding the variable response to drugs seems particularly pressing in the field of oncology, in which the stakes are high (failure to cure cancer usually leads to death), drugs commonly have a narrow therapeutic index, and toxicities can be severe (a significant frequency of toxic death is a feature of most acute myeloid leukemia (AML) protocols, for example). In one of the first examples of pharmacogenetics in oncology, Weinshilboum and Sladek identified polymorphic responses to the key antileukemic drug, 6-mercaptopurine (6-MP) in 1980, and polymorphism of the gene thiopourine S-methyl transferase (TPMT) remains one of the best understood examples of pharmacogenetic variation.6
Candidate Gene Approach to Pharmacogenetics: The Example of TPMT
TPMT is a cytosolic drug-metabolizing enzyme that catalyzes the S-methylation of 6-MP and azathioprine. In their original seminal study Weinshilboum and Sladek demonstrated a very clear tri-modal frequency of TPMT activity in red blood cells from 298 unrelated control adults.6 One in 300 subjects lacked TPMT activity, and 11% had intermediate levels. Family studies showed that the frequency distribution was due to inheritance. While phenotypic studies have shown a clear tri-modal distribution, the genetic basis of phenotypic variation has proved more complex. Seventeen variant TPMT alleles have been identified to date, although 3 variant alleles account for the majority (> 95%) of persons with intermediate (1 variant allele) or low (2 variant alleles) TPMT activity.7,8
Subsequent clinical studies have demonstrated very clearly that TPMT polymorphism can predict toxicity of 6-MP and consequences of therapy. Children with acute lymphocytic leukemia (ALL) with intermediate or absent TPMT activity are at higher risk of myelosuppression when prescribed standard doses of 6-MP.9 In addition, patients with low TPMT activity are at increased risk of secondary cancers.10,11
Showing a relationship between a polymorphism at a single locus and outcome of therapy including multiple drugs that exert cytotoxicity through a number of different pathways is a challenge. Despite this, a number of studies have shown that TPMT phenotype or genotype influences effectiveness of therapy, with low TPMT activity being associated with higher levels of cytotoxic 6-thioguanine nucleotides (6-TGN) and reduced relapse. Lennard et al described 104 children with ALL who had completed therapy.12 Children with 6-TGN concentrations below the group median had higher TPMT activities and a higher subsequent relapse rate. The group at St Jude Children’s Hospital has applied very careful focus to the optimization of 6-MP therapy in children with ALL. In a series of elegant and painstaking studies, these investigators have shown that TPMT phenotype predicts levels of 6-TGNs, TPMT genotype predicts phenotype, higher 6-TGNs are associated with increased myelosuppression and greater dose intensity of 6-MP is associated with reduced relapse.9,13–17 These observations have been applied to serial clinical studies that have shown that a combination of measurement of thiopurine metabolites, TPMT status and clinical tolerance of therapy can be used to selectively decrease the dose of 6-MP without decreasing the dose of other drugs in patients with low or intermediate TPMT activity, to counsel patients on compliance if levels of 6-TGNs are low, and to increase therapy in patients with persistently high white blood cell counts. This careful and rational approach has succeeded in eradicating any adverse effect of TPMT genotype; in the St Jude Total XIIIB study the incidence of relapse was not different in children with a wild-type or intermediate TPMT genotype.18
The generalizability of this labor-intensive and disciplined approach to other clinical settings (for example, cooperative groups involving many different institutions, and treatment regimens that use lower 6-MP doses than the St Jude studies [typically 75 mg/m2/day]) remains unclear. The NOPHO ALL-92 protocol randomized children with B-lineage ALL to have their 6-MP/methotrexate (MTX) dosage adjusted by blood counts alone (control group) or by a combination of blood counts and erythrocyte-TGN/MTX levels (pharmacology group).19 In the pharmacology group the doses of 6-MP and/or MTX were adjusted upwards if levels of erythrocyte TGN × MTX levels were lower than a prespecified target. In a surprising result, the dose adjustment did not improve outcome, and in girls pharmacologically driven dose adjustment notably increased the relapse rate. TPMT activity was the strongest predictor of risk of relapse for girls in the pharmacology group, and girls who relapsed off therapy had higher TPMT activity than those who did not relapse, although this was not the case for girls relapsing on therapy. The reason for these somewhat counter-intuitive outcomes is unclear. The investigators speculate that attempts to increase the dose of 6-MP lead to increased intracellular levels of methylated 6-MP metabolites, inhibition of de novo purine synthesis and cell cycle arrest of the leukemic blasts. The investigators conclude that dose adjustment on the basis of blood counts is sufficient to optimize outcomes for girls with ALL.
Pharmacogenetic studies commonly report data from studies performed as long as a decade previously, as clinical endpoints such as relapse must be reached before the importance of genotype can be analyzed. In an alternative approach, Stanulla et al have described the use of minimal residual disease as an interesting alternative endpoint for the analysis of pharmacogenetic variability in chemotherapy for ALL.20 This study genotyped TPMT in 814 children with ALL treated on the ALL-BFM (Berlin-Frankfurt-Munster) 2000 and showed significantly lower levels of minimal residual disease (MRD) in children with intermediate activity TPMT genotypes. This translated into a 2.9-fold reduction in risk of MRD at day 78 (after the start of 6-MP therapy) for patients with a low activity allele. In contrast, hematopoietic toxicity was not different in wild-type and low TPMT activity children, and Stanulla et al suggest that it is unnecessary to reduce the dose of 6-MP for heterozygous children. The prognostic importance of clearance of MRD is well established, and this study suggested that 6-MP in consolidation, as used in this BFM study, is an important component of therapy. MRD measurements are an early endpoint, and may be an effective surrogate endpoint of outcome to dissect the importance of pharmacogenetic variation and interaction with different components of therapy in a timely fashion.
The Hope: An Individualized Pharmacogenetic Profile for Every Patient
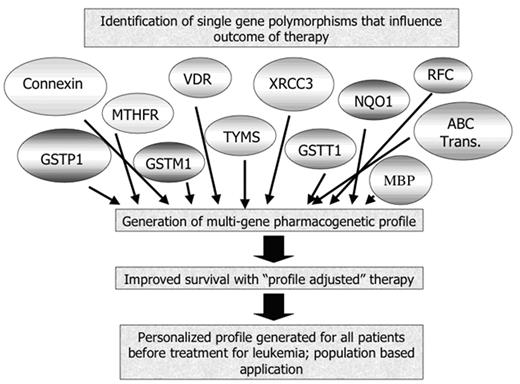
The identification of at least one genetic locus that modifies toxicity and efficacy of therapy has generated a great deal of optimism regarding personalization of cancer therapy. Pharmacogenetic research has a number of characteristics that are attractive to the media—the concept of individualized therapy is, at its most basic, simple and pleasing, there are ramifications of improved survival and reduced toxicity that are gratifying, and there are economic implications—it could be advantageous for investors to “get in at the start” of a revolution in drug treatment. Enthusiastic descriptions of the possibility of personal “bar codes” or routine generation of pharmacogenetic profiles prior to treatment with drugs have appeared in the scientific and popular press, driven by the expectation that investigators would move swiftly through the pathway from single gene studies to predictive profiling, as shown in Figure 1.
This model envisages four steps toward truly personalized medicine, including identification of significant genes, integration of multiple genes into a profile, demonstration that use of the profile to direct therapy improves outcomes then acceptance of the profile on a population-wide basis, not just in specialized research centers. The last 25 years of work have addressed the first step in this pathway, and have thus far identified only two genotypes, TPMT as described above and UGT1, which predicts toxicity of the drug irinotecan, that are commercially available and for which there is some understanding of the interpretation of the data, at least for the prediction of toxicity.
Clinical Utilization of Single Gene PharmacogeneticTesting
Despite extensive studies performed over 25 years, the relatively clear-cut clinical significance of this polymorphism and the commercial availability of TPMT genotyping in many laboratories in the US, TPMT genotyping is not universally used to regulate therapy. A recent review of current clinical practice in pharmacogenetic testing in Europe, including a relatively small number of practitioners, indicated that 53% of clinicians who treat with thiopurine drugs do so without pharmacogenetic testing and a further 35% do use the test but not in all patients who receive treatment.21 Respondents were asked about barriers to utilization of TPMT testing, and notably 25% of respondents thought the clinical utility of the test to be quite or very low and only 50% thought utility to be quite high. These data suggest reluctance to incorporate additional levels of testing into clinical management and a level of comfort with traditional “trial and error” based drug dosing. Data on improved outcomes will likely need to be compelling to overcome this hesitation in clinical practice.
Moving From Single Gene to Multiple Gene Profiles
Interpretation of pharmacogenetic data increases in complexity when more than one gene, or other nongenetic contributors to variable response, are considered. A single drug is commonly used for disorders such as seizures and hypertension, and such disorders may lend themselves to pharmacogenetic study analyzing one, or a small number of, genetic variants.22–25 In contrast, cancer, and in particular leukemia, is treated with multiple drugs with multiple different mechanisms of action, and many different genes are involved in response, each of which has the potential for gene-gene interactions. Rocha et al26 genotyped 16 polymorphic loci in 246 children with ALL, including 116 children with low risk (LR) and 130 with higher risk (HR) disease. The data were analyzed by using a classification and regression-tree (CART) analysis and showed that children in the HR group with the GSTM1 non-null genotype had an increased risk of relapse. The risk of relapse was further increased in the presence of the thymidylate synthetase 3/3 genotype, and both genotypes remained predictive in multivariate analysis. These data are of particular interest because previous single-gene studies, not analyzed according to leukemia risk group, had failed to identify any clinical importance of GSTM1 genotype.27,28 The study of Rocha et al illustrates the complexity of multiple gene analyses, requiring assessment of multiple genes, sophisticated statistical approaches to analysis and stratification of other biological factors known to influence outcome, such as NCI risk group.
Moving Forward: Broader Genomic Approaches
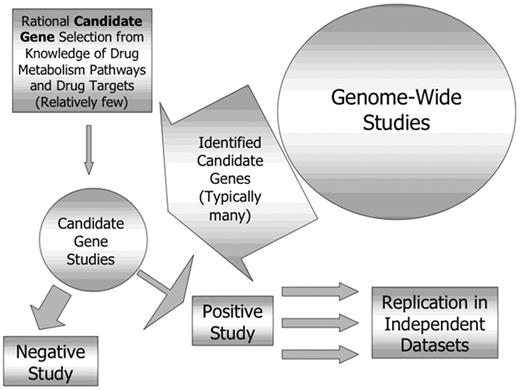
The studies discussed so far in this manuscript have used a candidate gene approach to select polymorphic loci for analysis. Genes are selected for study based on knowledge of the targets or metabolic pathways of the drugs used. An alternative to a candidate gene approach is the application of newer genome-wide approaches (pharmacogenomics rather than pharmacogenetics) to take a broad “discovery-based” approach to genes that may modify response.29,30 The advantages and disadvantages of these two approaches are compared in Table 1 .
The ability to identify novel genes not suspected to be involved in response to a particular drug is a very attractive feature of such an approach. It should be recognized, however, that these approaches are complementary, with genome-wide studies generating new candidate genes, each of which then need to be evaluated individually, and all positive findings need to be replicated in independent datasets (Figure 2 ). Genome-wide approaches include microarray-based comparisons of gene expression in tissues identified to be sensitive or resistant to a particular drug or combination. This approach has been used to compare gene expression in leukemia cells from children resistant or sensitive in vitro to prednisolone, vincristine, asparaginase and daunorubicin and succeeded in identifying 124 differentially expressed genes associated with the in vitro phenotype, only 3 of which were previously thought to be important.31
Are Pharmacogenetic Data Context Dependent?
Pharmacogenetic findings may be specific to the therapy protocol in which the observations were made, limiting the applicability of the findings to other treatment plans. This is a particular challenge in oncology when multiple drugs are being used and the whole “package” needs to be considered, not just metabolism of a single drug. In an example of context dependency, we have investigated polymorphisms in pathways of homologous recombination in children treated for AML.32 The data show that a polymorphism in the gene XRCC3 (C/T at codon 241) influenced post-induction outcomes, with superior 5-year disease-free survival (DFS) in heterozygotes (55.7 ± 4.7% for CT vs. 44.2 ± 4.5% for CC + TT genotypes; log-rank P = 0.05). The difference in outcome was due to an increased frequency of relapse in XRCC3 homozygotes (5-year relapse-free survival [RFS] 61.3 ± 4.8% for CT vs. 47.6% ± 4.7% for CC + TT genotypes; log-rank P = 0.02). The children in this study were enrolled in a clinical trial that included post-induction randomization to either idarubicin, dexa-methasone, cytarabine, thioguanine, etoposide and daunomycin (IDA-DCTER) or daunorubicin, fludarabine, cytarabine and granulocyte colony-stimulating factor (IDA-FLAG). We hypothesized that homologous recombination was likely to be important for repair in IDA-DCTER, but not for IDA-FLAG. In agreement with this, 5-year survival was significantly improved in heterozygous patients receiving IDA-DCTER compared with homozygous cases (68 ± 6.5% for CT vs. 55.6 ± 5.4% for CC + TT; log-rank P = 0.03). In contrast, in patients randomized to receive IDA-FLAG, survival was superior in heterozygotes, but the difference in outcome was not significant (55 ± 5.8% for CT vs. 46.7 ± 6.1% for CC + TT; log-rank P = 0.38). XRCC3 genotype also influenced RFS in the IDA-DCTER arm (62.6 ± 7.2% vs. 44 ± 6.1%; log-rank P = 0.03) but not in the IDA-FLAG arm (56.4 ± 6.3% vs 49.6 ± 7.2%; log-rank P = 0.59). These data demonstrate the likely challenges in identifying pharmacogenetic profiles that are applicable to more than one complex therapeutic protocol.
Different Contexts: Pharmacogenetics and Race
Considerable controversy surrounds the use of race as a surrogate for pharmacogenetic characteristics.33–36 Focus on this issue arose in 2005 after the US Food and Drug Administration (FDA) approved a pill called BiDil (a combination of hydralazine and isosorbide dinitrate) for the treatment of heart failure in self-identified black patients. Approval was granted based on a randomized study of BiDil in heart failure in black patients that showed a survival advantage compared with placebo; previous studies had failed to show benefit in whites.37 A new US patent was granted specifically for BiDil use in black patients, although previous patents for the two drugs already existed. Anxieties raised by these events are driven by uncertainty in the definition of race and the very clear demonstration that there is greater genetic variation within races than between races, showing clearly that race is a more powerful social than biological categorization, and includes a broad spectrum rather than discrete categories of phenotype. Discrimination in insurance and employment on the basis of perceived genetic differences between the races and a reluctance to investigate the biological basis of differential response to the drug once FDA approval has been granted are concerns. In this example, therapy is being assigned by race without any understanding of the biological differences that predict response in persons of any race.
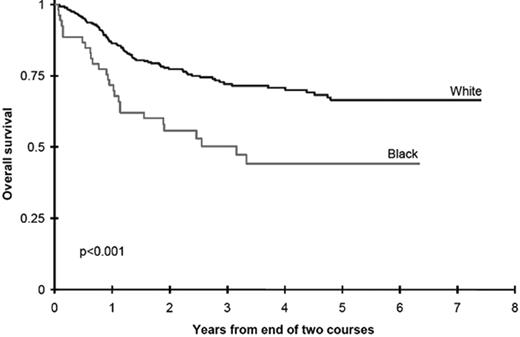
An alternative approach to incorporating racial and ethnic disparities into pharmacogenetics is the use of disparities in outcome as a “signpost” to direct investigators toward metabolic pathways in which genetic variants may influence survival. In an attempt to understand the role of race and ethnicity in childhood AML we evaluated outcomes in two Children’s Cancer Group (CCG) studies.38 Black children treated with chemotherapy in CCG-2891 had significantly inferior overall survival from study entry, compared with white children (34 ±10% vs. 48 ± 4%, p = 0.007). Analyses of the successor study, CCG-2961, confirmed that black children had a significantly decreased survival compared with white children (45 ± 12% vs. 60 ± 4%, p = 0.007). Induction outcomes were similar in children of all races and ethnicities; inferior survival in black children was due to an excess of relapse in black children receiving cytarabine-based postinduction consolidation chemotherapy (Figure 3 ). Children of all races receiving consolidation with allogeneic transplantation had equivalent outcomes, and biological characteristics of the leukemia were not different in children of different races.
Issues of access to healthcare, compliance with therapy and an increased frequency of high-risk disease noted in some studies have been implicated as explanations for disparities in outcome for children with ALL.39–40 In contrast, children with AML receive their therapy intravenously in an in-patient setting with prescribed supportive care and careful recording of drugs actually administered, removing these concerns. Our data suggest that pharmacogenetic differences in metabolism of cytarabine in black children with AML might underlie the disparity in outcome, and we are conducting an analysis of genetic variants in these pathways, with a focus on polymorphisms with different allele frequencies in persons of different race. It is also possible that biochemical differences, such as differences in the retention of intracellular AraCTP, may underlie these differences in outcome, and inclusion of such pharmacodynamic endpoints in future studies might assist in the interpretation of pharmacogenetic data.41 Understanding the genetic basis of the disparity in outcome will replace race-based prediction of outcome with genotype based risk assessment that will likely to benefit children of all races who may carry unfavorable alleles.
Realistic Expectations: What Can Be Achieved
Pharmacogenetics is facing the challenge of living up to enthusiastic and perhaps over-optimistic statements of the likely effectiveness of the technology and a lack of a realistic view of the likely timeline and applicability of the approach. Oncology therapy is complex and involves many drugs; variability in outcome has many contributors including the characteristics of the malignant cell, renal and hepatic function, diet, compliance with therapy and the therapy protocol employed. All of these variables are important to greater or lesser degrees in an individual case.
Candidate gene approaches have identified TPMT as an important polymorphic locus and the clinical consequences of variant genotype have been extensively described. Despite this work extending over 25 years and the easy availability of commercial genotyping since 2003, clinical application of the knowledge is patchy at best. Of note, clinical use of TPMT testing is perhaps greater among practitioners using azathioprine for treatment of inflammatory bowel disease, illustrating perhaps greater comfort in using a pharmacogenetic test in a drug regimen using a single drug (or small numbers of drugs). In the complex regimens used in leukemia, and with the low frequency of the severely affected genotype (1 in 300), there appears to be some skepticism, or perhaps lack of knowledge of the utility of routine testing.
These observations suggest that as pharmacogenetics moves forward with ever more powerful genome-wide technologies, the field will benefit from a cautious approach to describing applications that are still in the future; “personalized medicine for all” is not on the immediate horizon, more a distant goal. A systematic approach to the dissection of important metabolic pathways using a combination of strategies and determining the applicability and clinical importance of the knowledge gained will move us toward this goal.42 Surrogate endpoints such as minimal residual disease or pharmacodynamic endpoints may provide earlier data than waiting for a large randomized trial to mature and allow greater focus on single drugs within a complex therapeutic strategy.
Comparison of genome-wide and candidate gene approaches to pharmacogenetic studies.
| Candidate Gene Studies . | Genome-Wide Studies . |
|---|---|
| Rational selection of genes for study increases biological plausibility of findings, however novel genes cannot be identified. | Analysis of whole genome allows identification of genes not previously known to be important in response to drug. |
| Limited number of genes reduces possibility of false discovery. | Large number of analyses means significant risk of false discovery. |
| Expense reduced if candidate genes selected prove to be relevant to outcome. | Increased expense following up on large number of positive findings. |
| Moderate sample size may be adequate. | Large sample sizes are needed because of the large number of comparisons made. |
| Candidate Gene Studies . | Genome-Wide Studies . |
|---|---|
| Rational selection of genes for study increases biological plausibility of findings, however novel genes cannot be identified. | Analysis of whole genome allows identification of genes not previously known to be important in response to drug. |
| Limited number of genes reduces possibility of false discovery. | Large number of analyses means significant risk of false discovery. |
| Expense reduced if candidate genes selected prove to be relevant to outcome. | Increased expense following up on large number of positive findings. |
| Moderate sample size may be adequate. | Large sample sizes are needed because of the large number of comparisons made. |
The hoped-for pathway of development of personalized medicine for the treatment of leukemia.
The hoped-for pathway of development of personalized medicine for the treatment of leukemia.
Relationship between genome-wide and candidate gene pharmacogenetic studies.
Survival is inferior in black children treated with chemotherapy for acute myeloid leukemia (AML) compared with white children.
Survival is inferior in black children treated with chemotherapy for acute myeloid leukemia (AML) compared with white children.