Abstract
Families with multiple individuals affected with chronic lymphocytic leukemia (CLL) and other related B-cell tumors have been described in the literature. Familial CLL does not appear to differ from sporadic CLL in terms of prognostic markers and clinical outcome. While some environmental factors (such as farming-related exposures and occupational chemicals) may increase risk of CLL, results of epidemiologic studies have been generally inconsistent. Rates of CLL in the population show significant international variation, with the highest rates in the U.S. and Europe and the lowest rates in Asia. Migrants from Asia to the U.S. also have low rates of CLL, which supports a greater role for genetic compared with environmental risk factors. Large, population-based case-control and cohort studies have also shown significant familial aggregation of CLL and related conditions including non-Hodgkin and Hodgkin lymphoma. Monoclonal B-cell lymphocytosis also aggregates in families with CLL. However, the clinical implication of familial aggregation is minimal given the overall rarity of CLL. Linkage studies have been conducted in high-risk CLL families to screen the whole genome for loci that contribute to susceptibility, but no gene mutations have yet been identified by this method. Association studies of candidate genes have implicated immune function and other genes, but more studies are needed to verify these findings. The ability to conduct large-scale genomic studies will play an important role in detecting susceptibility genes for CLL over the next few years and thereby help to delineate etiologic pathways.
Introduction
B-cell chronic lymphocytic leukemia (CLL) is a neoplastic disease characterized by the accumulation of small, mature-appearing lymphocytes in the blood, bone marrow and lymphoid tissues. CLL accounts for 30% of all leukemia and is the most common form of leukemia among older adults in Western countries. The latest World Health Organization (WHO) classification scheme considers CLL as a mature B-cell neoplasm and does not distinguish it from small lymphocytic lymphoma (SLL).1 Data from the United States Surveillance, Epidemiology, and End Results (SEER) Registry estimate the U.S. incidence in the period 2000 through 2004 to be 3.9 per 100,000 people, with a median age at diagnosis of 72 years.2 Incidence rates in men are nearly twice as high as in women. Although advanced age, white ancestry, and family history of hematologic malignancies are risk factors, the etiology of CLL is unknown.
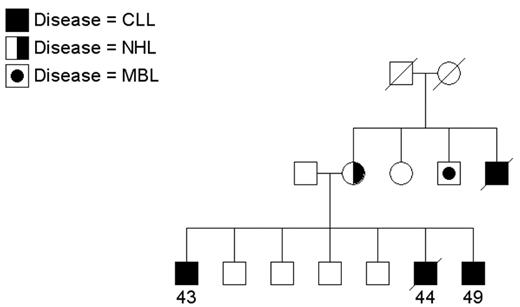
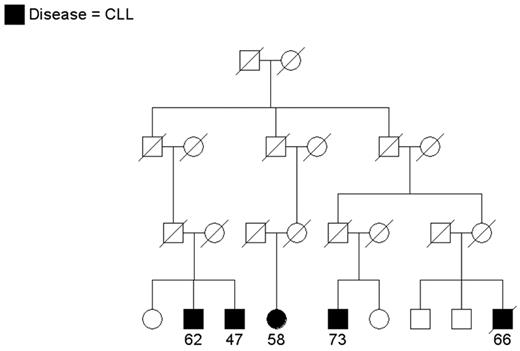
Since CLL is an uncommon cancer, patients who have at least one relative with CLL are considered “familial” and others are considered “sporadic.” In population-based samples, approximately 5% of patients with CLL reported a family history of leukemia (reviewed in Houlston et al3). Clinical descriptions of CLL families have appeared in the literature for a number of years (reviewed in Caporaso et al4 and Sellick et al5). Since the 1970s, the National Cancer Institute group has been recruiting CLL families for genetic studies. While most referrals have been small families with 2 patients, some show quite a striking aggregation of CLL or of CLL and other lymphoproliferative (LP) tumors. Two interesting families are shown in Figures 1 and 2 . Figure 1 shows a family that includes 4 patients with CLL and 1 patient with non-Hodgkin lymphoma (NHL) spread across two generations. Another individual in this family has a monoclonal B-cell lymphocytosis (MBL) as detected by flow cytometry studies. Other families have been recruited aggregating both CLL and Waldenström macroglobulinemia or CLL and Hodgkin lymphoma (HL), suggesting a common etiology to these variant B-cell tumors. In Figure 2 , CLL is present in siblings and in distant cousins. These families illustrate that there is no consistent pattern of illness that can be explained by a simple mode of genetic transmission.
What is the explanation for familial aggregation of CLL and related conditions? What does it tell us about the etiology of CLL? Is it due to genes and/or environment? Which genes? Which environmental factors? Finally, what is the clinical significance of familial aggregation?
Environmental Risk Factors
CLL shows substantial geographic variation worldwide. Rates show as much as a 40-fold difference, the highest among whites in North America and Europe and the lowest in Asians.6 Within the U.S., CLL is more frequent in the north central part of the country and lower in the southern states. Based on U.S. SEER rates per 100,000 for 2000 to 2004, Whites have the highest rates (4.14), followed by Blacks (3.03), Hispanics (1.94), Native Americans (1.44), and Asians (0.84). The rates of CLL in the U.S. have been relatively stable over time.2 Studies of Asian populations in the U.S. have reported their rates to be very similar to those of similar populations in Asia.7 Thus, unlike other cancers, risk of CLL does not seem to increase in individuals who come to the U.S. from low-risk countries. This supports a stronger role for genetic factors than environmental factors in disease etiology.
Case-control and cohort studies have tested for the effects of environmental risk factors in CLL. Some studies have found that farming exposures (pesticides, herbicides, exposure to animals) are significant.6 Studies of occupational cohorts and case-control studies have found increased risk of CLL due to exposure of rubber industry chemicals and benzene,8 although results are not consistent.6 In studies of atomic bomb survivors, CLL was not found to be elevated and thus has been thought to be a nonradiogenic cancer. However, CLL is very rare in Asia, and thus its lack of association with A-bomb survivors is not surprising. Studies of other occupational radiation–exposed cohorts have not found an increased risk, but there are several methodologic difficulties relevant to CLL that plague these studies, including the long latency of this tumor, the varying classifications of leukemias among studies, and the difficulty in obtaining accurate outcome data for cohort studies relying on death certificates.9 There is some evidence that radiation due to magnetic fields is associated with risk of CLL.6
Clearly, more data are needed to evaluate the role of environmental risk factors. However, these exposures are unlikely to account for the familial aggregation of CLL. The evidence is currently weak and inconsistent, and the relative rarity of CLL and other methodologic difficulties in evaluating the risk for CLL have limited the conclusions of these studies.
Evidence for a Familial Component for CLL
Sporadic CLL cases versus familial CLL cases
As mentioned earlier, multiple-case families have been described in the literature for several decades,4 and some studies have compared the clinical features between sporadic and familial CLL cases. Generally, familial cases have an earlier age of onset than sporadic cases.10 Some investigators have noted that there is anticipation in age of onset in multigenerational pedigrees, where younger generations have an earlier age of onset than older generations.11 However, it is hard to eliminate ascertainment bias in these studies. There have been some attempts to look for other features that distinguish familial CLL from sporadic CLL. Both subtypes appear with mutated and nonmutated IgVH status.12 Further, studies have reported shorter telomeres13 or CD38 positivity14 in a proportion of familial cases, similar to findings in sporadic CLL cases. Ng et al15 found that 12 of 14 familial cases studied by FISH had a chromosome 13q deletion, which contrasts with a rate of 50 to 60% in large case series.16 However, families had to have 2 or more living patients with CLL in order to participate in these studies, which may have preferentially selected more indolent patients (and therefore those more likely to carry the 13q deletion). A recent study found that levels of B-lymphocyte stimulator were higher in patients with familial CLL than in sporadic patients or controls.17 Another recent study of 1449 patients with CLL concluded that a family history of a hematologic malignancy did not indicate an adverse prognosis.18 The literature to date is limited, but overall there do not appear to be striking or consistent differences between familial and sporadic CLL.
Case-control studies of familial risk
There are data from case-control and cohort studies addressing the familiality of CLL. Early population-based case-control studies of CLL reported family history of hematolymphoproliferative (HLP) tumors to be a significant risk factor.19 More recently, a family history analysis of case-control samples pooled from the InterLymph Consortium examined family history of HLP by subtype of lymphoma in the patients. Because of the large number of studies that were pooled, they were able to break-out patients with CLL/SLL and found that family history of “any HLP” or family history of “leukemia only” were significant predictors of risk for CLL/SLL.20
Swedish-Danish registry studies of familial risk
A recently published a series of studies based on linked registry data from Sweden and Denmark quantified familial aggregation of CLL, NHL, HL, and multiple myeloma (MM).21–24 These studies were conducted by linking population-based registries that contain parent-offspring links to the cancer registries. These are the largest studies to date that are population-based and quantify risks to relatives of LP tumors in a comprehensive manner. In Sweden, the multigeneration register (contains individuals born in 1932 and later linked to parents) was linked with the Swedish Cancer Registry. In Denmark, the Danish Central Population Register, which contains parent-offspring links starting in 1968, was linked with the Danish Cancer Register. Controls and their relatives was also chosen from each population registry. In the CLL study,21 the risk of specific cancer outcomes were quantified in 14,336 relatives of 5918 patients with CLL compared to that of relatives of matched controls. First-degree relatives of patients with CLL were found to have a significantly increased risk for CLL (relative risk [RR] = 7.5, 95% confidence interval [CI]: 3.63–15.56). Risks were also increased for NHL (RR = 1.45, 95% CI:0.98–2.16) and HL (RR = 2.35, 95% CI:1.08–5.08). CLL risks were similar in parents, siblings, and offspring of cases and in male and female relatives, and were not affected by the patient’s age at diagnosis. There was no increase in other leukemias or solid tumors among relatives of patients with CLL compared with relatives of controls.
Spectrum of CLL-related conditions
Table 1 summarizes the data from all four Swedish- Danish Registry studies, showing that there is specific aggregation for each LP tumor type but there is also co-aggregation among CLL, NHL, and HL. However, MM appears to aggregate as a separate entity. These data suggest that the variant LP phenotypes seen within multigenerational pedigrees are not simply due to a referral bias but do reflect common risk factors for these conditions. The Swedish-Danish data also do not support anticipation in age of onset of LP tumors.25
In the last several years, three- and four-color flow cytometry studies have been able to detect a monoclonal B-cell lymphoctyosis (MBL) in individuals who do not have CLL.26 Current consensus definition of MBL has been described: specifically, these individuals have a CLL phenotype based on flow cytometry; however, they do not meet the WHO criteria of an absolute lymphocyte count higher than 5.0 × 109/L. In population studies, these clones have been reported to occur in 3.5% of individuals older than 40 years27 and in 5.5% of individuals older than 65 years.26 Rawstron et al28 and Marti et al29 conducted studies in unaffected first-degree relatives in CLL families. Both groups found a significantly higher rate of MBL (13.5% and 18%, respectively) in first-degree relatives of patients with CLL than in control populations. The significance of these clones is not known. MBL as a precursor to CLL may be analogous to monoclonal gammopathy of undetermined significance (MGUS) as a precursor to MM.31 Further, deletions of 13q14 have been described in MBL.27 In a follow-up study of 111 patients with MBL, Rawstron et al32 found that 42% had an increase in their lymphocyte counts during a median of follow-up of 3.7 years, with 5% requiring treatment for disease. More studies are needed to assess long-term outcome in individuals with MBL and how the outcome is affected by the specific phenotype.
Given that CLL is associated with autoimmunity, Landgren et al33 analyzed the CLL case-control data from the Swedish Registry for the occurrence of autoimmune diseases. Autoimmune disease outcomes were obtained by linking individuals to the Swedish Inpatient Hospital Registry. The authors found that having a relative with an autoimmune condition did not affect the risk of CLL. Other than pernicious anemia, a previous personal history of an autoimmune disease was not a risk factor for CLL in this study.
The fact that a broad spectrum of LP conditions is associated with CLL in families should inform the search for susceptibility genes using both the family linkage and association approaches. While there may be genes that predispose only to CLL, the data support the existence of genes that predispose to a broader range of phenotypes.
Approaches to Finding Genes
Linkage studies
Linkage studies localize genes using the co-inheritance of genetic markers and disease (e.g., CLL) in families. They are based on the idea that the occurrence of multiple individuals with CLL from the same family share genetic risks or a shared “genetic” exposure. Using the knowledge of the family relationship and known CLL status, one searches the genome for shared genes that are shared more often than what is expected by chance given the familial relationship. To date, two genome-wide linkage studies of CLL families have been conducted. Goldin et al34 genotyped 359 microsatellite markers in 94 individuals (38 with CLL) from 18 families that had 2 or more individuals with CLL. Using both nonparametric and parametric linkage analyses, regions on chromosomes 1q, 3q, 6q, 12q, and 13q showed increased sharing among affected relatives. Of particular interest is the finding on chromosome 13q, which overlaps with a region that is commonly associated with cytogenetic changes. However, further studies were not able to identify a germline mutation in this region.15 In 2005, Sellick et al35 genotyped 11,560 single nucleotide polymorphisms (SNPs) in 228 individuals from 115 families. Evidence for linkage was found on chromosome 11p11, and additional regions of interest were identified on chromosomes 5q22–23, 6p22, 10q25, and 14q32. Unfortunately, none of the regions identified by these two studies were statistically significant. Further, despite the general similarity of methodology between these two linkage studies, no overlap of genomic regions was observed. The most likely reason for this is that both studies had limited power to detect linkage due to genetic heterogeneity of CLL and the fact that most of the families in each study had only 2 individuals affected with CLL genotyped. Larger studies with greater statistical power are needed to identify genetic susceptibility loci. These studies will only be possible through multicenter collaborations, such the Genetic Epidemiology of CLL consortium (principal investigators: Drs Susan Slager and Neil Caporaso) and the International CLL Consortium (principal investigator: Dr Richard Houlston).
Association studies
Candidate-gene studies are epidemiologic studies that focus on a gene or a set of genes that have biological plausibility in the pathophysiology of the disease. They are powerful study designs for detecting common genetic variants of modest risk. The basic study design consists of comparing the frequency of alleles (or genotypes) in cases to that of unrelated controls. A list of association studies that have been conducted are summarized in Table 2 , and these studies are expected to increase considerably over the next few years. The number of genes evaluated in these studies range from a few genes to sets of genes from a defined disease pathway. Recently, several promising genes have been identified, particularly for immunoregulatory genes36 and Toll-like receptor genes.37 Due to advances in technology, larger candidate genes studies can be performed. Cerhan et al44 investigated 7670 SNPs from 1253 inflammation and immune genes. Interesting findings were located in TRAF1 gene (P = .0001) and FGG (P = .0004). An alternative to candidate genes studies is large genomic studies. These studies are agnostic in that one looks for association across the entire genome without any biologic assumptions. Rudd et al38 evaluated 1467 nonsynonymous SNPs from 865 genes in a large case-control study. They found genes in the ATM-BRCA2-CHEK2 pathway to be associated with CLL. For example, genetic variants in the ATM gene that were associated with CLL risk with dominant odds ratios of 2.28 (95% CI: 1.53–3.40) for the rs1800056 SNP and 1.68 (95% CI: 1.25–2.28) for the rs1800057 SNP. In the near future, results from whole-genome association studies using 500,000 SNPs or more will be available, as these studies have become a feasible approach to finding genes.
Although the findings from these genetic association studies are of interest, they need to be validated in an independent sample before any firm conclusions can be reached. Ioannidis et al39 have shown that most reported findings from genetic association studies are refuted. This is in part because of relatively small sample sizes of the initial study reporting the finding. As shown in Table 2 , many of the genetic association studies of CLL risk fall under this situation, and a clear example of this problem is with the P2X7 receptor story. The initial study40 included only 36 patients with CLL and 46 controls. Subsequent studies with larger sample sizes failed to find any association, and a meta-analysis of this gene also provided no evidence of an association.41 Note that many of the more recent studies reported in Table 2 are statistically powered to identify low-penetrance genes for risk of non-Hodgkin lymphoma (NHL). However, they are inadequately powered for detecting these genes within the NHL subtypes, such as CLL. Large sample sizes will be needed, which could be easily obtained through collaborations and consortiums such as InterLymph.
Other genetic approaches
The role of other types of genetic mechanisms (involving differences in gene regulation) have been studied in relation to familial CLL. Calin et al42 have described germline mutations in a microRNA gene (miR-16-1) located in the commonly deleted region on 13q14 in 2 of 75 patients with CLL tested (1 of 2 had a family history of CLL). Raval et al43 have recently described a germline change near the DAPK1 gene on chromosome 9 that was associated with decreased allelic expression in CLL cases from 1 family. This change was associated with increased methylation of DAPK1, a change that leads to decreased apoptosis. However, the variant was not found in 75 other patients with familial CLL. Linkage studies in CLL families have not found a signal in this region of chromosome 9, so this locus is not likely to be a common cause of familial CLL. The role of epigenetic changes in CLL needs to be further explored.
Clinical Applications
At this point, the clinical implications of familial aggregation and of the genetic findings are minimal. There are several predictors of progression of CLL (e.g., cytogenetic abnormalities, ZAP-70 levels, mutated vs unmutated IgVH) that affect treatment decisions, but there are no data to suggest that family history of CLL or another LP predicts a more aggressive course. The other relevant question is, should close relatives of patients with CLL worry about their own risk of developing the disease? What if they have the subclinical phenotype, MBL? The studies reviewed here do indicate that the risk of CLL in first-degree relatives is elevated severalfold compared with that of the population, and the risk of other LP tumors is also increased. However, since the baseline risk of CLL in the population is low (lifetime risk estimated to be 0.45%2), the absolute excess risk to relatives of cases is still low and not of clinical significance. Likewise, the absolute risks of a relative developing NHL or HL are also low. MBL, as determined by flow cytometry, occurs at an increased rate in relatives of patients with CLL. However, the criteria for MBL are evolving,27 and while some patients will progress to a more significant disease, more data are needed to better characterize long-term outcome.
Conclusions
It is clear that there is significant familial aggregation of CLL, with the evidence pointing to a greater importance of genes rather than shared environments. There are likely to be specific genes associated with CLL but also genes common to CLL and other related LP tumors. However, the failure to identify any specific mutation with a large effect (like BRCA1 for breast cancer) suggests that multiple genes with smaller effects cause much of the familial aggregation. These genes may be harder to identify, but the advances in large-scale genomics methods that can be applied to large population samples offer promise that specific genes causing susceptibility to CLL and other LP will be identified in the near future.
As our knowledge of germline changes that lead to susceptibility for CLL expands, we will obtain a better understanding of the etiologic pathways relevant to both familial and sporadic CLL. This will therefore lead to better treatment approaches to this still incurable tumor.
Risk of lymhoproliferative (LP) tumor in relatives by LP tumor type in probands.
| . | Lymphoproliferative tumor type of relative . | ||||
|---|---|---|---|---|---|
| Proband . | CLL . | NHL . | HL . | MM . | All LP . |
| Bold signifies same tumor type in proband and relative. | |||||
| Abbreviations: CLL, chronic lymphocytic leukemia; NHL, non-Hodgkin lymphoma; HL, Hodgkin lymphoma; MM, multiple myeloma. | |||||
| CLL | 7.5 | 1.4 | 2.3 | 1.1 | 2.1 |
| NHL | 1.3 | 1.8 | 1.5 | 1.1 | 1.5 |
| HL | 2.3* | 1.3 | 3.5 | 1.0 | 1.7 |
| MM | 0.9 | 1.2 | 0.7 | 1.7 | 1.2 |
| . | Lymphoproliferative tumor type of relative . | ||||
|---|---|---|---|---|---|
| Proband . | CLL . | NHL . | HL . | MM . | All LP . |
| Bold signifies same tumor type in proband and relative. | |||||
| Abbreviations: CLL, chronic lymphocytic leukemia; NHL, non-Hodgkin lymphoma; HL, Hodgkin lymphoma; MM, multiple myeloma. | |||||
| CLL | 7.5 | 1.4 | 2.3 | 1.1 | 2.1 |
| NHL | 1.3 | 1.8 | 1.5 | 1.1 | 1.5 |
| HL | 2.3* | 1.3 | 3.5 | 1.0 | 1.7 |
| MM | 0.9 | 1.2 | 0.7 | 1.7 | 1.2 |
List of genetic association studies of chronic lymphocytic leukemia (CLL).
| Study design . | Reference . | NHL,*n . | CLL/SLL, n . | Control, n . | No. of genes . | No. of markers . | Notes . |
|---|---|---|---|---|---|---|---|
| Abbreviations: NHL, non-Hodgkin lymphoma; SLL, small lymphocytic lymphoma; SNPs, single nucleotide polymorphisms. | |||||||
| Candidate gene | Machulla et al. Int J Cancer . 2001 ;92 :203 | 101 | 157 | 1 | HLA class 1 and 2 loci | ||
Wiley et al. Lancet . 2002 ; 359 :1114 | 36 | 46 | 1 | 1 | P2X7 receptor | ||
Thunberg et al. Lancet . 2002 ;360 :1935 | 170 | 200 | 1 | 1 | P2X7 receptor | ||
Starczynski et al. Br J Haematol . 2003 ;123 :66 | 121 | 95 | 1 | 1 | P2X7 receptor | ||
Zhang et al. Leukemia . 2003 ; 2097 | 144 | 348 | 1 | 1 | P2X7 receptor | ||
Nückel et al. Eur J Haematol . 2004 ;72 :259 | 111 | 97 | 1 | 1 | P2X7 receptor | ||
Sellick et al. Cancer Epidemiol Biomarkers Prev . 2004 ;13 :1065 | 424 | 428 | 1 | 1 | P2X7 receptor | ||
Bogunia-Kubik et al. Int J Immunogeneti . 2005 ;33 :21 –24 | 61 | 180 | 1 | 1 | TNF-alpha -308 SNP | ||
Guzowski et al. J Biomolecular Techniques . 2005 ;16 :154 | 17 | 25 | 1 | 3 | IL10 promoter SNPs | ||
Pérez-Chacón et a. Am J Clin Pathol . 2005 ;132 :646 | 134 | 102 | 1 | 1 | CD5 microsattellite | ||
Zhang et al. J Natl Cancer Inst . 2005 ;97 :1616 | 461 | 59 | 535 | 1 | 1 | BCL6 | |
Jamroziak et al. Pharmacological Reports . 2006 ;58 :720 | 110 | 201 | 1 | 1 | MDR1 | ||
Morton et al. Pharmacogenet Genomics . 2006 ;16 :537 | 1172 | 146 | 982 | 2 | 10 | NAT1 and NAT2 | |
Lan et al. Carcinogenesis . 2007 ;28 :823 | 461 | 59 | 535 | 4 | 5 | Caspase genes | |
Nückel et al. Blood . 2007 ;109 :290 | 123 | 120 | 1 | 1 | BCL2 | ||
| Disease pathway | De Roos et al. Cancer Epidemiol Biomarkers Prev . 2006 ;15 :1647 | 1172 | 148 | 982 | 11 | 15 | Metabolic gene variants |
Hill et al. Blood . 2006 ;108 :3161 | 1172 | 148 | 982 | 19 | 34 | DNA repair and related genes | |
Shen et al. Hum Genet . 2006 ;119 :659 | 461 | 59 | 535 | 18 | 32 | DNA repair genes | |
Nieters et al. Genes Immun . 2006 ;7 :615 | 710 | 104 | 710 | 7 | 11 | Toll-like receptor, IL10, IL10RA | |
Wang et al. Hum Genet . 2006 ;120 :297 | 1172 | 148 | 982 | 7 | 12 | Cell cycle genes | |
Wang et al. Cancer Res . 2006 ;66 :9771 | 1172 | 148 | 982 | 36 | 57 | Proinflammatory and other immunoregulatory genes | |
Wang et al. Carcinogenesis . 2007 ;27 :1828 | 1172 | 148 | 982 | 10 | 13 | Oxidative stress pathway | |
Lan et al. Hum Genet . 2007 ;121 :161 | 461 | 59 | 535 | 10 | 14 | Oxidative stress pathway | |
Lim et al. Blood . 2007 ;109 :3050 | 1172 | 147 | 982 | 18 | 30 | Folate and 1 carbon metabolism | |
Cerhan et al. Blood . 2007 ; Sept 7 [Epub ahead of print] | 458 | 126 | 484 | 1450 | 7670 | Inflammation and immune genes | |
| Genomic studies | Rudd et al. Blood . 2006 ;108 :638 | 992 | 2707 | 865 | 1467 | Nonsynonymous SNPs | |
| Study design . | Reference . | NHL,*n . | CLL/SLL, n . | Control, n . | No. of genes . | No. of markers . | Notes . |
|---|---|---|---|---|---|---|---|
| Abbreviations: NHL, non-Hodgkin lymphoma; SLL, small lymphocytic lymphoma; SNPs, single nucleotide polymorphisms. | |||||||
| Candidate gene | Machulla et al. Int J Cancer . 2001 ;92 :203 | 101 | 157 | 1 | HLA class 1 and 2 loci | ||
Wiley et al. Lancet . 2002 ; 359 :1114 | 36 | 46 | 1 | 1 | P2X7 receptor | ||
Thunberg et al. Lancet . 2002 ;360 :1935 | 170 | 200 | 1 | 1 | P2X7 receptor | ||
Starczynski et al. Br J Haematol . 2003 ;123 :66 | 121 | 95 | 1 | 1 | P2X7 receptor | ||
Zhang et al. Leukemia . 2003 ; 2097 | 144 | 348 | 1 | 1 | P2X7 receptor | ||
Nückel et al. Eur J Haematol . 2004 ;72 :259 | 111 | 97 | 1 | 1 | P2X7 receptor | ||
Sellick et al. Cancer Epidemiol Biomarkers Prev . 2004 ;13 :1065 | 424 | 428 | 1 | 1 | P2X7 receptor | ||
Bogunia-Kubik et al. Int J Immunogeneti . 2005 ;33 :21 –24 | 61 | 180 | 1 | 1 | TNF-alpha -308 SNP | ||
Guzowski et al. J Biomolecular Techniques . 2005 ;16 :154 | 17 | 25 | 1 | 3 | IL10 promoter SNPs | ||
Pérez-Chacón et a. Am J Clin Pathol . 2005 ;132 :646 | 134 | 102 | 1 | 1 | CD5 microsattellite | ||
Zhang et al. J Natl Cancer Inst . 2005 ;97 :1616 | 461 | 59 | 535 | 1 | 1 | BCL6 | |
Jamroziak et al. Pharmacological Reports . 2006 ;58 :720 | 110 | 201 | 1 | 1 | MDR1 | ||
Morton et al. Pharmacogenet Genomics . 2006 ;16 :537 | 1172 | 146 | 982 | 2 | 10 | NAT1 and NAT2 | |
Lan et al. Carcinogenesis . 2007 ;28 :823 | 461 | 59 | 535 | 4 | 5 | Caspase genes | |
Nückel et al. Blood . 2007 ;109 :290 | 123 | 120 | 1 | 1 | BCL2 | ||
| Disease pathway | De Roos et al. Cancer Epidemiol Biomarkers Prev . 2006 ;15 :1647 | 1172 | 148 | 982 | 11 | 15 | Metabolic gene variants |
Hill et al. Blood . 2006 ;108 :3161 | 1172 | 148 | 982 | 19 | 34 | DNA repair and related genes | |
Shen et al. Hum Genet . 2006 ;119 :659 | 461 | 59 | 535 | 18 | 32 | DNA repair genes | |
Nieters et al. Genes Immun . 2006 ;7 :615 | 710 | 104 | 710 | 7 | 11 | Toll-like receptor, IL10, IL10RA | |
Wang et al. Hum Genet . 2006 ;120 :297 | 1172 | 148 | 982 | 7 | 12 | Cell cycle genes | |
Wang et al. Cancer Res . 2006 ;66 :9771 | 1172 | 148 | 982 | 36 | 57 | Proinflammatory and other immunoregulatory genes | |
Wang et al. Carcinogenesis . 2007 ;27 :1828 | 1172 | 148 | 982 | 10 | 13 | Oxidative stress pathway | |
Lan et al. Hum Genet . 2007 ;121 :161 | 461 | 59 | 535 | 10 | 14 | Oxidative stress pathway | |
Lim et al. Blood . 2007 ;109 :3050 | 1172 | 147 | 982 | 18 | 30 | Folate and 1 carbon metabolism | |
Cerhan et al. Blood . 2007 ; Sept 7 [Epub ahead of print] | 458 | 126 | 484 | 1450 | 7670 | Inflammation and immune genes | |
| Genomic studies | Rudd et al. Blood . 2006 ;108 :638 | 992 | 2707 | 865 | 1467 | Nonsynonymous SNPs | |
A multigenerational family with individuals affected with chronic lymphocytic leukemia (CLL), non-Hodgkin lymphoma (NHL), and monoclonal B-cell lymphocytosis (MBL). Age at diagnosis is shown.
A multigenerational family with individuals affected with chronic lymphocytic leukemia (CLL), non-Hodgkin lymphoma (NHL), and monoclonal B-cell lymphocytosis (MBL). Age at diagnosis is shown.
A multigenerational family with cases of chronic lymphocytic leukemia (CLL) in siblings and distant cousins. Age at diagnosis is shown.
A multigenerational family with cases of chronic lymphocytic leukemia (CLL) in siblings and distant cousins. Age at diagnosis is shown.
Acknowledgments
This material is based upon work supported by the Intramural Program of the National Cancer Institute, National Institutes of Health, Bethesda, Maryland, and by the grants CA118444 and CA097274 from the National Cancer Institute.
References
Author notes
Genetic Epidemiology Branch, Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, MD
Mayo Clinic College of Medicine, Mayo Clinic, Rochester, MN