Abstract
The role of molecular monitoring for patients with chronic myeloid leukemia (CML) is multifaceted. Milestone measurements up to 18 months of first-line imatinib therapy are prognostic and provide warning signals of suboptimal response. Serial measurements for patients with a complete cytogenetic response determine ongoing treatment efficacy or signal pending relapse. The pattern of molecular and cytogenetic response is generally comparable, but only cytogenetic analysis can monitor for the acquisition of clonal abnormalities and has an important role in case of loss of molecular response. For patients treated with imatinib, a rising level of BCR-ABL is a trigger for kinase domain mutation analysis. The characterization of BCR-ABL inhibitor-resistant mutations is important to direct therapeutic intervention because it is now apparent that each resistant mutation functions as a distinct protein with unique biological properties that may confer a gain or loss of function. The benefit to patients of regular molecular analysis is a reassurance of ongoing response using the most sensitive of techniques or a potential improvement in outcome for those where relapse is indicated early. However, despite the obvious benefits of molecular analysis, the measurement techniques may not be quite ready for acceptance into the routine clinical monitoring practices of all clinicians. The challenge now is to standardize and simplify the method so that it can be readily and reliably incorporated into routine laboratory testing procedures.
In the years since the IRIS trial (International Randomized Study of Interferon versus STI571) commenced in 2000, molecular monitoring of BCR-ABL transcripts using real-time quantitative polymerase chain reaction (RQ-PCR) has become an integral component for management of patients with chronic myeloid leukemia (CML) who were treated with imatinib.1 This is because the frequency of chronic-phase patients achieving a complete cytogenetic response (undetectable Philadelphia chromosome; CCR) is very high2 and, therefore, cytogenetic analysis provides limited information on the kinetics of residual disease. Conversely, for patients with a CCR, molecular analysis can indicate pending relapse by rising levels of BCR-ABL transcripts,3,4 and the degree of reduction of BCR-ABL correlates with progression-free survival.2 Indeed, failure to achieve a major molecular response (MMR) by 18 months of imatinib therapy is now considered suboptimal, and reassessment of therapy is recommended.1
Molecular analysis for the IRIS trial was not a comprehensive assessment since patients were only tested once a CCR was achieved. Responses considered as trial endpoints were the hematologic and cytogenetic responses. However, subsequent recognition of the importance of the molecular response to BCR-ABL inhibitor therapy is evidenced by the inclusion of molecular endpoints in current clinical trials of kinase inhibitors. Furthermore, BCR-ABL kinase domain mutation analysis is now included in the testing regimen, since such mutations are the major mechanism of imatinib resistance. Molecular endpoints have also been used to guide treatment algorithms in a clinical trial for patients with relapse following allogeneic transplantation.5 Clinical trials have served to demonstrate the value of molecular monitoring as performed in specialized laboratories. However, outside of the clinical trial setting clinicians need reassurance that individual patients can gain benefit from molecular monitoring when performed in their local laboratory. To this end, a concerted effort is under way towards method optimization, standardization, and an international reporting scale.6,7 To demonstrate how molecular analysis can impart patient-specific benefit, examples from our institution are described using peripheral blood (PB) RQ-PCR analysis, and direct sequencing of the BCR-ABL kinase domain.
Molecular Analysis Provides a Sensitive Indication of Response to BCR-ABL Kinase Inhibitor Therapy
Molecular analysis may be used for a patient’s first diagnosis of CML. The vast majority of patients have high levels of p210 BCR-ABL transcripts. If these are absent, it is necessary to search for atypical transcripts, since such rare patients may be precluded from analysis by the standard quantitative PCR assays. Otherwise, false-negative BCR-ABL values could be reported during therapeutic monitoring. For patients treated with imatinib upon diagnosis of CML, the achievement of an MMR is highly significant. An MMR is achieved when the BCR-ABL level falls approximately 1 log below the level at which a CCR is achieved. For the IRIS trial, this level was 3 log below a standardized baseline BCR-ABL level for untreated patients. For those with an MMR at 12 months, the probability of remaining free of progression to advanced phase at 5 years of imatinib treatment was 100%.2 The importance of achieving an MMR has been confirmed in other clinical studies. An MMR within the first year of imatinib treatment for chronic-phase patients, either without prior therapy or after interferon-αfailure, was predictive of a durable cytogenetic remission8 and was a marker of progression-free survival.9
A higher imatinib dose of 800 mg per day compared to the standard dose of 400 mg may allow molecular responses to be achieved earlier,10 but whether earlier achievement of these responses is important for progression-free survival remains largely unknown. The improved molecular response observed with a higher dose included a higher rate of complete molecular response (undetectable BCR-ABL).10 This may well prove to be significant. In a recent study of 59 chronic-phase patients with a CCR, there was no case of subsequent loss of CCR among the patients with undetectable BCR-ABL.11 Conversely, 30% of the patients with detectable BCR-ABL by RQ-PCR lost their CCR. We have also found that undetectable BCR-ABL using strict PCR sensitivity criteria is a durable response and the incidence of complete molecular response increased with prolonged imatinib therapy.12 While recognizing that undetectable BCR-ABL does not equate to eradication of leukemia, future clinical trials of BCR-ABL kinase inhibitor therapy may indeed include a complete molecular response as a clinical endpoint.
It has not been resolved whether patients with undetectable BCR-ABL will be able to cease imatinib treatment and maintain response. A recent report indicates that this may be the case for selected patients, at least in the short term. Rousselot et al reported that 6 of 12 patients who ceased imatinib after a period of at least 2 years of undetectable BCR-ABL relapsed within 6 months, while the remainder maintained response.13 Frequent, sensitive molecular monitoring may be critical for such patients so that treatment can be recommenced if BCR-ABL becomes detectable.
Molecular Response Corresponds to the Cytogenetic Response
The pattern of molecular and cytogenetic response is generally comparable, which raises the question of whether molecular analysis alone is sufficient to monitor patients responding to imatinib therapy. Only cytogenetic analysis can monitor for the acquisition of clonal abnormalities in bone marrow (BM), and such clonal evolution may be associated with disease progression. Furthermore, clonal abnormalities in Philadelphia-negative cells may infrequently be associated with myelodysplasia and acute leukemia.14 To address the benefit of simultaneous RQ-PCR and cytogenetic analysis, we studied 183 patients with chronic-phase CML and found limited value of cytogenetic analysis for patients with an MMR, which represented approximately half of the patients studied.15 Cytogenetic abnormalities were not detected in any of these patients and indicate that regular RQ-PCR monitoring alone may be sufficient in certain circumstances. A rational approach to spare most patients from regular BM aspirate is to perform cytogenetic analysis for those who have not achieved an MMR or who have lost an MMR.1
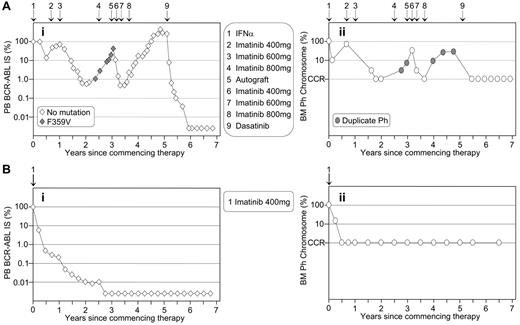
The similarity of the pattern of molecular and cytogenetic response is particularly striking in the patients illustrated in Figure 1 , which contrasts the disease course of two patients enrolled in the IRIS trial at our institution on the same day in July 2000. Patient A was randomized to interferon-α plus cytarabine but failed interferon-α and subsequent imatinib therapy. The patient relapsed after autologous transplantation but responded very well to dasatinib therapy. Molecular analysis demonstrated a significantly greater depth of response to dasatinib treatment than was experienced when the patient had achieved a CCR with imatinib therapy. Patient B was randomized to imatinib treatment and had a typical rapid and sustained response. Both patients demonstrate the superior sensitivity of molecular analysis for determining the depth of response when CCR is achieved. However, patient A highlights the benefit of cytogenetic analysis for detecting additional chromosomal abnormalities upon treatment failure.
Regular Molecular Monitoring Following Allogeneic Stem Cell Transplantation Can Identify Early Relapse
Serial molecular analysis following allogeneic transplantation remains a powerful tool for detecting early relapse in patients who maintain a cytogenetic response. The Hammersmith Hospital group defined molecular relapse based on the percentage ratio level of BCR-ABL/ABL and the frequency of detection of these levels.16 Positive values below a certain low level were not deemed to indicate molecular relapse. However, patients with persistently low levels of BCR-ABL had the highest risk of subsequent relapse, and these patients warrant close monitoring.16
In cases of relapse after allogeneic transplantation, imatinib has been used successfully to re-induce response.5,17 Suppression of the residual clone by imatinib may delay or avoid the requirement for donor lymphocyte infusion in patients at risk of significant graft-versus-host disease. It is yet to be determined whether imatinib will produce durable responses in patients with molecular relapse and whether discontinuation of imatinib is safe in patients with stable molecular response. Imatinib alone may not cure patients with relapse after transplantation since a higher relapse rate was reported for imatinib treatment compared with donor lymphocyte infusion in this situation.18
Towards Standardization of RQ-PCR Techniques
Outside of academic reference laboratories, patients may already be receiving reliable molecular monitoring. BCR-ABL measurements in one laboratory of experience will certainly be beneficial for patients and provide predictive information in the absence of international standardization. However, since the achievement of an MMR has prognostic significance and now guides therapeutic decisions, it is necessary that laboratories establish the level of BCR-ABL by their method that corresponds to an MMR as established in the IRIS trial. To facilitate this, an international reporting scale (IS) was proposed where the absolute BCR-ABL value representing MMR is standardized at 0.10%.6 A value of 1.0% is approximately equivalent to the achievement of a CCR. The establishment of an IS may facilitate more consistent adherence to treatment guidelines.
Standardization is usually achieved with the aid of certified reference material. Reference material is currently not available for BCR-ABL quantitation but may be a critical factor for generation of comparable quantitative data worldwide. The proposed IS for reporting BCR-ABL values aims to eliminate the terms “standardized baseline” and “log reduction” that currently define the molecular response. In their place is the more readily standardized BCR-ABL/control gene percentage ratio value. The scale is fixed to an MMR as defined by the laboratories that performed the molecular analysis in the IRIS trial.
Conversion to the IS is achieved by application of laboratory-specific conversion factors. In the absence of certified reference material, an alternative method to determine conversion factors is to calculate the bias between patient BCR-ABL values of a particular laboratory to those generated in an IRIS trial–participating laboratory with an established value for MMR (reference laboratory). If the comparison of values is consistent across the dynamic range, then the mean bias between the specific laboratory and the reference laboratory defines the conversion factor. To ensure ongoing validity of the conversion factors, it is essential that a quality assurance system is in place in each laboratory to maintain measurement reliability.7
The patient bias method of determining calculation factors is impractical in the long term due to the large number of laboratories requiring conversion factors and underscores the necessity for certified reference material. Such material is under assessment and development by an international collaborative group. However, the concept of reference material for BCR-ABL that closely mimics patient samples is difficult and must be carefully considered. Their development and certification is especially challenging because of the complexity of the unstable RNA matrix and the requirement that it contain both BCR-ABL and various control gene sequences (at physiologic levels), the required sensitivity, and the large dynamic range. Lyophilization of material may overcome the issue of stability.19
The future promises to simplify the standardization process by the launch of standardized kits into routine clinical use with high-quality performance characteristics, ideally ones that produce results on an international scale. The choices for minimal residual disease testing may be expanded with the use of novel cartridge-based microfluidic systems that incorporate RNA extraction, reverse transcription, and quantitative PCR.20
Detecting BCR-ABL Kinase Domain Mutations as They Emerge is Central to Guiding Clinical Management
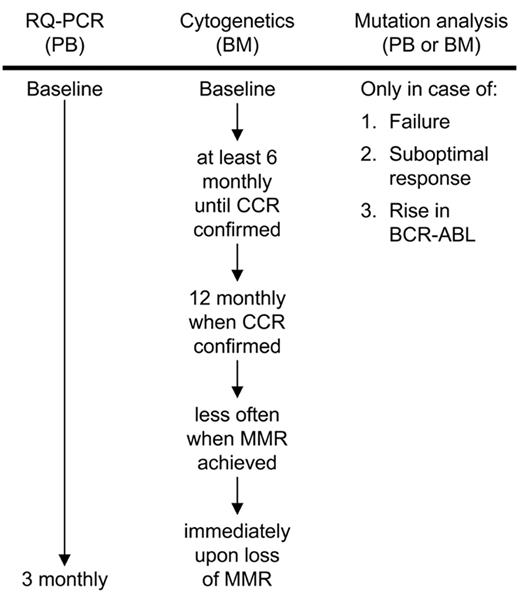
Recent recommendations by an expert panel suggest that the molecular status of a patient should not be limited to the measurement of BCR-ABL transcripts, but should also include kinase domain mutational analysis in case of imatinib failure, suboptimal response, and rising BCR-ABL levels.1 Since mutations are the major mechanism of imatinib resistance, it is highly recommended that molecular laboratories incorporate this analysis into their testing procedures. Figure 2 provides a molecular and cytogenetic workflow according to the recommendations of Baccarani et al.1
The number of reported mutations detected in patients treated with imatinib is still increasing, with approximately 90 now described at 57 residues. Most of these are listed in Melo and Chuah,21 and the relative frequency and imatinib insensitivity of some of the more common mutants are described in recent reviews.1,6 Most of the mutations are rare, with a frequency less than 1% to 2% of all patients with mutations, whereas fewer than 10 common mutants account for 60% to 85%.22–24 The regions of the kinase domain where mutations are detected most commonly are the P-loop and at residues T315 and M351. In a study by Jabbour et al of 171 patients with imatinib failure, mutations occurred more frequently in the later disease stages and were associated with older age, prior interferon therapy, development of clonal evolution, high-risk Sokal score at diagnosis, and failure to achieve a CCR with imatinib.22 In a comparable study by Soverini et al of 297 patients with imatinib failure, mutations were detected most frequently with acquired rather than primary resistance and in those who commenced imatinib when in advanced phase. Similar to the study of Jabbour et al, failure to achieve a CCR by 12 months of imatinib therapy was associated with the detection of mutations (37 of 134 patients, 28%).23
Among the most common imatinib-resistant mutants, those that occur at residues Y253, E255 and T315 confer high-level imatinib resistance, and the detection of these mutations indicates imatinib failure.1 Recommendation for patients with such mutations is allogeneic transplant, another kinase inhibitor, or investigational treatments.1 Mutations at residues M244, F317 and M351 confer low-level resistance and indicate suboptimal response. These mutations may respond to an increased imatinib dose. However, since the majority of mutations are rare, their insensitivity to imatinib is unknown. Appropriate therapeutic decisions for patients with such rare, uncharacterized mutations may be problematic.
Undoubtedly, detection of the T315I mutation influences therapeutic decisions since it is highly resistant to the BCR-ABL kinase inhibitors where amino acid 315 is critical for binding, such as imatinib, nilotinib and dasatinib. However, are the other mutants amenable to the more potent inhibitor therapy? Data suggest that most of those tested so far are sensitive to these inhibitors.25–27 Nevertheless, recent in vitro studies of the more potent inhibitors nilotinib and dasatinib have identified a limited spectrum of resistant mutations.28–31 Most of these resistant mutations are frequently detected in patients treated with imatinib, such as T315I, E255V/K and Y253H, detected in nilotinib resistance screens, and T315I and F317L detected in dasatinib resistance screens.
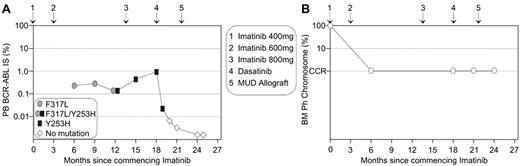
This new in vitro information may provide an aid for therapeutic decisions for patients who develop imatinib resistant mutations and a case in point from our institution is illustrated in Figure 3 . Upon diagnosis of chronic-phase CML, the 17-year-old male commenced imatinib on a fluctuating dose of 400 and 600 mg. A significant rise in BCR-ABL transcript level triggered mutation analysis and the highly imatinib resistant Y253H mutant was detected, which indicated imatinib failure.1 Interestingly, retrospective analysis identified the low-insensitivity mutant, F317L. It is predicted that CCR was maintained in this patient on a higher imatinib dose and BCR-ABL levels did not begin to rise until Y253H emerged. The type of mutations guided therapeutic options for this patient. In mutagenesis and sensitivity assays, Y253H was consistently identified as conferring moderate resistance to nilotinib.26,29–31 Potentially, the presence of the Y253H mutation could lead to a delayed response to nilotinib. F317L confers moderate resistance to dasatinib25,28 and was identified in resistance assays.28,29 Potentially, F317L could re-emerge on dasatinib therapy in our patient due to its reduced sensitivity compared to that of Y253H. Allogeneic transplantation is also an important option for patients who develop imatinib-resistant mutations.32 For our patient, a matched unrelated allogeneic transplantation was undertaken after a short course of dasatinib therapy. The patient maintained CCR throughout, and it was the molecular analyses that identified imatinib failure in a timely manner. This case highlights that patients in a CCR are still at risk of developing mutations.3,33
Recent clinical studies indicate that kinase domain mutations will also be highly relevant for acquired resistance to the second-generation inhibitors, albeit with a limited spectrum of mutations compared with those that arise during imatinib therapy. In two studies, mutations emerged in all 30 patients with acquired dasatinib resistance following imatinib failure.34,35 These mutations were not detectable prior to commencing dasatinib. As predicted, T315I was the most frequent mutant to emerge (18 of 30 patients, 60%) but T315A, V299L, and F317L/I were also detected. Each of these mutations were previously shown to confer dasatinib resistance in vitro28,29 and occur at dasatinib contact residues.36 T315A, V299L, and F317I are predicted to retain sensitivity to imatinib and potentially nilotinib.34 Interestingly, Shah et al demonstrated that the Aurora kinase inhibitor MK-0457, which is under clinical trial for its activity against T315I,37 was also effective against V299L.34 These data suggest a role for combination of ABL kinase inhibitors that retain sensitivity to a broad range of mutations. However, sequential kinase inhibitor therapy can select for cells harboring multiple mutations within the same molecule (compound mutations) that confer resistance to both inhibitors.34
Varying Transformation Potency of BCR-ABL Kinase Domain Mutations
An association between mutations in the P-loop of the BCR-ABL kinase domain and a poorer prognosis for patients who were treated with imatinib has been reported in some clinical studies,24,38,39 but not all.22 It was clear that additional evaluation of the biological activity of mutations within the P-loop was required to determine whether some of these mutants actually confer an enhanced transforming capacity. Several recent studies have now demonstrated that imatinib-resistant mutants can substantially alter kinase function and confer unanticipated biological properties that may impact disease progression.34,40,41
Griswold et al determined that some mutations may alter the transformation potency of BCR-ABL by altering substrate specificity and signal transduction pathway activation.40 The transformation potency of 5 common mutants was compared in various biological assays. The 2 P-loop mutations that were tested, E255K and Y253F, displayed increased transformation potency compared with wild-type and the other mutants, including T315I. Another common mutant, M351T, was the least transforming of the five mutants investigated.40 Shah et al demonstrated that two or more weaker mutants arising in the same BCR-ABL clone, compound mutations, could enhance oncogenic potency.34 Skaggs et al studied 8 mutations and found a range of transformation potencies that were greater or weaker than unmutated BCR-ABL.41 This group used a mass spectrometry–based phosphotyrosine profiling technique and confirmed that each mutation has a unique phosphoprotein signature, which effects substrate recognition. Consistent with the study of Griswold et al,40 M351T showed substantially weaker transforming activity. E255K and T315I showed an increase in potency compared with unmutated BCR-ABL.41 Similar to P-loop mutations, T315I has been reported to impair the clinical outcome of patients.23,24 Interestingly, Skaggs et al found that another common P-loop mutation, Y253H, conferred a substantially lower transformation potency relative to wild-type, which may be inconsistent with recent clinical findings.42
In light of these studies, one could speculate that some patients who acquire a mutation during imatinib treatment may indeed acquire a less-transforming form of BCR-ABL, for example, those patients who acquire the M351T mutation. Whether this translates into a prolonged survival with imatinib treatment would require analysis of a large cohort of patients with various mutations. Evidently, the clinical outcome upon acquisition of BCR-ABL mutations is a complex issue and may vary depending on the specific mutation, whether mutations are compound, and the disease phase. Phosphoproteome profiling and transformation assays for individual or compound mutations may prove useful tools to determine functional alterations. Searching for mutations that increase oncogenicity in selected patients by sensitive mutation detection techniques may allow early intervention before rapid expansion of the resistant clone.
Molecular Analysis as an Aid to Identify Imatinib Resistance Unrelated to the Development of BCR-ABL Mutations
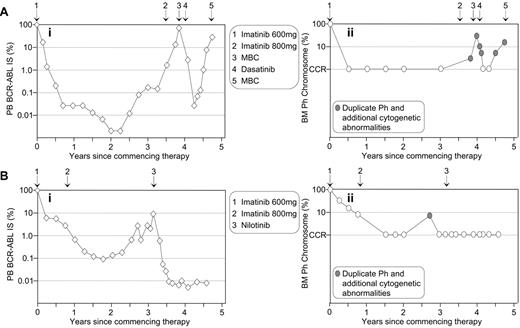
We previously reported a strong association between a significant rise in BCR-ABL levels and acquired imatinib resistance.3 However, resistance was not always associated with the detection of BCR-ABL mutations. For patients treated with BCR-ABL kinase inhibitor therapy, it is important to investigate the cause of rising BCR-ABL levels by both mutation and cytogenetic analyses. Cytogenetic abnormalities in the Ph-positive clone have been detected in 58% of imatinib-resistant patients.43 The detection of a second Ph chromosome indicates amplification of the BCR-ABL gene, which has been reported as a mechanism of imatinib resistance with an overall frequency of 18% (reviewed in Melo and Chuah21). Of course, rising BCR-ABL levels may be related to inconsistent therapy compliance, which should be considered.33Figure 4 describes 2 patients with rising BCR-ABL values while in a CCR. In both cases, mutations were not detectable. Cytogenetic analysis at the time of loss of CCR indicated that both patients had duplication of the Ph chromosome and additional chromosomal abnormalities, which indicated imatinib failure or suboptimal response.1
Conclusion
In the era of highly successful BCR-ABL kinase inhibitor therapy for patients with CML, molecular monitoring is essential to establish response and to guide mutation and cytogenetic analysis for the investigation of resistance. The challenge in the next few years is to enable more widespread adoption of molecular techniques. The generation of comparable molecular results when tested in laboratories anywhere in the world will be facilitated through the standardization of assays, the introduction of certified reference material, and an international reporting scale. It is envisioned that collaboration between clinical laboratories and commercial entities will ultimately see this become a reality. Characterization of the cause of loss of response to BCR-ABL inhibitors is essential to guide therapeutic intervention decisions, since some BCR-ABL kinase domain mutations may not respond optimally to alternate inhibitor therapy, and some create more biologically aggressive forms of BCR-ABL.
The pattern of molecular response is mirrored by the corresponding cytogenetic response. (A) The patient crossed over to imatinib after interferon-α failure and a rise in BCR-ABL (Ai) was associated with the detection of a consistently minor mutated clone. Corresponding bone marrow (BM) cytogenetic analysis (Aii) revealed loss of complete cytogenetic response (CCR) and a duplicate Ph. Interestingly, after an autologous transplantation the mutation was not detected. However, the duplicate Ph was present at loss of response, which possibly indicates that the minor mutated clone was eradicated at transplantation. This case illustrates the importance of cytogenetic analysis after loss of molecular response. (B) The patient had a typical rapid and sustained response to first-line imatinib. Molecular analysis (Bi) showed that BCR-ABL remained detectable for more than 2 years after CCR was documented at 6 months (Bii). This case illustrates that cytogenetic analysis may be of no clinical benefit while the patient maintains molecular response.
The pattern of molecular response is mirrored by the corresponding cytogenetic response. (A) The patient crossed over to imatinib after interferon-α failure and a rise in BCR-ABL (Ai) was associated with the detection of a consistently minor mutated clone. Corresponding bone marrow (BM) cytogenetic analysis (Aii) revealed loss of complete cytogenetic response (CCR) and a duplicate Ph. Interestingly, after an autologous transplantation the mutation was not detected. However, the duplicate Ph was present at loss of response, which possibly indicates that the minor mutated clone was eradicated at transplantation. This case illustrates the importance of cytogenetic analysis after loss of molecular response. (B) The patient had a typical rapid and sustained response to first-line imatinib. Molecular analysis (Bi) showed that BCR-ABL remained detectable for more than 2 years after CCR was documented at 6 months (Bii). This case illustrates that cytogenetic analysis may be of no clinical benefit while the patient maintains molecular response.
Molecular and cytogenetic workflow for imatinib-treated patients according to the recommendations of an expert panel. Mutation analysis is recommended in case of failure to achieve a complete hematologic response after 3 months of imatinib therapy, a major cytogenetic response (≤35% Ph chromosome) after 6 months, a complete cytogenetic response (CCR) after 12 months and a major molecular response (MMR) after 18 months. Loss of these responses, progression to accelerated phase or blast crisis or a significant rise in BCR-ABL according to the measurement reliability of the RQ-PCR assay should also initiate mutation analysis.1
Molecular and cytogenetic workflow for imatinib-treated patients according to the recommendations of an expert panel. Mutation analysis is recommended in case of failure to achieve a complete hematologic response after 3 months of imatinib therapy, a major cytogenetic response (≤35% Ph chromosome) after 6 months, a complete cytogenetic response (CCR) after 12 months and a major molecular response (MMR) after 18 months. Loss of these responses, progression to accelerated phase or blast crisis or a significant rise in BCR-ABL according to the measurement reliability of the RQ-PCR assay should also initiate mutation analysis.1
Molecular analysis and the characterization of imatinib-resistant mutations can aid timely therapeutic decisions.
(A) A 3-fold rise in BCR-ABL prompted mutation analysis. Imatinib failure was confirmed by the detection of the highly imatinib-resistant Y253H mutation, which became detectable after F317L. Y253H is one of the least sensitive mutations to nilotinib and F317L has been identified in dasatinib resistance screens. Dasatinib was commenced prior to matched unrelated donor (MUD) transplantation to maintain response. (B) The corresponding bone marrow (BM) cytogenetic analyses indicated maintenance of complete cytogenetic response (CCR), and these analyses did not contribute to early warning of imatinib failure that was evident by molecular analysis.
Molecular analysis and the characterization of imatinib-resistant mutations can aid timely therapeutic decisions.
(A) A 3-fold rise in BCR-ABL prompted mutation analysis. Imatinib failure was confirmed by the detection of the highly imatinib-resistant Y253H mutation, which became detectable after F317L. Y253H is one of the least sensitive mutations to nilotinib and F317L has been identified in dasatinib resistance screens. Dasatinib was commenced prior to matched unrelated donor (MUD) transplantation to maintain response. (B) The corresponding bone marrow (BM) cytogenetic analyses indicated maintenance of complete cytogenetic response (CCR), and these analyses did not contribute to early warning of imatinib failure that was evident by molecular analysis.
Molecular analysis as an aid to identify imatinib resistance unrelated to BCR-ABL mutations. Rising BCR-ABL levels of two patients (Ai and Bi) indicated potential imatinib resistance. No BCR-ABL mutations were detected.
Cytogenetic analysis indicated duplication of the Ph chromosome and the presence of additional cytogenetic abnormalities at loss of CCR for both patients (Aii and Bii). (A) The 57-year-old female was diagnosed in accelerated phase CML and commenced imatinib at 600 mg. An increased imatinib dose of 800 mg upon the rise in BCR-ABL failed to prevent progression to myeloid blast crisis (MBC). Dasatinib therapy reestablished CCR, but the patient relapsed. (B) The 42-year-old male commenced 600 mg imatinib upon diagnosis of chronic-phase CML. CCR was maintained after commencing nilotinib therapy.
Molecular analysis as an aid to identify imatinib resistance unrelated to BCR-ABL mutations. Rising BCR-ABL levels of two patients (Ai and Bi) indicated potential imatinib resistance. No BCR-ABL mutations were detected.
Cytogenetic analysis indicated duplication of the Ph chromosome and the presence of additional cytogenetic abnormalities at loss of CCR for both patients (Aii and Bii). (A) The 57-year-old female was diagnosed in accelerated phase CML and commenced imatinib at 600 mg. An increased imatinib dose of 800 mg upon the rise in BCR-ABL failed to prevent progression to myeloid blast crisis (MBC). Dasatinib therapy reestablished CCR, but the patient relapsed. (B) The 42-year-old male commenced 600 mg imatinib upon diagnosis of chronic-phase CML. CCR was maintained after commencing nilotinib therapy.
Division of Molecular Pathology, Institute of Medical and Veterinary Science, South Australia, Australia