Abstract
The identification of molecular genetic alterations such as gene mutations or deregulated gene expression in acute myeloid leukemia (AML) has greatly advanced our understanding of leukemogenesis. These markers now allow us to unravel the enormous heterogeneity seen within cytogenetically defined subgroups of AML. Furthermore, the molecular alterations are providing targets for molecular therapies. In this article, major molecular findings of prognostic and predictive significance are reviewed, with an emphasis on the discussion of gene mutations found in two major AML subgroups, cytogenetically normal and core-binding factor AML.
Acute myeloid leukemia (AML) is a genetically heterogeneous disease with accumulation of acquired genetic alterations in hematopoietic progenitor cells that disturb normal mechanisms of cell growth, proliferation and differentiation.1 Clonal chromosome alterations are detected in approximately 55% of adults with AML, and presenting cytogenetic alterations have long been recognized as the strongest prognostic factor for response to therapy and survival. Based on these findings, patients with AML are currently categorized into three broad risk groups: favorable, intermediate, or adverse.2,3 In recent years, molecular genetic abnormalities such as gene mutations or deregulated gene expression have been identified, unravelling the enormous heterogeneity among such cytogenetically defined subgroups.
There are several lines of evidence for a multistep pathogenesis of AML in most cases.1,4 First, data from murine models have shown that one mutation is not sufficient to cause overt leukemia. As an example, RUNX1-RUNX1T1 or CBFB-MYH11 fusion genes, resulting from t(8;21) and inv(16)t(16;16), respectively, are able to impair myeloid differentiation but do not lead to a leukemic phenotype. Second, there are rare inherited gene mutations predisposing to the development of leukemia, e.g., heterozygous loss-of-function mutations in the transcription factor RUNX1 associated with the familial platelet disorder with propensity to AML, or mutations in the transcription factor CCAAT/enhancer-binding protein alpha (CEBPA) that have been reported in rare cases of familial AML. Third, in many sporadic AML more than one mutation can be detected. On the other hand, for leukemias involving the MLL gene there is evidence that these leukemias may be the result of a single genetic hit. Barabe et al recently showed that the sole expression of MLL-AF9 or MLL-ENL in lineage-depleted human cord blood cells induces leukemias in NOD-SCID recipients.5
Genetic alterations appear to fall into two broadly defined complementation groups (Figure 1; see Color Figures pg 519).4 One group (class I) comprises mutations that activate signal transduction pathways, resulting in enhanced proliferation and/or survival of leukemic progenitor cells. Mutations leading to activation of the receptor tyrosine kinase FLT3 or the RAS signaling pathway would be considered class I mutations. Another complementation group (class II) comprises mutations that affect transcription factors or components of the transcriptional co-activation complex, resulting in impaired differentiation. Recurring gene fusions resulting from t(8;21), inv(16)/t(16;16), t(15;17), or mutations in CEBPA, MLL, and possibly also NPM1 would fall into this group. These different types of mutations cooperate in leukemogenesis. This is supported by murine models and by the distribution of mutations in human disease.
Cytogenetically Normal AML
In recent years, a number of somatically acquired mutational changes have been identified in cytogenetically normal (CN) patients with AML, including partial tandem duplications (PTD) of the MLL gene, internal tandem duplications (ITD) or tyrosine kinase domain (TKD) mutations of the FLT3 gene, and mutations in the NPM1, CEBPA, NRAS and WT1 genes (Table 1 ).6,7 In part, these gene mutations have emerged as important prognostic markers and they now allow us to dissect CN-AML in prognostic subgroups. Furthermore, these mutant molecules may eventually become targets for rational molecular therapies.
Mutations in Nucleophosmin (NPM1)
An important discovery made by the group of Falini et al in 2005 was that of abnormal cytoplasmic localization of NPM1 that is caused by mutations in exon 12 of the gene.8 Cytoplasmic accumulation of NPM1 mutants is caused by two major alterations acting in concert: loss of tryptophan residues normally required for NPM1 binding to the nucleoli, and generation of an additional nuclear export signal motif at the C-terminus by the exon 12 mutation.9 Moreover, NPM1 leukemic mutants were shown to recruit wild-type NPM1 from nucleoli to nucleoplasm and cytoplasm through dimerization. NPM1 mutations can be identified in 45% to 62% of patients with CN-AML and thus are the most frequent genetic change in this patient subset.8,10–15 They appear to be less frequent in pediatric AML.16,17
NPM1 is an abundant and highly conserved phospho-protein that physiologically resides in nucleoli and shuttles between nucleus and cytoplasm. NPM1 has a role in various cellular processes, including ribosome biogenesis, response to stress stimuli such as UV irradiation and hypoxia, maintenance of genomic stability, regulation of activity and stability of tumor suppressor genes such as p53 and ARF, and transcriptional regulation.18 These activities are related to proliferation and growth suppression pathways, but also to cellular differentiation. Recently, NPM1 was shown to act as a corepressor in retinoic acid (RA)–associated transcriptional regulation.19 This study showed that during RA-induced cellular differentiation, activating protein transcription factor 2 α (AP2α) recruits NPM1 to the promoter of certain RA-responsive genes. NPM1 seems to exert its repressive effect by inducing changes in chromatin structure that involve histone deacetylases. An interesting hypothesis is that through cytoplasmic dislocation, the NPM1 protein pool is depleted from the AP2α-mediated transcriptional network, leading to de-repression of RA target genes. Thus, pharmacologic modulation of the RA-signaling pathway might be one option for a targeted therapeutic approach.
NPM1 mutations are associated with several pretreatment characteristics of patients with CN-AML, including predominance of female sex, higher bone marrow blast percentages, lactate dehydrogenase levels, white blood cell (WBC) and platelet counts, as well as high CD33-antigen expression but low or absent CD34-antigen expression.8,10–15 In univariate analysis, data on the prognostic impact of NPM1 mutations have been somewhat controversial.8,10–15 Whereas in some studies there was a significant effect on complete remission (CR) rate, relapse-free survival (RFS), and event-free survival (EFS), other studies revealed no significant differences in these parameters.
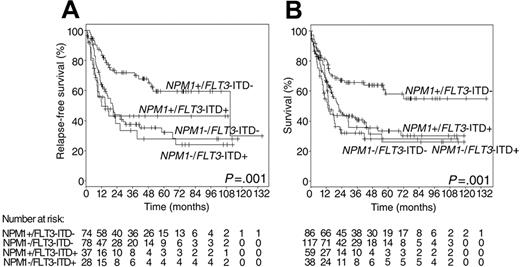
Approximately 40% of patients having NPM1 mutations also carry FLT-ITD.8,10,11,12,15 Of note, four large studies of more than 250 patients each independently showed that NPM1 mutation is a favorable prognostic marker, but only in the absence of FLT3-ITD.10–12,15 This could be shown for CR achievement, EFS, RFS, disease-free survival (DFS) and overall survival (OS) (Figure 2 ). NPM1 mutation had no influence on the adverse prognosis in patients with FLT3-ITD.
Approximately 11% to 15% of NPM1 mutations occur outside the group of CN-AML, in combination with various recurring cytogenetic abnormalities.11,13,15 These findings raise the question whether NPM1mut AML defines a distinct biological and clinical entity or not. Nonetheless, once a targeted therapy becomes available, one could speculate that all NPM1mut AML, and not just NPM1mut CN-AML, would benefit from such a therapy.
Mutations in FLT3
The receptor tyrosine kinase FLT3 and its ligand are important for the proliferation and differentiation of early hematopoietic progenitor cells. In patients with AML, somatic mutations that result in the constitutive activation of FLT3 have been identified in two functional domains of the receptor, the juxtamembrane (JM) domain and the split TKD.20,21 The JM domain that has been shown to be crucial for kinase autoinhibition is disrupted by ITDs of various size and insertion sites in 28% to 34% of CN-AML,22–25 whereas JM domain point mutations are rare. FLT3-ITDs result in ligand-independent dimerization and tyrosine autophosphorylation as well as activation of the RAS/MAPK, STAT5 and PI3K/AKT pathways.26
The activation loop (AL) in the carboxy-terminal lobe of the TKD is affected by point mutations, small insertions, or deletions mainly involving codon 835 and 836 in 11% to 14% of CN-AML.24,25,30 Rare point mutations or insertions have been reported at other codons in the TKD. In vitro studies and results from global gene expression profiling revealed that there are similarities but also important differences in signal transduction properties between FLT3-ITDs and FLT3 TKD mutations that may explain differences in clinical phenotypes. Similarily, in a mouse model transplanting retroviral-transduced hematopoietic progenitor cells, the phenotype differed significantly according to the transplanted mutation type.27
From a clinical perspective, FLT3 mutations are relevant because of their prognostic impact and because constitutively active FLT3 is an attractive target for molecular therapy. Numerous studies have shown that patients with CN-AML harboring FLT3-ITD have a significantly inferior outcome compared to patients without FLT3-ITD.6,22–25 This has been shown for EFS, RFS and OS. More recent data provide evidence that outcome is related more to the level of mutant allele and not just to its mere presence.22,23,25 First evidence came from a study by Whitman et al showing that only patients having lost wild-type FLT3 have a dismal outcome and not those with heterozygous mutations.23 Recent data from single nucleotide polymorphism (SNP) chip analysis indicate that loss of the wild-type allele results from mitotic recombination leading to partial uniparental disomy (UPD).28 Thiede et al were able to identify a threshold that distinguished prognostic subgroups by using Genescan analysis to quantify the level of mutant allele.25
The prognostic relevance of FLT3 AL mutations has been discussed controversially. A previous meta-analysis on 1160 cases (FLT3wt, n = 833; FLT3-ITDpos, n = 243; FLT3-TKDpos, n = 84) including all AML subsets showed a negative prognostic impact.29 No subset analysis for patients with CN-AML was performed. In contrast, a recent study by the British Medical Research Council (MRC) suggests a positive impact on survival.30 In this study, 1107 unselected young adults treated within the MRC AML10 and AML12 trials were screened for FLT3 TKD mutations using a denaturing high-performance liquid chromatography (DHPLC)–based assay. Of 1107 patients with AML, 127 (11%) had FLT3 TKD mutations; 51 of 437 patients with CN-AML (12%) had FLT3 TKD mutations. Quantification of the relative mutant level was performed, and patients were classified into TKD high-level and low-level groups. Patients with levels greater than the median level of 25% of total FLT3 alleles had a significantly superior OS compared with patients with low levels of mutant alleles. In the subset of CN-AML, this effect was of borderline significance. These data need to be interpreted with caution, since the effect of other gene mutations (e.g., NPM1) was not taken into consideration in multivariate analysis.
Currently, there are a number of FLT3 inhibitors at various stages of clinical development, such as PKC412 (midostaurin), CEP-701 (lestaurtinib), or MLN518 (tandutinib). When used as single agents, these compounds have limited activity in AML with mutant FLT3. These data look more promising when combining these inhibitors with conventional chemotherapy. Results from phase 3 clinical trials that are about to start will tell us whether inhibition of mutant FLT3 will be a successful targeted approach for these patients.
Mutations in CEBPA
The transcription factor CEBPA is a key molecule in the mediation of lineage specification and differentiation of multipotent myeloid progenitors into mature neutrophils.31 Two major types of heterozygous CEBPA mutations have been identified in sporadic and familial AML. Nonsense mutations affecting the N-terminal region of the molecule prevent expression of the full-length CEBPA protein, thereby upregulating the formation of a truncated isoform with dominant-negative properties and in-frame mutations in the C-terminal basic region-leucine zipper domain, resulting in CEBPA proteins with decreased DNA-binding or dimerization activity. N- and C-terminal mutations often occur simultaneously. Four current studies could show that CEBPA mutations are associated with a favorable outcome.3,6
Mutations in MLL, RAS, WT1
Partial tandem duplication (PTD) of the MLL gene was the first gene mutation shown to affect prognosis in patients with CN-AML.3,6 The incidence in CN-AML ranges from 5% to 11%. MLL-PTD most commonly involves duplication of a genomic region spanning exons 5 through 11 and insertion of the duplicated segment into intron 4 of the gene; in a few cases the duplication encompasses exons 5 through 12. Unlike MLL chimeric fusion proteins, MLL-PTD retains all functional protein domains. The presence of MLL-PTD is concurrent with silencing of the MLL wild-type allele, possibly through epigenetic mechanisms.
MLL-PTD has been associated with shorter CR duration or worse RFS and EFS; however, in these studies, MLL-PTD had no effect on OS. Cancer and Leukemia Group B (CALGB) recently reported on the impact of MLL-PTD in 238 young adult patients who received autologous blood stem cell transplantation (SCT) in first CR.32 Neither DFS nor OS differed between the groups with or without MLL-PTD, suggesting that intensive consolidation therapy using autologous SCT may improve outcome in this subgroup of patients. Whitman et al have shown that the combination of decitabine, a DNA methyltransferase inhibitor, and depsipeptide, a histone deacetylase inhibitor, can reactivate the transcription of the wild-type allele in MLL-PTD+ blasts.33 The induction of MLL wild-type expression was associated with enhanced cell death of the blasts. These data may provide a rationale to evaluate DNA methyltransferase and/or histone decetylase inhibitors in patients with MLL-PTD.
Two recent large studies detected NRAS mutations in 9% of adult patients with CN-AML and 14% of patients with CN-AML who were younger than 56 to 60 years.3,6 Consistent with previous reports, there was no prognostic impact of NRAS mutations in either CN-AML patients or in those with intermediate-risk cytogenetics. Although NRAS mutations have not been shown to be a prognostic factor, these mutations may still provide a target for molecular therapy.
Mutations in Wilms’ Tumor 1 gene (WT1) in AML were first reported by King-Underwood and Pritchard-Jones in 1998.7 It has been shown that WT1 directs both quiescence in hematopoietic progenitor cells and differentiation of myelomonocytic cells. Therefore, disruption of WT1 function by mutation of the gene could either promote proliferation or induce a block in differentiation. In a recent study of 70 patients with CN-AML by Summers et al, WT1 mutations were detected in 10% of the patients.34 Mutations consisted of insertions or deletions that mainly clustered in exons 7 and 9. The two published small studies on heterogeneous patient population suggest that WT1 mutations may be associated with induction failure; however, the impact of WT1 mutations needs to be evaluated in larger series.
The prevalence and clinical relevance, if any, of other gene mutations, such as inactivating mutations of the ubiquitin ligase CBL, an important negative regulator of receptor tyrosine kinase signaling, remain to be determined.35,36
The meaningfulness of previous gene mutation studies in AML is limited by the fact that mostly single genetic markers have been analyzed, by small patient numbers, or by lack of homogeneity in treatment. The German-Austrian AML Study Group (AMLSG) recently presented a large individual patient data meta-analysis on 872 younger adults with CN-AML.37 Patients had received intensive, state-of-the-art induction and postremission therapy, including a genetic randomization for allogeneic SCT from a matched related donor (MRD). Leukemia cells were analyzed for mutations in NPM1, FLT3, CEBPA, MLL, and NRAS. Significant factors for induction success were the genotypes NPM1mut/FLT3-ITDneg and CEBPAmut, de novo AML, lower WBC count, and younger age. Cox proportional hazard models on RFS revealed the genotypes NPM1mut/FLT3-ITDneg and CEBPAmut as well as the availability of a MRD as favorable prognostic markers. Subgroup analyses showed that patients with the favorable genotypes did not benefit from allogeneic SCT in terms of RFS or OS. The 4-year OS probabilities for patients with the genotypes NPM1mut/FLT3-ITDneg and CEBPAmut were 60% and 62%, respectively. These outcome data are comparable with those of patients with core-binding factor (CBF) leukemias.
Deregulated Gene Expression
Overexpression of a number of genes, such as BAALC, ERG, MN1 or EVI1, has been shown to be of prognostic significance.6 The mechanisms by which these genes become deregulated remain elusive. Translocations, inversions or gene amplifications are found in AML involving EVI1, ERG and MN1, for example, but in general overexpression occurs in the absence of such rearrangements. No activating mutations have so far been identified in these genes.
BAALC is expressed primarily in neuroectoderm-derived tissues and hematopoietic progenitors but not in more mature hematopoietic cells, and encodes a protein with no homology to any known proteins or functional domains. In the initial study that was performed on younger adults with CN-AML, high BAALC expression in blood was associated with inferior EFS, DFS, and OS in patients with wild-type FLT3 or with FLT3-ITD (retaining the wild-type FLT3 allele).38
ERG is a member of the ETS family of transcription factors that are involved in regulation of cell proliferation, differentiation, and apoptosis. Overexpression was initially found in AML with complex karyotypes harboring cryptic amplification of chromosome 21.39 In a recent study on younger adults with CN-AML, ERG overexpression in pre-treatment blood was validated as an adverse predictor in CN-AML.40 Interestingly, within the subset of NPM1mut/ FLT3-ITDneg patients, high ERG predicted shorter EFS.
The meningioma 1 (MN1) gene has been found rearranged in a t(12;22)(p13;q11–12), a balanced translocation in AML leading to a fusion of MN1 with ETV6. Using global gene expression profiling, MN1 was identified as part of a signature associated with induction failure in patients with AML. In a subsequent study on younger adults with CN-AML, high MN1 expression was associated with a higher relapse rate and inferior RFS and OS.41
Finally, EVI1 overexpression has been shown to be associated with inferior outcome in patients with intermediate-risk cytogenetics.42 All of these studies will need independent corroboration. Furthermore, in order to demonstrate that these single-gene expression markers provide independent information, they will have to be studied in the context of the powerful gene mutation markers such as NPM1, FLT3, and CEBPA.
Core-Binding Factor AML
Cytogenetically, the group of CBF AML is defined by the presence of the t(8;21)(q22;q22) or the inv(16)(p13q22)/ t(16;16)(p13;q22). Both subgroups are considered favorable-risk diseases based on high CR rates and high survival probabilities.2,3 Nevertheless, about 40% to 50% of patients with CBF leukemias relapse. Currently, research in CBF leukemias focuses on the identification of additional genetic markers underlying the clinical heterogeneity. Candidates include secondary cytogenetic abnormalities, for example, trisomy 22 in patients with inv(16)/t(16;16); various gene mutations (see below); or the relative amount of an alternatively spliced isoform of the RUNX1-RUNX1T1 fusion transcript in patients with t(8;21).43
The study of secondary gene mutations in CBF leukemias has been of much interest. Mutations have been identified in the genes encoding KIT, FLT3, NRAS, KRAS, and JAK2. KIT exon 8 mutations located in the extracellular portion of the receptor or mutations at codon 816 in the AL are detected in approximately 25% of CBF AML.44,45 Such mutations occur rarely outside the group of CBF AML. Extracellular KIT mutations have been shown in vitro to result in spontaneous receptor dimerization and activation, leading to activation of downstream effectors such as MAPK and PI3K in response to SCF stimulation. Mutations at codon 816 induce constitutive activation of STAT3 and upregulation of downstream targets such as BCL-XL and MYC, as well as activation of the PI3K/AKT pathway.
A number of studies evaluated the prognostic significance of KIT mutations in CBF leukemias.44–46 In t(8;21) AML KIT mutations, and in some studies specifically mutations at codon 816 in exon 17, have been associated with inferior EFS, RFS, cumulative incidence of relapse (CIR), and OS. In contrast, in inv(16)/t(16;16), two smaller studies did not detect any difference in outcome measure, whereas in the study by Care et al KIT mutations were associated with a higher relapse rate but not with inferior survival.46 In the study by CALGB, KIT mutations were associated with a higher CIR; this difference was mainly due to effect of KIT exon 17 mutations.45 In multivariable analysis, KIT mutation was an adverse prognostic factor for OS. All these results await further independent confirmation in large patient cohorts that have received uniform treatment.
Irrespective of the prognostic significance, mutant KIT alleles are potential targets for molecular therapies. Imatinib is active against exon 17 mutations involving N822 and exon 8 mutations, but not against the D816 mutations in exon 17. The latter mutations have been shown to be sensitive to inhibition, for example, by the staurosporine-derived tyrosine kinase inhibitor PKC412 or by the dual SRC/ABL kinase inhibitor dasatinib. The efficacy of TK inhibitors combined with conventional chemotherapy needs to be tested in future clinical trials of CBF AML.
Identification of Novel Genetic Changes by Using Modern Genomics Techniques
Recent progress in genomics technology has resulted in the identification of several novel genetic abnormalities and holds the promise of making the systematic characterization of leukemia genomes feasible.
For instance, the introduction of genome-wide SNP-based mapping arrays, providing both copy number and allele specific information, led to the identification of a novel mechanism of leukemogenesis involved in AML (i.e. partial UPD). UPD is caused by non-disjunction or by homologous mitotic recombination leading to partial duplication of the maternal or paternal chromosome. By using low resolution 10k SNP-mapping arrays, Raghavan et al detected UPDs in 20% of cytogenetically unselected AMLs.47 The distribution of genomic regions affected was non-random, and some of the regions encompassed genes known to be mutated in AML. Fitzgibbon et al reported on the association between mutant alleles and UPD for the following marker constellations: FLT3-ITD/UPD13q, CEBPAmut/UPD19q, WT1mut/UPD11p, and RUNX1mut/ UPD21q.28 In addition to the detection of UPDs, array-based SNP analysis allows the identification of submicroscopic copy-number alterations. Using the low-resolution Affymetrix 10k mapping array (with a mean intermarker distance of 258 kb), Gorletta et al detected submicroscopic deletions in 10% of CN-AML, mapping to 9q34.1, 12p21.3–13.2, 15q14–15.3, 16q24, 21q21.3, and 21q22.1–q22.2.48 Schön et al analyzed paired samples from 56 patients with de novo CN-AML using a 100k mapping array.49 In their study, 5 submicroscopic deletions were identified mapping to 3p14.1 (n = 3), 12p13, and 12q23. It can be anticipated that by using even higher resolution mapping arrays, a number of novel changes will be identified. The power of such a high-resolution, genome-wide approach has been impressively shown in the study of 242 pediatric patients with ALL, which revealed previously unknown deletions, amplification, point mutation and structural rearrangements in genes encoding principal regulators of B lymphocyte development and differentiation in 40% of B-progenitor ALL patients.50
Another technique that has proven useful for the identification of genetic abnormalities of pathogenetic or clinical relevance is array-based comparative genomic hybridization. For example, the recent discovery that the homeo-box transcription factor CDX2 is aberrantly expressed in most patients with AML was guided by the identification of genomic amplification of the CDX2 locus in a subset of patients with complex cytogenetic abnormalities.51,52 Similarly, increased expression of the ETS transcription factor ERG, which is associated with inferior clinical outcome in CN-AML as well as in T-cell acute lymphoblastic leukemia, was first detected in AML with complex karyotype and partial amplification of chromosome 21.39
While analyses of genomic copy number using these high-density approaches will continue to be informative with regard to selection of candidate leukemia genes, it is also hoped that high-throughput DNA sequence analysis in large numbers of primary patient samples will become possible at an affordable cost, which may ultimately result in the development of comprehensive disease- and allele-specific oncogene mutation profiling strategies. Finally, innovative functional genetic approaches, such as large-scale RNA interference screens, have great potential for the identification of novel cancer genes.
Conclusion and Perspectives
For a long time, pretreatment karyotype has been one of the most important prognostic factors for response to therapy and for survival. In recent years, various molecular markers have been identified that now allow us to dissect cytogenetically defined patient subsets. A number of problems are inherent to many of these studies. They relate to the fact that in most cases only one or two genetic markers have been studied, not taking into consideration important biological interactions between markers. Additional limitations relate to often low numbers of patients studied, or to non-homogeneities in treatment. Therefore, many of the reported findings need confirmation by larger studies evaluating as many markers as possible. Since most of the AML subsets occur at only low frequency, intergroup collaboration will be necessary to answer relevant questions.
Notwithstanding, a few markers are about to enter clinical practice, in particular in the context of clinical trials. For instance, KIT mutations in CBF leukemias as well as activating FLT3 mutations, in particular in CN-AML, have prognostic significance and, importantly, are attractive targets for molecularly targeted therapy. The first randomized studies using TK inhibitors in these patient subsets have started or will soon start. Furthermore, two genotypes, NPM1mut/FLT3-ITDneg and CEBPAmut, have been identified that are associated with a favorable risk profile, comparable with that of CBF AML. Patients whose AML harbor such genotypes may not benefit from allogeneic SCT in first-line treatment. These data have important implications since the two genotypes represent approximately 45% of patients with CN-AML and 20% of all patients with AML. As a consequence, NPM1, FLT3 and CEBPA mutational screening should become part of the initial work-up of a newly diagnosed AML.
We are only beginning to unravel the biological heterogeneity of AML. By using novel genomics technologies such as high-resolution SNP-mapping arrays or high-throughput sequencing of coding sequences, novel changes will likely be identified that will contribute to a better understanding of the disease biology. This information eventually will lead to a refined disease classification and to the development of rational therapies.
Gene mutations predominantly occurring in cytogenetically normal acute myeloid leukemia (AML).
| Gene . | Biological/clinical features . | Selected references . |
|---|---|---|
| Abbreviations: BM, bone marrow; LDH, lactate dehydrogenase; CN, cytogenetically normal; ITD, internal tandem duplication; TKD, tyrosine kinase domain; MRD, matched related donor; CR, complete remission; JM, juxtamembrane domain; UPD, uniparenteral disomy; OS, overall survival; RFS, relapse-free survival; EFS, event-free survival; PTD, partial tandem duplication. | ||
| NPM1 | Protein with pleiotropic functions | 18 |
| Associated with presenting clinical and laboratory features such as female sex, higher BM blast counts and LDH levels, as well as high CD33 but low or absent CD34 levels | 6,10–15 | |
| Found in 25% to 35% of AML; predominantly in CN-AML (45% to 62%) Associated with FLT3-ITD and TKD mutations | 6,11,13,15 | |
| NPM1mut/FLT3-ITDneg genotype associated with a favorable prognosis | 10–12,15 | |
| Patients with NPM1mut/FLT3-ITDneg genotype may not benefit from MRD allogeneic transplantation in first CR | 10,37 | |
| FLT3 | Member of the class III receptor tyrosine kinase family Constitutively active FLT3 molecules are targets for molecular therapy | 26 |
| ITD | Found in 28% to 34% of CN-AML In-frame mutations, mostly in exons 14 and 15 of the JM domain | 3,6 |
| Consistently associated with inferior outcome | 22–25 | |
| Level of mutant allele likely of importance | 23,25 | |
| Homozygous FLT3 mutations as a result of mitotic recombination leading to partial UPD | 28,49 | |
| TKD | TKD point mutations found in 11% to 14% of CN-AML | 24,25,30 |
| More recently associated with better OS | 30 | |
| Level of mutant allele may be of importance: high-level mutants associated with improved OS | 30 | |
| CEBPA | Transcription factor mediating lineage specification and differentiation of multipotent myeloid progenitors into mature neutrophils | 31 |
| Found predominantly in CN-AML and AML with 9q deletion | 3,6 | |
| Associated with higher CR rate and better RFS and OS | 3,6 | |
| MLL | PTD found in 5% to 11% of CN-AML Associated with shorter CR duration, or inferior RFS and EFS | 6 |
| Autologous transplantation may improve outcome | 32 | |
| Rationale for the use of DNA methyltransferase and/or histone deacetylase inhibitors based on in vitro data | 33 | |
| RAS | NRAS mutations found in ~9% of CN-AML No prognostic significance | 6 |
| WT1 | Mutations found in ~10% of CN-AML Initial studies on small patient cohorts suggest association with induction failure | 7,34 |
| Gene . | Biological/clinical features . | Selected references . |
|---|---|---|
| Abbreviations: BM, bone marrow; LDH, lactate dehydrogenase; CN, cytogenetically normal; ITD, internal tandem duplication; TKD, tyrosine kinase domain; MRD, matched related donor; CR, complete remission; JM, juxtamembrane domain; UPD, uniparenteral disomy; OS, overall survival; RFS, relapse-free survival; EFS, event-free survival; PTD, partial tandem duplication. | ||
| NPM1 | Protein with pleiotropic functions | 18 |
| Associated with presenting clinical and laboratory features such as female sex, higher BM blast counts and LDH levels, as well as high CD33 but low or absent CD34 levels | 6,10–15 | |
| Found in 25% to 35% of AML; predominantly in CN-AML (45% to 62%) Associated with FLT3-ITD and TKD mutations | 6,11,13,15 | |
| NPM1mut/FLT3-ITDneg genotype associated with a favorable prognosis | 10–12,15 | |
| Patients with NPM1mut/FLT3-ITDneg genotype may not benefit from MRD allogeneic transplantation in first CR | 10,37 | |
| FLT3 | Member of the class III receptor tyrosine kinase family Constitutively active FLT3 molecules are targets for molecular therapy | 26 |
| ITD | Found in 28% to 34% of CN-AML In-frame mutations, mostly in exons 14 and 15 of the JM domain | 3,6 |
| Consistently associated with inferior outcome | 22–25 | |
| Level of mutant allele likely of importance | 23,25 | |
| Homozygous FLT3 mutations as a result of mitotic recombination leading to partial UPD | 28,49 | |
| TKD | TKD point mutations found in 11% to 14% of CN-AML | 24,25,30 |
| More recently associated with better OS | 30 | |
| Level of mutant allele may be of importance: high-level mutants associated with improved OS | 30 | |
| CEBPA | Transcription factor mediating lineage specification and differentiation of multipotent myeloid progenitors into mature neutrophils | 31 |
| Found predominantly in CN-AML and AML with 9q deletion | 3,6 | |
| Associated with higher CR rate and better RFS and OS | 3,6 | |
| MLL | PTD found in 5% to 11% of CN-AML Associated with shorter CR duration, or inferior RFS and EFS | 6 |
| Autologous transplantation may improve outcome | 32 | |
| Rationale for the use of DNA methyltransferase and/or histone deacetylase inhibitors based on in vitro data | 33 | |
| RAS | NRAS mutations found in ~9% of CN-AML No prognostic significance | 6 |
| WT1 | Mutations found in ~10% of CN-AML Initial studies on small patient cohorts suggest association with induction failure | 7,34 |
Treatment results according to the combined NPM1 and FLT3 ITD mutation status. (A) Relapse-free survival. (B) Overall survival (overall P values are given).
Reprinted with permission from
Treatment results according to the combined NPM1 and FLT3 ITD mutation status. (A) Relapse-free survival. (B) Overall survival (overall P values are given).
Reprinted with permission from
University of Ulm, Ulm, Germany
Acknowledgments
Supported in part by grants 01GI9981 (Network of Competence Acute and Chronic Leukemias) and 01KG0605 (IPD-Meta-Analysis: a model-based hierarchical prognostic system for adult patients with acute myeloid leukemia (AML)) from the Bundesministerium für Bildung und Forschung (BMBF), Germany.