Abstract
Despite the various abnormalities identified in the immune system or the bone marrow microenvironment in patients with myelodysplastic syndrome (MDS), most of the investigation of this disorder has centered on the hematopoietic stem/progenitor compartment. It is generally written that MDS is a stem cell disorder, and there is certainly evidence supporting this view. However, whether it occurs in a cell with only myeloid multipotentiality (i.e., that involves megakaryocytic, erythroid and granulocytic/monocytic lineages) or occurs in a true stem cell is open to debate. The absence of an assay for human stem cells necessitates the use of surrogate markers for such cells, such as gene expression profiles, or the identification of specific genetic or epigenetic abnormalities that are found in multiple lineages. Clearly, the common cytogenetic and genetic abnormalities found in MDS are most indicative of a clonal myeloid disease similar to AML, rather than a lymphoid disease, and the often tri-lineage ineffective hematopoiesis and dysplasia are generally not found within the lymphoid compartment. Recent studies, using modern molecular detection techniques, have identified new recurring molecular lesions in these disorders but have not really unraveled its pathogenesis.
Introduction
Even though numerous abnormalities have been identified in the immune system and bone marrow microenvironment of patients with MDS, most attention has (rightly?) centered on identifying defects in the hematopoietic stem/progenitor compartment. These defects can give rise to secondary abnormalities in stem cell-stem cell niche interactions or in immune function, although the stem/progenitor cell defects may also arise due to an abnormal environment in which they reside (the seed vs. soil argument). Evidence exists both for and against the idea that MDS is a true “stem cell” disorder, as it may more commonly occur in a cell with only myeloid (i.e., megakaryocytic, erythroid and granulocytic/monocytic lineages) multipotentiality. Clearly, MDS stem/progenitor cells must be capable of sufficient self-renewal to perpetuate the disease. Understanding the genetic defects found in MDS patients, how they affect the immune and non-immune components of the bone marrow microenvironment, and how these abnormalities lead to the replacement of normal HSCs with MDS HSCs, will be critical to developing new treatments for this disease.
Defining Human Hematopoietic Stem and Progenitor Cells
We know quite a bit about how murine hematopoietic stem cells differentiate and lose lineage capability but not as much about how human hematopoietic stem cells lose their multipotentiality. While the definition of a murine hematopoietic stem cell (HSC) is quite clear (namely, a single cell that has the ability to repopulate the hematopoietic and immune system of a lethally irradiated mouse), surrogate assays are required to identify the human counterpart of this cell, as no such assay exists for human HSCs. Thus, a variety of in vitro culture systems are used to establish the hierarchy of human stem/progenitor cells and to grow lymphoid or myeloid progenitor cells in culture. Xenotransplantation of human cells into immunodeficient mice has been a major surrogate assay for identifying the leukemia “stem cell” (actually the NOD/SCID leukemia-initiating cell)1 and also for identifying normal hematopoietic stem/progenitor cells (the so-called SCID repopulating cell [SRC]).2
Over time, the generation of increasingly immunodeficient mice has allowed for more efficient engraftment of both malignant and normal human hematopoietic cells (including cells from the bone marrow of patients with MDS3). The spectrum of cells capable of engrafting various types of immunodeficient mice is different: only CD34+, CD38− cells will engraft NOD/SCID mice, while more mature cells (with greater restrictions in proliferative and differentiation capability) can engraft NOD/SCID β2-microglobulin null or NOD/SCID/IL-2R γc null immunodeficient mice. Clearly, engraftment itself can no longer be taken as a sign of transformation. Thus, this assay needs to be combined with genetic analysis of the human cells present in the chimeric mice in order to specifically identify and characterize the MDS (or AML) cells.
The absence of an assay for human stem cells should necessitate the use of the term “primitive hematopoietic progenitor (PHP)” to describe these cells. However, because the term “stem cell” is so widely (mis-) used in this context, the term “hematopoietic stem/progenitor cells” will be used throughout this article. Based on in vitro stem/progenitor cell assays and a series of antibodies that recognize specific cell surface markers such as CD34, an immunophenotype of cells highly enriched for primitive human hematopoietic progenitor cells can be obtained. It is clear that CD34+ CD38− human bone marrow or cord blood cells are more primitive than the CD34+ CD38+ cell population, and recent work suggests that the use of a Thy1 marker (CD90), various human cell lineage markers (Lin) and the CD45RA marker can define the most primitive human cells in cord blood (they are Lin−, CD34+, CD90+, CD45RA−). These markers can also be used to establish a hierarchy of primitiveness, based on in vitro assays and engraftment in NOD/SCID/IL-2Rγ null mice. The CD90+ CD45RA− cells first become CD90−CD45RA− and then CD90−CD45RA+.4
Differences exist between CD34+ cells isolated from various sources, in terms of their functional properties. In normal human bone marrow only 0.1% of the mononuclear cells are CD34+, and of these only 1% are CD38−. In umbilical cord blood collections, approximately 1% of the mononuclear cells are CD34+, and 4% of these are CD38−. Thus, using the term HSCs to describe CD34+ CD38− cells should be avoided; even if CD34+ CD38− cells are 99.9% enriched for HSCs, they likely contain only ~3% true stem cells.5
The classic model for murine hematopoiesis has identified long-term and short-term repopulating HSCs and multi-potent progenitor cells, dividing LSK cells (lineage−, Scal+, c-kit+) cells into three subsets (based on expression of the CD34 and/or FLT3 cell surface markers). The LT-HSC (which is CD34− and FLT3−) gives rise to the ST-HSC (which is CD34+ FLT3−), which gives rise to multipotent progenitors (MPPs; which are CD34+ and FLT3+). The SLAM (signaling lymphocyte activation molecule) cell surface receptor family, which contains CD150, CD244, CD48 can also be used to isolate various populations of murine hematopoietic stem/progenitor cells. However, the use of antibodies that recognize SLAM receptors to define human HSPCs has not been well studied. CD133 (also known as AC133 or prominin-1) is expressed on human HSCs, and in fact some CD133+ CD34− CD38− cells appear to give rise to CD34+ cells. Expression of CD133 is seen in the more slowly dividing fraction of CD34+ cells, which may be associated with more primitive cells within this heterogeneous cell population. HSCs mark as CD150+ CD244− CD48− cells, while MPPs are CD150− CD244+ CD48− and more differentiated progenitors CD150− CD244+ CD48+.6 It has been reported that MPPs give rise to common myeloid progenitors (CMP) or common lymphoid progenitors (CLP). However, isolating a cell with both B and T lineage capacity and no myeloid capacity has been challenging, and recent studies have challenged the notion that murine stem cells (fetal or adult) break off into “lymphoid only” and “myeloid only” progenitors. These studies suggest that HSCs in fact possess a myeloid lineage bias, and that early thymic precursor cells (ETPs) have T lineage and myeloid capability (macrophage, dendritic cell, NK cell and granulocyte), but no B-cell capability.7,8
The properties of single fetal and adult murine primitive progenitor cells are being defined using a variety of sophisticated in vitro and in vivo systems; however, it is unknown how precisely this applies to human adult hematopoietic cells. Thus, the multipotentiality of MDS stem cells (stem cells that are altered by genetic abnormalities that lead to MDS) or MDS-initiating cells (the cell of origin that is transformed into an MDS-propagating cell) may remain difficult to fully define. What is not disputed is the clonal nature of MDS, established by historically by isoenzyme analysis9 or by cytogenetic analysis. Fluorescence in situ hybridization (FISH) studies represent an advance on metaphase analysis; they can be very useful to document clonality in patient with normal cytogenetics (or with no analyzable metaphases). The recent use of high-density genomic analysis to identify previously unknown sites of genomic DNA loss or amplification10,11 in samples from patients with MDS may also be helpful in cases with normal cytogenetics (and in those with karyotypic abnormalities). In this chapter, I will use the term MDS stem/progenitor cells to refer to transformed cells, and cell of origin to refer to normal hematopoietic cells.
Cell Surface Marker Expression
Although immunophenotyping of AML or MDS can be extremely useful in identifying aberrant cell surface marker expression and detecting minimal residual disease, it has not been possible to isolate leukemia- or MDS-initiating cells or stem cells based on simple immunophenotyping. The most widely accepted marker for distinguishing malignant hematopoietic stem-progenitor cells from normal ones has been the IL-3 receptor alpha chain (IL-3Rα), which together with the granulocyte-macrophage colony-stimulating factor/IL-3/IL-5 common β receptor generates a high affinity receptor for IL-3 (an early acting cytokine that is expressed by T cells, mast cells, and some leukemia cells, e.g., megakaryocytic leukemias). Several studies have suggested that expression of this receptor subunit (which is designated CD123) can distinguish the leukemic stem cell from its normal hematopoietic stem cell counter part.12 Enhanced IL-3Rα expression on AML blasts correlates with poor prognosis,13 which may relate to the blasts possessing more “stem-cell like” properties, including possibly being more quiescent.
Several strategies have been studied to target the CD123+ malignant cell compartment. Conjugation of IL-3 to diphtheria toxin generates a fusion protein capable of killing AML blasts that are able to engraft immunodeficient mice (NOD/SCID leukemia-initiating cells).14 The expression level of CD123 on CD34+ CD38− CD71− cells correlates with the cytotoxicity of this agent,15 suggesting that it may kill leukemia “stem cell-like” cells. In contrast to studies in AML, only one limited study of IL-3Rα expression on MDS cells has been conducted;16 no real difference was found in its expression in MDS marrow cells versus normal marrow cells. While IL-3 itself has shown little clinical activity in patients with MDS, IL-3R expression on the MDS “stem cell” could be a reasonable therapeutic target. In a recently published phase I trial, one of the 5 patients with MDS treated with the diphtheria toxin-IL-3 fusion protein had a partial response.17
In addition to CD123, the CLL-1 marker (C type lectin-like molecule) has recently been proposed to identify leukemic blast (stem?) cells18 with greater accuracy than CD123, which may also be expressed on cells within a regenerating bone marrow. Another cell surface marker on human HSCs was recently identified, CD143, which is the angiotensin converting enzyme.19 CD143 may be over-expressed in some AMLs; its expression in HSPCs suggests that the renin-angiotensin system may play a role in regulating hematopoietic stem/progenitor cell biology. Whether it too could be a target for therapy of MDS (or AML) is unknown.
Are Nonmyeloid Cells Commonly Clonal in Patients with MDS?
The cytogenetic and genetic abnormalities found in MDS are rarely found in the lymphoid cell compartment, and the often tri-lineage ineffective hematopoiesis and dysplasia seen in this disorder are not generally found in the lymphoid compartment. While this was widely thought to reflect the long-lived nature of the immune system, which can mask involvement of small numbers of dysplastic mature B cells or T cells, modern molecular detection techniques, especially PCR-based methods, have not generally identified recurring molecular lesions in these cellular compartments. While this challenges the assumption that the MDS-initiating cell is a pluripotent stem cell with both myeloid and lymphoid capability, it is also possible that the genetic (and epigenetic) abnormalities that occur in the MDS stem cell block the ability of these cells to commit to the B cell lineage. Thus, although, the genetic abnormalities found in patients with MDS are more indicative of a clonal myeloid disease and MDS rarely evolves into a lymphoid acute leukemia, the possibility that MDS arises in a cell with lymphoid potential cannot be excluded.
Indeed, there are a few reports where an identical cytogenetic marker was found in both the lymphoid (B cell) and myeloid compartments of the patient. For instance in a xenotransplantation model trisomy 8 was found in both lymphoid and myeloid human cells, demonstrating that a self-renewing cell with both myeloid and lymphoid potential was present in the patient.2 In another example, peripheral blood mononuclear cells from a patient with MDS and a 20q deletion were transformed with Epstein-Barr virus (EBV) and a B-lymphoblastoid cell line was generated that contained a 20q deletion identical to that seen in myeloid cell metaphases.20 PHA-stimulated bone marrow cells from this patient did not show a 20q deletion (and in fact, there really is no evidence that clonal T cells are generated from MDS clonal cell populations). PCR-based assays were performed to assess various peripheral blood cell compartments of the patient; no B or T cells were found with the 20q deletion, even though the granulocytes and monocytes clearly carried the deletion. Thus, although the 20q-can arise in a progenitor cell with both myeloid and B-cell capacity the abnormalities it generates may not permit B-cell differentiation. In general, a variety of reports describe significant defects in B-cell populations and B-lineage development in MDS patients, which is of unclear pathogenetic importance. Given reports that 20q- cells in the bone marrow may contribute minimally to the circulating myeloid cell compartment,21 it may not be surprising that they are not detected in the circulating lymphoid compartment. Indeed, others have reported B-cell lineage involvement in MDS; for instance, the 13q deletion found in two patients with acquired sideroblastic anemia was also found in EBV-transformed B cells22 from these patients.
Another issue is whether the non-hematopoietic cells found in MDS patients may be derived from the malignant MDS clone. Mesenchymal stem cells from MDS patients have been shown to contain the same cytogenetic abnormalities as the myeloid cells in some instances.23,24 Stromal cells that carry the BCR-ABL fusion gene have been identified in CML; however, they appear to be macrophage derived.25 Similar findings could certainly exist for the “stromal cells” of patients with MDS, and macrophage defects have been postulated to play a key role in MDS pathogenesis. NK cell and NK T-cell deficiencies in numbers or function have been found in MDS patients, but these deficiencies generally improve when the MDS is “ successfully” treated. This suggests that these defects may be secondary to the MDS rather than primary; however, this is really not knowable. Abnormalities in dendritic cell numbers and function have been also reported in MDS. Clonal cytogenetic abnormalities have been found within this population of cells26 (monocyte-derived dendritic cells for the most part). However, defects in dendritic cell maturation and aberrant cytokine expression have also been reported in the non-clonal populations.27
Studies of MDS HSPCs
Thanapoulou and colleagues isolated bone marrow cells from 11 different MDS patients and showed that in most cases the cells could at least transiently repopulate the marrow of NOD/SCID β2-microglobulin null mice.2 Because several murine cytokines have no effect on human cells, this group used mice engineered to express human IL-3, GM-CSF and SCF to improve the engraftment of human cells. While these mice had superior engraftment of normal human bone marrow cells, there was no real difference in the growth of MDS bone marrow cells. In many instances the MDS cells only transiently engrafted; while this did not correlate with the FAB subtype of MDS (i.e., with the blast percentage), only 9 samples were studied and several samples were only transplanted into a single mouse. Nonetheless, there was clear engraftment of MDS cells as cytogenetic markers found in the patient’s MDS cells could be detected in the engrafted cells. Abnormal B cells were generally not found in mice receiving MDS cells, but neither are CD41+ cells, which suggests that the microenvironment may not be conducive to the growth of these lineages from MDS marrow (and not necessarily that the cells infused did not have the ability to generate these specific lineages, although that is also a real possibility). Of note, none of the recipient mice developed acute leukemia.
Studies of 5q- MDS
5q- MDS has attracted a lot of attention due to the efficacy (and largely unknown mechanism of action) of lenalidomide in abrogating red blood cell transfusion–dependence in patients with International Prognostic Scoring System low-risk or int-1 disease, and also inducing significant numbers of major or complete cytogenetic remissions. A series of studies of bone marrow cells from 5q- patients has demonstrated the frequent involvement of the HSPC in the malignant process. Early on, Nilsson and colleagues used FISH assays to examine CD34+, CD38−, Thy1+ (CD90) cells from 11 patients with a 5q- cytogenetic abnormality: 92% to 100% of the cells had the 5q- deletion.28,29 The 5q deletion was often detected in the CD34+ CD19+ pro-B cells isolated from these 5q- MDS patients, reflecting involvement of the lymphoid compartment. Nonetheless, LTC-IC activity, which is contained within this cell population, could be demonstrated in only 2 of 4 patients, reflecting the intrinsic growth abnormalities present in the MDS cells and also the replacement of normal marrow cells by 5q- cells (or suppression of their growth). In these patients, only 0.14% of the bone marrow cells were CD34+, CD38−, Thy1+ cells, which is only slightly higher than normal. This suggests that HSC expansion is minimal, despite the efficient manner in which the 5q- cells take over the bone marrow. Perhaps this relates to the number of available stem cell niches. Yet, the basis for the ability of these cells to out-compete the normal HSC populations in MDS patients remains a mystery.
A molecular signature for 5q- MDS “stem cells” (CD34+, CD38−, Thy1+ cells), has been recently reported, identifying genes that are either up- or down-regulated in the 5q- cells versus normal CD34+, CD38−, Thy1+ cells.29 By using the published gene expression data in normal CD34+CD38+ and CD34+CD38− cells (reported by Georgantas et al30), Nilsson and colleagues identified a number of interesting candidate genes in the 5q- cells, including the MEF2C, FBXL5, HLF, RPS21 genes, which were all downregulated. Upregulated genes include BMI-1, which is known to play a role in HSC (and neural stem cell) self-renewal, Id1 (which is involved in preventing the premature differentiation of HSCs and ES cells31), DNMT3 (which is involved in DNA methylation), CMYC (which plays a role in stem cell regulation) and others. Expression of CEBPA, a gene required for neutrophilic differentiation, was upregulated in the CD38− cell fraction, but down-regulated in the CD38+ progenitor cells. This study was focused on the level of expression of genes that may influence the behavior of the MDS stem cell rather than examining changes in the expression of genes contained within the 5q- commonly deleted region. Nonetheless, careful descriptions of these gene signatures will hopefully lead to a better understanding of what is awry in MDS.
Do Gene Expression Profiles Identify the Cell of Origin in MDS?
Gene expression profiles of CD34 CD133 selected cord blood cells, and granulocyte colony-stimulating factor–mobilized peripheral blood progenitor cells, have been reported.32,33 Many groups have also published analyses of the expression profiles of the CD34+ fraction of MDS patient samples, obtained from patients with specific cytogenetic features, or specific FAB or WHO subtypes. Lastly, expression profiles of cells from distinct subtypes of AML have also been reported.30,31,34 Overall, these studies, and studies of the genetics of MDS versus AML, demonstrate that MDS is a truly distinct disease.
Studies in AML have shown that transformed progenitor cells may attain a stem cell phenotype, for example in MLL-AF9 driven murine leukemia, the gene expression profile of the transformed progenitors, the so called L-GMP (leukemic granulocyte macrophage progenitor) more closely resembles a normal GMP than a normal LT-HSC.35 One issue with using a specific pattern of gene expression for identifying stem cells is that the “stem cell” gene signature may depend very much on the local milieu; thus, “the HSC” may have a variable signature depending on where it is isolated from. Nonetheless, for the purpose of identifying normal versus MDS stem cells, MDS stem cells from MDS progenitor cells, or even deciding on therapeutic options gene expression profiles can be useful.
Why Transcript Profiling Is Not Enough to Decipher MDS
The incredible heterogeneity in MDS, including the observed differences in behavior between the hematopoietic stem cells and the progenitor cells, makes it particularly challenging to decipher MDS by looking at the relative levels of total cellular RNAs that are expressed. There are also many limitations to using microarray data to study the “functioning genome” of any cell. Our new understanding of the critical roles that microRNAs (miRNAs) can play in cell behavior has clearly illustrated that even without changes in the level of RNA, a cell can suddenly begin to express a functional protein. A recent, elegant demonstration of this came from the Peschle lab, who showed that downregulation of a miRNA directed against the 3′ UTR of the AML1 RNA occurs with monocytic differentiation. This downregulation allows translation of the AML1 mRNA into AML1 protein, which can then activate expression of the M-CSF receptor, allowing the cell to respond to M-CSF and develop along the monocytic lineage.36
Beyond this regulatory step are a variety of post-translational modifications that may also be critical for proper hematopoietic cell development. In addition to phosphorylation events, these modifications include ubiquitination, acetylation, methylation and sumoylation. Acetylation of the p53 protein by enzymes historically called histone acetyltransferases (now assigned the name protein acetyltransferases37) alters its transcriptional activating properties and is essential for its ability to trigger apoptosis in the cell.38,39 Methylation on arginine and/or lysine residues can regulate the function of transcription factors such as AML1 (RUNX1),40 elongation factors such as SPT541 and various splicing factors. Involvement of nuclear pore proteins (NUP98 and NUP214) in myeloid malignancies suggests that defective nuclear transport (of mRNAs or proteins) may also contribute to malignant cell behavior. Furthermore, genes that regulate ribosomal biogenesis may play a role in the pathogenesis of MDS, based on studies of Diamond-Blackfan anemia42 and 5q- MDS,43 which have implicated the ribosomal proteins RPS19 (or RPS24) and RPS14, respectively.
Aberrant Growth Properties within the Hematopoietic Stem/Progenitor Cell Compartment in MDS
The increased apoptosis seen in the early hematopoietic cell compartment in MDS, as well as the decreased proliferation and survival, has led to studies of interferon (IFN) signaling, TNFα signaling, and TGFβ signaling (three negative regulators of hematopoiesis) and also of the oxidative and DNA damage stress responses. Gene expression profiles have identified a number of IFN-inducible genes that are upregulated in MDS bone marrow cells.44 TGFβ has potent erythroid potentiating effects and anti-proliferative effects, which have been shown to be mediated via distinct signaling molecules (TIF1γ and Smad4),45 and inhibiting TGFβ signaling can improve hematopoiesis in MDS.46
Depletion of early hematopoietic cells is seen in MDS patients,47 which suggests defective self-renewal. Clearly, abnormalities in the genes regulating HSC self-renewal, differentiation and quiescence could contribute to the observed hematopoietic abnormalities found in MDS patients. A variety of genes have been shown to play important regulatory roles in HSC self-renewal in mice (Table 1 ), including the polycomb gene BMI-1, the cdk inhibitor p18, the PTEN phosphatase, zfx48 and the ETS protein MEF.49 Similarly, a variety of genes have been implicated in controlling HSC quiescence, including Fbw7, TPO, angiopoietin-1, GATA-2 and MEF. However, these genes have not been extensively studied in MDS patients.
The stem cell niche provides signals that keep the HSC quiescent and largely protected from oxidative stress. Defects in this signaling (stem cell–niche signals) could cause the steady depletion of HSCs. The prolonged myelo-suppression seen in patients with MDS, and in patients with AML following MDS, suggests a limited hematopoietic reserve and implies that patients with MDS likely lose normal HSCs over time (perhaps due to defective self-renewal) and accumulate MDS HSCs (which may be better suited for competing for stem cell niches). Implicated in the loss of HSCs due to ROS generation and oxidative stress in murine models are the FoxO transcription factors,50 p38 MAPK and the ATM protein.51 The p38 kinase pathway is particularly interesting as p38 kinase levels are elevated in MDS bone marrow cells and p38 inhibitors have been shown to decrease the level of apoptosis observed in MDS progenitor cells.52
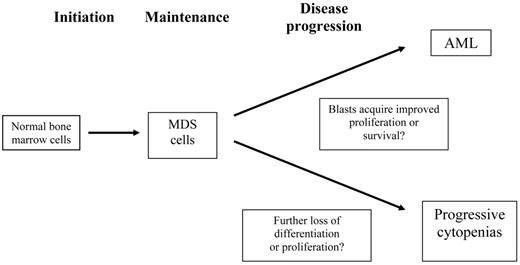
There are three (or four) phases of MDS, including the progression to acute leukemia and the loss of proliferation or differentiation capability leading to progressive cytopenias (Figure 1 ). While many investigators have studied the transition of MDS to AML, little is known about the mechanisms that lead to progressive cytopenias in patients who do not have a progressive increase in blasts or worsening dysplasia. In particular, whether defective proliferation or survival occurs, and whether this occurs in the earliest cells or in the ‘transient amplifying cell compartment” is not known. MDS is known to occur in patients with aplastic anemia, especially in those with an incomplete response to immunosuppressive therapy. Such MDS is commonly accompanied by trisomy 8 or partial or complete loss of chromosome 7. The basis for this is not known, but it suggests “survival of the fittest” selection, i.e., the outgrowth of cells that can survive in an unfavorable environment.
A number of genes that regulate the commitment of cells to specific lineages or to myeloid versus lymphoid lineages have been identified. Impaired differentiation can certainly occur due to defective activating properties of these TFs (CEBPA, PU.1, GATA-1). Other proteins, such as Id1, which blocks E box protein DNA binding, can prevent premature commitment of HSCs to the myeloid lineage.53 In the absence of Id1, HSCs cannot self-renew and they exhaust with serial transplantation. Cell-cycle regulating genes and genes in the p53 pathway also contribute to this process. The role of specific pathways in the self-renewal of aging HSCs has not been well studied as yet.
Are There Recurring Microenvironmental Abnormalities in MDS?
There is an intimate relationship between HSCs and the stem cell “niche,” yet transplant experiments show that upon successful engraftment of recipient mice, donor bone marrow cells that show enhanced self-renewal will repopulate the bone marrow to the same enhanced degree. This implies that HSCs can send signals to open up the right number of niches needed to support their self-renewal. Both endosteal niches and vascular niches have been identified in murine model systems. The endosteal niches are sites where osteoblasts play an important role in regulating stem cell behavior. But the absence of osteoblasts in the liver, spleen and lymph nodes, sites of extramedullary hematopoiesis in certain disorders, argues in favor of there also being vascular stem cell niches, where endothelial cells maintain HSC integrity.
It may be possible to interfere with the adhesion or homing of MDS stem cells to the marrow niche. Targeting CD44 has been shown to eliminate AML1 “stem cells” by affecting homing of the cells and also by triggering differentiation.54 Likewise, targeting of the interaction between CXCR4 and its chemokine ligand the SDF-1 (CXCL12) may trigger mobilization of MDS HSPCs from the marrow. Other ligand-receptor interactions, such as the angio-poietin-1/Tie-2 interaction, may be targetable. For instance, angiopoietin-1 interacts with Tie-2 to regulate stem cell behavior (i.e., quiescence). Angiopoietin-1 is expressed by osteoblasts and by perivascular mesenchymal cells,55 whereas Tie-2 is expressed on HSCs.
Can Mouse Models of MDS Help Us Understand This Disease?
Based on the identification of specific molecular lesions in MDS an increasing number of genetically modified mice are being reported that have features of a myelodysplastic syndrome (Table 2 ). Perhaps the most common and best studied is the t(3;21), which is seen in MDS and in CML blast crisis and generates the AML1-MDS1-EVI1 protein, via fusion of the AML1 gene and the MDS1-EVI1 gene.
Retroviral transduction of bone marrow cells followed by transplantation of the genetically modified bone marrow cells has been used to study EVI-1 overexpression. Ten months post-transplantation the mice get sick, and erythroid dysplasia is seen, although the mice do not get AML.56
Mutations in the AML1 gene are found in approximately 15% of MDS/AML and in 46% of radiation-induced MDS. Another murine MDS model has been generated by introducing mutant forms of AML1 (RUNX1) found in MDS patients into bone marrow cells that are then used for transplantation into wild-type recipient mice. Most of the recipient mice develop MDS or MDS/AML-like symptoms within 4 to 13 months post-BMT and for one specific AML1 mutant protein (AML1 D171N), overexpression of EVI-1 due to retroviral insertional activation is commonly seen when the mice develop full-blown AML.57 Co-expression of EVI-1 with this mutant AML1 protein results in AML, further suggesting that these two proteins can cooperate in leukemogenesis (likely due to enhanced EVI-1 function and deficient AML1 function). In contrast, a C terminally truncated form of AML1 induces pancytopenia with eryth-roid dysplasia that progresses to RAEB and then AML, without a specific identified additional hit.
Perhaps the most faithful model at this time is the NUP98-HoxD13 (NHD) transgenic mouse, where the NHD transgene is expressed from the VAV promoter. These mice have an MDS phase, with multilineage dysplasia and cytopenias. This phase can be fatal, but if not, it is followed by evolution to acute leukemia.58
Ras mutations, p53 mutations and AML1 (RUNX1) mutations are probably the most common, recurring genetic abnormalities found in MDS patients. Activating tyrosine kinase mutations are rarely found in MDS patients, unless the patient’s disease possesses more of a proliferative nature. TEL-PDGFRβ is found in CMML with eosinophilia, but this entity is often more of a myeloproliferative disease than an MDS. Activated JAK2 (V617F) is found in RARS-T patients59 and in some patients with 5q- and thrombocytosis.
An association between Ras mutations and AML1 mutations has been reported in MDS and AML, but not yet modeled in mice. One mouse model for MDS, using mutant N-RAS and BCL2 overexpression, was recently reported.60 Another mouse model of MDS is the NPM+/− mouse, which shows significant dysplasia, without anemia or development of AML.61 However, several features of these models are less than ideal, as mutant RAS and BCL-2 may not be found together and NPM mutations are not commonly found in MDS. Thus, all mouse models need to be evaluated carefully to assess their accuracy in mimicking the human condition.
Conclusions
Whether MDS arises in a true hematopoietic stem cell or in a somewhat more committed cell remains an academic issue at this time. Little is known about the cellular or molecular basis of MDS or about the progression of “early” MDS to “late” (or advanced) MDS or to AML. It is clear that MDS has features that are quite distinct from those that characterize AML. As we better define these features we may be able to rationally choose therapies based on the genetic or gene expression profiles of the MDS cells. Acquiring a better understanding of the nature of the interaction of MDS HSPCs with the bone marrow microenvironment will be critical as well.
Genes or pathways that affect hematopoietic stem cell (HSC) behavior.
| Gene/Pathway . | HSC Behavior . | Reference . |
|---|---|---|
| c-myc | HSC/niche | Wilson et al. Genes & Dev 2006 |
| Mef | Self-renewal and quiescence | Lacorazza et al. Cancer Cell 2006 |
| Foxo TFs | Resistance to ROS | Tothova et al. Cell 2007; Miyamoto et al. Cell Stem Cell 2007 |
| Gfi-1 | Proliferation | Hock et al. Nature 2004 |
| GATA-2 | Quiescence | Ling et al. J Exp Med 2004 |
| MAD1 | Proliferation | Walkley et al. Nat Cell Biol 2005 |
| p21 | Quiescence | Cheng et al. Science 2000 |
| p27 | HPC proliferation | Cheng et al. Nat Med 2000 |
| p18 | Self-renewal | Yuan et al. Nat Cell Biol 2004 |
| p16 | Aging | Janzen et al. Nature 2006 |
| Rb | HSC/niche | Walkley et al. Cell 2007 |
| Bmi-1 | Self-renewal and proliferation | Park et al. Nature 2003; Molofsky et al. Nature 2003 |
| HoxB4 | Self-renewal | Krosl et al. Nat Med 2003 |
| Fbw7 | Quiescence | Thompson et al. J Exp Med 2008 |
| Cdc42 | HSC/niche | Yang et al. PNAS 2007 |
| Ang-1/Tie2 | Quiescence | Arai et al. Cell 2004 |
| TPO/MPL | Quiescence | Qian et al. Cell Stem Cell 2007; Yoshihara et al. Cell Stem Cell 2007 |
| PTEN | Self-renewal and quiescence | Zhang et al. Nature 2006; Yilmaz et al. Nature 2006 |
| GSK-3 | HSC maintenance | Trowbridge et al. Nat Med. 2006 |
| LNK | Self-renewal | Seita et al. PNAS 2007 |
| Notch | Self-renewal | Duncan et al. Nature Immunol 2005 |
| Wnt signaling | Self-renewal | Reya et al. Nature 2003 |
| Hedgehog | Self-renewal | Trowbridge et al. PNAS 2006 |
| Zfz | Self-renewal | Galan-Caridad et al. Cell 2007 |
| Gene/Pathway . | HSC Behavior . | Reference . |
|---|---|---|
| c-myc | HSC/niche | Wilson et al. Genes & Dev 2006 |
| Mef | Self-renewal and quiescence | Lacorazza et al. Cancer Cell 2006 |
| Foxo TFs | Resistance to ROS | Tothova et al. Cell 2007; Miyamoto et al. Cell Stem Cell 2007 |
| Gfi-1 | Proliferation | Hock et al. Nature 2004 |
| GATA-2 | Quiescence | Ling et al. J Exp Med 2004 |
| MAD1 | Proliferation | Walkley et al. Nat Cell Biol 2005 |
| p21 | Quiescence | Cheng et al. Science 2000 |
| p27 | HPC proliferation | Cheng et al. Nat Med 2000 |
| p18 | Self-renewal | Yuan et al. Nat Cell Biol 2004 |
| p16 | Aging | Janzen et al. Nature 2006 |
| Rb | HSC/niche | Walkley et al. Cell 2007 |
| Bmi-1 | Self-renewal and proliferation | Park et al. Nature 2003; Molofsky et al. Nature 2003 |
| HoxB4 | Self-renewal | Krosl et al. Nat Med 2003 |
| Fbw7 | Quiescence | Thompson et al. J Exp Med 2008 |
| Cdc42 | HSC/niche | Yang et al. PNAS 2007 |
| Ang-1/Tie2 | Quiescence | Arai et al. Cell 2004 |
| TPO/MPL | Quiescence | Qian et al. Cell Stem Cell 2007; Yoshihara et al. Cell Stem Cell 2007 |
| PTEN | Self-renewal and quiescence | Zhang et al. Nature 2006; Yilmaz et al. Nature 2006 |
| GSK-3 | HSC maintenance | Trowbridge et al. Nat Med. 2006 |
| LNK | Self-renewal | Seita et al. PNAS 2007 |
| Notch | Self-renewal | Duncan et al. Nature Immunol 2005 |
| Wnt signaling | Self-renewal | Reya et al. Nature 2003 |
| Hedgehog | Self-renewal | Trowbridge et al. PNAS 2006 |
| Zfz | Self-renewal | Galan-Caridad et al. Cell 2007 |
Mouse models of myelodysplastic syndrome (MDS).
| Mouse model . | Macrocytic anemia . | Neutropenia . | Thrombocytopenia . | Bone marrow dysplasia . | Time to MDS . | Evolution to AML . | References . |
|---|---|---|---|---|---|---|---|
| NPM +/− mouse | Yes | No | No | Yes | Immediately | Yes | Grisendi et al 200561 Sportoletti et al 200862 |
| NUP98-HOXD13 | |||||||
| TG Mouse | Yes | Yes | Yes | Yes | Immediately | Yes | Lin et al 200558 |
| EVI-1 BMT | Yes | No | Yes | Yes | 8-10 months | No | Buonamici et al 200459 |
| AML1 mutations | Watanabe-Okochi et al 200857 | ||||||
| D171N | Yes | No | Yes | Yes | 4-13 months | MDS/AML 13 of 16 | |
| S291fsx 300 | Yes | Yes | Yes | Yes | 4-13 months | MDS/RAEB 2 of 8 MDS/AML 5 0f 8 | |
| N-Ras + BCL-2 TG mouse | No | No | Yes | Yes | 1 month | MDS/AML 5 of 8 | Omidvar et al 200760 |
| Mouse model . | Macrocytic anemia . | Neutropenia . | Thrombocytopenia . | Bone marrow dysplasia . | Time to MDS . | Evolution to AML . | References . |
|---|---|---|---|---|---|---|---|
| NPM +/− mouse | Yes | No | No | Yes | Immediately | Yes | Grisendi et al 200561 Sportoletti et al 200862 |
| NUP98-HOXD13 | |||||||
| TG Mouse | Yes | Yes | Yes | Yes | Immediately | Yes | Lin et al 200558 |
| EVI-1 BMT | Yes | No | Yes | Yes | 8-10 months | No | Buonamici et al 200459 |
| AML1 mutations | Watanabe-Okochi et al 200857 | ||||||
| D171N | Yes | No | Yes | Yes | 4-13 months | MDS/AML 13 of 16 | |
| S291fsx 300 | Yes | Yes | Yes | Yes | 4-13 months | MDS/RAEB 2 of 8 MDS/AML 5 0f 8 | |
| N-Ras + BCL-2 TG mouse | No | No | Yes | Yes | 1 month | MDS/AML 5 of 8 | Omidvar et al 200760 |
Distinct phases of myelodysplastic syndrome (MDS) that require modeling
Disclosures Conflict-of-interest disclosure: The author declares no competing financial interests Off-label drug use: None disclosed.
Acknowledments
The author is grateful for support from a Leukemia & Lymphoma Society Specialized Center of Research (SCOR) grant, NIC R01 CA102202, NIH R01 DK52208 and R01 DK52621, and the Starr Foundation Stem Cell Initiative Award.
References
Author notes
Sloan-Kettering Institute, New York, NY