Abstract
Over the last three decades there have been dramatic advances in deciphering the cytogenetic and molecular lesions underlying the pathogenesis of acute myeloid leukemia (AML). These have not only afforded greater insights into disease biology, but also provided useful information predicting the likelihood of any given patient achieving and maintaining remission following conventional chemotherapy, leading to the development of risk-stratified treatment approaches. However, it is becoming increasingly apparent that AML is highly heterogeneous at the molecular level. Defining the individual genetic abnormalities or combinations of markers that provide significant independent prognostic information and establishing their respective relationships to other pre-treatment characteristics that impact on outcome, such as age and presenting white blood cell count, presents a major ongoing challenge. Moreover, there is increasing evidence that risk of relapse and overall survival can be predicted by assessment of kinetics and depth of response following front-line therapy and monitoring of the leukemic burden using molecular or immunological approaches to minimal residual disease (MRD) detection. These advances present the exciting prospect that panels of pre-treatment parameters affording independent prognostic information can be integrated with precise measurement of treatment response using MRD technologies to provide greater refinement in risk-adapted management of AML. This could lead to further improvements in outcome and serve to identify in a more reliable fashion those patients most likely to benefit from allogeneic transplant in first remission.
Current management of patients with acute myeloid leukemia (AML) is determined by a number of parameters, including age, performance status and the cytogenetic/molecular genetic characteristics of the leukemic clone. Together, these factors have an important bearing on treatment strategy, identifying potential candidates for molecularly targeted therapies (eg, all trans retinoic acid [ATRA] and arsenic trioxide in PML-RARA+ acute promyelocytic leukemia, or FLT3 inhibitors in AML with FLT3 mutations) and informing decisions on allogeneic transplantation.
In older adults (defined as older than 60 years) presenting with AML, who account for the majority with the disease and who generally have a poor prognosis, the major goal of pre-treatment assessment is to distinguish those patients who may benefit from intensive chemotherapy. Rapid cytogenetic analysis can be particularly helpful, identifying patients with AML with adverse karyotypic features, who have a particularly poor prognosis with conventional chemotherapy (less than 5% survival at 5 years). Such patients may therefore be considered as candidates for experimental treatment approaches, non-intensive therapy or supportive care (reviewed in Grimwade1).
In children and younger adults, one key objective of the diagnostic work-up is to distinguish patients at differing risk of relapse to guide the use of allogeneic transplantation in first complete remission (CR). Conventional allografting has been clearly shown to reduce rates of relapse as compared to standard intensive chemotherapy (reviewed in Rowe2), although the situation regarding overall survival (OS) is less clear. Given the procedure-related mortality associated with allogeneic transplantation, which is influenced by a range of factors, particularly patient age and type of transplant, one could argue that only those individuals at significant risk of relapse might be expected to gain any potential survival benefit from a transplant delivered in first CR. Accurate prediction of disease prognosis is therefore invaluable to help inform such treatment decisions, given the considerable expense associated with allogeneic transplantation as well as its longer term impact on health and quality of life.
Pre-treatment Prognostic Factors
Karyotype
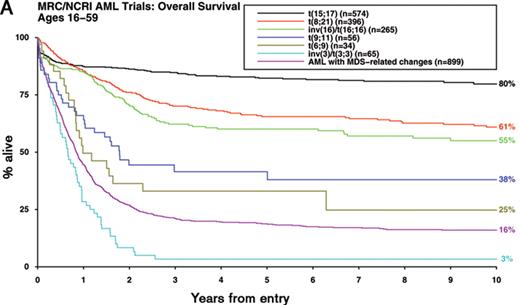
In approximately 60% of patients with AML, pre-treatment cytogenetic analysis reveals an abnormal karyotype.1 While there has been uncertainty as to which abnormalities represent primary events in the pathogenesis of AML, which provide critical second hits that are required for progression to full-blown leukemia and which are merely markers of the leukemic process, it is nevertheless clear that diagnostic karyotype is a key determinant of outcome.1 Moreover, there is mounting evidence that cytogenetic analysis can identify biologically distinct subsets of AML requiring tailored therapeutic approaches.1 Accordingly, the updated World Health Organisation classification of AML3 specifies a greater number of cytogenetic entities, which exhibit marked differences in disease outcome (Figure 1A ).
In multivariable analyses that take into account age, type of AML (de novo or secondary) and presenting white blood cell count (WBC), diagnostic karyotype emerges as the most significant prognostic factor and accordingly provides the framework for current risk-stratified treatment approaches. Large multicenter studies have consistently reported that patients with acute promyelocytic leukemia (APL) with the t(15;17)(q22;q12~21) treated on ATRA- and anthracycline-based protocols together with the core binding factor (CBF) leukemias with t(8;21)(q22;q22) or inv(16)(p13q22)/t(16;16)(p13;q22) treated with intensive chemotherapy involving cytarabine at a range of doses are characterized by relatively favorable prognoses1 (Figure 1A, Table 1 ). In this favorable risk group, relapse rates are too low and salvage rates too high for there to be any survival benefit for allogeneic transplantation in first remission.1,4 Conversely, adults presenting with AML and abnormalities of 3q [abn(3q)], deletions of 5q [del(5q)], monosomies of chromosome 5 and/or 7 (−5/−7) or complex karyotype have a very poor prognosis with conventional chemotherapy and are therefore considered candidates for allogeneic transplant and experimental treatment approaches.1
Although karyotype analysis provides a powerful independent prognostic factor for rates of CR, relapse risk and OS in multivariable analyses, there is uncertainty concerning a number of miscellaneous cytogenetic abnormalities that together account for ~10% of AML.1 The relative rarity of some of these abnormalities has presented a challenge in determining their prognostic significance, reflected in inconsistencies in risk assignment between different trial groups (Table 1 ). There has also been a lack of consensus regarding the prognostic impact of additional abnormalities accompanying the t(8;21), the outcome of AML with the various translocations involving the MLL locus at 11q23 and the level of cytogenetic complexity considered to confer adverse risk. Moreover, a recent study involving 1975 adults (aged 15–60 years) with AML has highlighted poor prognosis in the group with an autosomal monosomy in conjunction with at least one other autosomal monosomy or structural abnormality (denoted monosomal karyotype positive, MK+).5
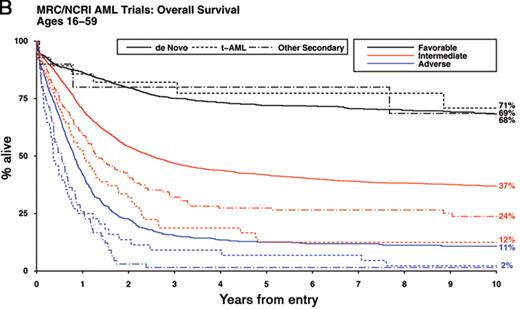
To address these issues with a view to achieving further refinement of cytogenetic classification of AML, which could ultimately facilitate comparison of clinical trial data from different groups, the Medical Research Council (MRC) has recently considered the outcome of a series of 5635 adult cases (aged 16–59 years) with successful karyotypic examination.6 First, in order to reconcile differences in the risk group assignment of t(8;21)-associated CBF leukemia (Table 1 ), the impact of additional abnormalities was considered in a cohort of 380 patients. No significant difference in OS was observed according to whether the t(8;21) was accompanied by deletions of the long arm of chromosome 9 [del(9q)] or by two or more unrelated abnormalities, as compared to those with t(8;21) alone. Subsequently, having excluded 1462 cases with t(15;17), t(8;21) and inv(16)/t(16;16) and after adjusting for age, presenting WBC and type of AML (de novo/secondary), Cox regression analyses revealed no other cytogenetic abnormalities conferring a relatively favorable prognosis. However, in these multivariable analyses, various abnormalities were found to predict a significantly poorer outcome, namely: abn(3q) [excluding t(3;5)(q21~25;q31~35)], inv(3)(q21q26)/t(3;3)(q21;q26), add(5q), del(5q), −5, −7, add(7q), t(6;11)(q27;q23), t(10;11)(p11~13;q23), t(9;22)(q34;q11), −17 and abn(17p) with other changes. Patients lacking these aberrations but with more than 3 unrelated abnormalities (designated “complex karyotype”) also exhibited a significantly poorer prognosis.6 These analyses conducted in this large study cohort have allowed further refinement of the original MRC cytogenetic classification,7 effectively reconciling a number of differences in the classification systems employed by other cooperative groups (Table 1 ). The revised MRC classification retains its prognostic significance in patients with secondary AML, who within each cytogenetic risk group had significantly poorer survival than that associated with de novo disease (Figure 1B ). In this large cohort, age, WBC, performance status and cytogenetics all retain independent prognostic significance in patients with secondary AML; the prognostic impact in this group of other presenting factors, such as prior chemotherapy, or organ damage is difficult to determine reliably. Although the proportion of patients falling within each cytogenetic risk group differs with age, with higher numbers of patients with adverse karyotype particularly beyond about 60 years of age (incidence 23% vs 15% in adults less than 60 years, P < .00001), the revised MRC classification was independently validated in a cohort of 2129 older adults (> 60 years) as well as in 895 children treated in the MRC AML trials (P < .00001 for each age group). Of note, in pediatric patients, outcomes for the intermediate and adverse risk groups are generally better than in adults, such that relatively few children are considered candidates for transplant in first CR.8 Despite a varying cytogenetic profile by age, age remains an important independent prognostic factor in AML.
Molecular Genetics
There are several limitations in the use of karyotype as a risk stratification tool; these include failed cytogenetic analyses, presence of cryptic chromosomal rearrangements, and notably the fact that a substantial proportion of AML cases (~40%) have a normal karyotype.1 However, in recent years there have been significant advances in defining genomic variations of pathogenic and independent prognostic relevance in AML, with mutation status of the genes encoding Fms-like tyrosine kinase 3 (FLT3), nucleophosmin (NPM1) and CCAAT/enhancer binding protein alpha (CEBPA) shown to be of major importance (reviewed in Mrózek et al9). The discovery of such mutations has meant that it is becoming increasingly possible to distinguish subsets of patients with differing outcomes from the large cohort with normal karyotype AML or miscellaneous cytogenetic abnormalities, which according to commonly applied cytogenetic classification systems would formerly have been grouped together, classed as “intermediate risk.”
FLT3 is a receptor tyrosine kinase expressed on hematopoietic progenitors, which is mutated in approximately a third of AML, including those with normal karyotype (reviewed in Scholl et al10). The majority of mutations are internal tandem duplications (ITDs) that lead to in-frame insertions within the juxtamembrane region of the receptor. Less frequent are mutations involving the region encoding the activation loop (tyrosine kinase domain, TKD), which most commonly affect codons aspartate 835 and isoleucine 836 (D835/I836), found in approximately 7% of patients with AML.10 Both classes of mutation lead to constitutive activation of the receptor and are associated with higher presenting WBC.10,11 While there are conflicting data concerning the prognostic implications of the TKD mutations,11–13 studies have consistently found that presence of FLT3-ITD provides a major independent adverse prognostic indicator, associated with an increased risk of relapse and poorer overall survival.9 Worse outcomes are observed in those patients with high FLT3-ITD allelic ratios,14–16 indicative of homozygous mutations generated by acquired uniparental disomy (UPD).17
CEBPA is a transcription factor involved in normal myelopoiesis, which is subject to mutation in ~10% of AML, being most frequently found in patients with normal karyotype.9 Mutations commonly involve the amino-terminal region of CEBPA, which lead to expression of a truncated protein or occur within the carboxy-terminus, disrupting regions required for dimerization and/or DNA binding. In a significant proportion of patients with AML with CEBPA mutations, both alleles are involved. These may be homozygous mutations resulting from acquired UPD involving the CEPBA locus on chromosome 1917 or biallelic compound heterozygous mutations. Interestingly, some patients harbor a germline mutation in CEBPA, with acquisition of a second mutation involved in progression to AML18. A number of studies suggested that CEBPA mutations predict a relatively favorable outcome in AML,9,19,20 with more recent reports considering larger patient cohorts suggesting that this effect relates to the group with biallelic mutations, who tend to have a normal karyotype and lack FLT3-ITD mutations.21,22
A major advance in understanding the molecular basis of AML was the discovery by Falini and colleagues that a third of cases, including approximately 50% with normal karyotype, harbor heterozygous mutations in the carboxy-terminus of the nucleolar phosphoprotein, nucleophosmin (NPM1), which lead to delocalization of the protein to the cytoplasm.23 There is increasing evidence that NPM1 mutations represent primary lesions in leukemogenesis, being generally mutually exclusive of balanced translocations.24 Interestingly, NPM1 mutation and FLT3-ITD commonly co-exist in normal karyotype AML, suggesting that they may cooperate in generating the leukemic phenotype. Presence of an NPM1 mutation is associated with higher presenting WBC, even allowing for FLT3 mutation status, but nevertheless has been consistently shown to predict a better outcome in terms of higher CR rate and reduced relapse risk compared to patients lacking the mutation.9,16
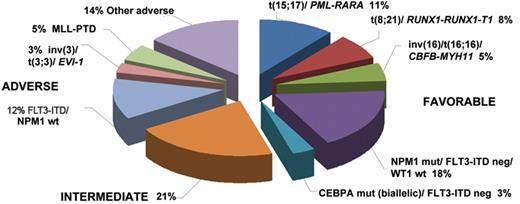
Recent studies have drawn attention to the complex interplay between mutations and their prognostic relevance, allowing further dissection of the intermediate karyotypic risk group (Figure 2 ). In particular, cases of AML with unmutated FLT3 in the presence of NPM1 or biallelic CEBPA mutations (which between them account for 21% of younger adults with AML) exhibit a prognosis akin to CBF leukemia.19–22,24,25 This has prompted the proposal that patients with AML with these configurations of molecular markers are unlikely to benefit from allogeneic transplantation in first CR owing to their relatively low risk of relapse.19–22,24,25 Conversely, molecular genetics can identify patients with intermediate risk cytogenetics, who have a very poor prognosis comparable to that observed in patients with adverse karyotype. This group includes those with partial tandem duplication in the MLL gene, detected in approximately 10% of patients with AML, particularly those with normal karyotype or trisomy 11 (reviewed in Grimwade1 and Mrózek et al9). Mutations in the Wilms’ tumor (WT1) gene, found in ~10% of patients with normal karyotype AML, have also been found to be associated with poorer prognosis in some studies,26–28 with a particularly bad outcome in combination with FLT3-ITD.26,27,29 Whether the outlook of patients with these molecularly defined subsets of AML can be improved by novel experimental approaches and whether they benefit from allogeneic transplant in first remission remains to be established.
A number of other recurring mutations have been identified in AML patients. RAS mutations are relatively common occurring in ~15% of cases, with evidence to date suggesting that they are prognostically neutral (reviewed in Grimwade1 and Mrózek et al9). Mutations in the TET2 gene have been identified in ~10% of patients with AML,30 including a quarter with secondary disease,31 and may confer a poor prognosis.30 The IDH1 gene has recently been reported to be mutated in a similar proportion of AML cases.44 Other mutations, such as those involving KIT, PTPN11, RUNX1 and CBL, are relatively rare (identified in < 5% of cases), making their relevance to risk-stratified treatment approaches uncertain at the present time.
Impact of Over-expressed Genes on Disease Outcome
There have been a number of reports linking over-expression of particular genes and prognosis in AML, which in some instances have been shown to discriminate patients at differing risk of relapse within cytogenetic risk groups. These include the EVI1 (ecotropic virus integration-1) gene located at 3q26, which is upregulated as a result of the inv(3)(q21q26) and t(3;3)(q21;q26) rearrangements.1 A recent study lends support to routine screening by real-time quantitative polymerase chain reaction (RQ-PCR) for EVI1 expression, which can be associated with cytogenetically cryptic 3q26 abnormalities, to identify a subgroup of patients with a particularly poor prognosis.32 The molecular basis of up-regulation of other genes that have been associated with poorer prognosis such as MN1, BAALC, FLT3 and ERG remains poorly understood (reviewed in Mrózek et al9). Since the level of expression of these markers appears to be a continuum in AML, “up-regulation” has been defined variously on the basis of expression levels falling within the upper quartile or above the median. As such, cases with very similar levels of expression and comparable outcomes could just straddle the cut-point and end up being assigned to different risk groups. Given this limitation, establishing the clinical utility of such markers as tools to achieve greater individualization of patient care will necessitate the setting of robust validated thresholds of expression relative to standard housekeeping genes (eg, ABL) and importantly evidence that these defined thresholds refine outcome prediction after allowing for cytogenetics and the mutation status of NPM1, FLT3-ITD and CEBPA. It is possible that the relative expression of some of these markers reflect differences in the nature of the hematopoietic progenitors subject to leukemic transformation and/or the characteristics and relative size of the leukemic stem cell compartment. Immunophenotypic profiles of the leukemic population may provide similar prognostic information. Previous studies have shown that BCL2/BAX ratio and drug resistance protein expression are predictive of treatment response and risk of relapse (reviewed in Grimwade1). Similarly, poor outcomes have been observed in AML with a relatively large early progenitor population defined by a CD34+/CD38− phenotype.33 These parameters may become increasingly relevant as newer agents modulating drug resistance or targeting the leukemic stem cell pool become available. Expression of factors that may relate to interactions of leukemic cells with the bone marrow microenvironment (eg, VEGF, CXCR4, reviewed in Lane et al34) or gene variants that affect drug handling have also been found to impact on outcome. However, their relationship to other well-established prognostic factors is poorly defined and hence their clinical utility remains to be established.
A number of studies have highlighted the potential of microarrays to evaluate expression of mRNA transcripts or microRNAs as a tool to distinguish subgroups of patients with differing prognosis (reviewed in Wouters et al35). These platforms have already shown their capacity to identify currently well-recognized cytogenetically or molecularly defined subsets of leukemia, such as those with t(15;17)/PML-RARA.35 A key question is whether these approaches ultimately realize their promise of identifying particular targets or discrete constellations of novel markers distinguishing patients at differing risk of relapse that are independent of well-established prognostic factors, which can be accurately measured in a robust fashion and validated in independent data sets. However, given the considerable challenges relating to the performance of microarrays and conduct of the downstream bioinformatic analyses in a standardized fashion, these technologies appear somewhat unlikely to shape the way in which individual patients with AML are treated in the very near future.
Post-treatment Prognostic Factors: Residual Disease Detection
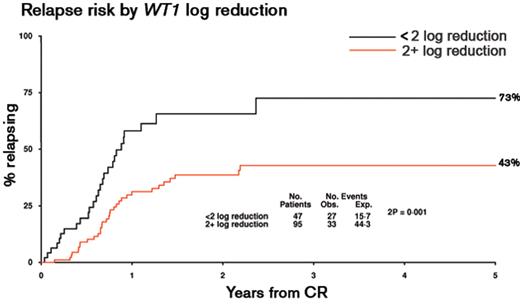
Response to induction therapy has long been recognized as a major independent prognostic factor in AML, predicting risk of relapse and overall survival, leading to the development of standardized response criteria.36 While the percentage of residual leukemic blasts following induction has been used to refine risk stratification, morphological appearances can be difficult to interpret. To provide a more objective and sensitive approach to assess treatment response to induction therapy, numerous studies have applied flow cytometry or RQ-PCR to detect minimal residual disease (MRD). The former depends upon the characterization of an aberrant “leukemia associated immunophenotype” (LAIP) of the blast population at diagnosis, which can be defined in over 90% of patients with AML (reviewed in Freeman et al37). The RQ-PCR approach relies on the detection of leukemia-specific targets (ie, fusion genes [eg, PML-RARA, RUNX1-RUNX1T1, CBFB-MYH11] or mutations [eg, mutant NPM1]) or genes that are commonly over-expressed in AML, particularly WT1.37 The sensitivity limit for identification of MRD is technique and target dependent. For flow cytometry, typically 1 AML cell in 103–104 non-leukemic cells can be detected.37 For RQ-PCR detection of leukemia-specific targets, maximal sensitivities range between 1 in 103 to 106 depending upon the relative level of expression (as compared with a housekeeping gene, eg, ABL) that may vary, not only between molecular subtypes of AML, but also between cases with the same target (reviewed in Freeman et al37). The sensitivity of the WT1 assay (evaluable in ~45% of patients with AML) is limited by background levels of expression in normal blood and marrow, such that it is most appropriately used to measure early treatment responses (Figure 3 ) rather than for sequential MRD monitoring.38
Studies using RQ-PCR detection of WT1 transcripts or flow cytometry to assess reduction in leukemic burden following induction and consolidation therapy have shown that both approaches can distinguish patients at differing risk of relapse within cytogenetic risk groups; they retain their prognostic significance when other factors such as age and type of AML (de novo/secondary) are taken into account.37–39 Studies using RQ-PCR have also consistently shown that response kinetics are predictive of risk of subsequent relapse in CBF leukemias, although the relationship to other prognostic factors such as presenting WBC and KIT mutation status remains unclear (reviewed in Freeman et al37).
In addition to investigating the potential of MRD detection to give a risk stratification tool, longitudinal monitoring may provide early warning of impending relapse, allowing the opportunity for pre-emptive therapy. This approach has been most extensively studied in APL in which sequential monitoring of PML-RARA fusion transcripts by RQ-PCR provides the strongest predictor of relapse, far superior in multivariable analyses to presenting WBC,40 which has traditionally been used as the basis for risk-stratified treatment approaches in this subtype of AML.41 Evidence to date suggests that sequential MRD monitoring to direct preemptive therapy can reduce rates of frank relapse and improve survival in APL.40,41 Whether sequential MRD monitoring is useful in other subsets of AML remains an important question for ongoing studies.
Prospects for Refining Outcome Prediction in AML
The last 15 years have seen major advances in predicting the outcome of patients with newly diagnosed AML, and these have informed treatment approach. Greater understanding of the cytogenetic and molecular basis of AML and the biology of the subtypes of disease that predominate in different age groups have provided major insights into the parameters underlying a number of long-established prognostic factors such as age, presenting WBC and whether AML arises de novo or following an antecedent hematologic disorder or chemotherapy/radiation exposure. The evidence suggesting that cytogenetics can distinguish biologically distinct subsets of AML has had a major impact upon clinical practice, being used to guide consolidation therapy, particularly with respect to allogeneic transplantation. While there is increasing consensus that there is no net benefit for routine use of allogeneic transplantation in patients with PML-RARA+ APL or the CBF leukemias, lack of consistency in the cytogenetic classification of AML has hampered the reliable determination of those groups most likely to benefit. The recent meta-analysis of data from four study groups by Cornelissen et al has suggested a benefit for allogeneic transplant in first CR in younger adults presenting with AML with intermediate and adverse risk karyotypic features.4 This study proposed that a predicted relapse risk exceeding 35% serves as a useful threshold to identify younger adults who may gain a net benefit from allogeneic transplant in first CR.4 However, it is important to note that the cytogenetic risk grouping used in this study differed in some respects from those employed by other cooperative groups (Table 1 ). Additionally, these data came from trials run some time ago; the effect of transplantation compared with more modern chemotherapy regimens (including, for example, gemtuzumab ozogamicin) needs to be established. The issue of assessment of allogeneic transplantation in first remission is the matter of current debate: the advent of transplants from unrelated donors as well as the well-known benefits of allograft in second remission means that traditional “sibling donor versus no sibling donor” analyses may no longer be relevant. As an alternative, the MRC has used Mantel-Byar analysis to adjust for time to transplant: in retrospective analyses, a newly developed risk index, taking into account age, presenting WBC, type of AML (de novo/secondary), karyotype and blast percentage in the post-induction bone marrow identified a group of poor-risk patients who were the only group to show a survival benefit for allogeneic transplantation.42 The additional impact of molecular markers, particularly mutation status of NPM1, CEBPA and FLT3 still needs to be assessed in this context. While the role of allogeneic transplantation in patients with intermediate risk cytogenetics and FLT3-ITD remains somewhat controversial (reviewed in Rowe2), a recent large study conducted by the German AMLSG that was limited to patients with normal karyotype AML suggested on the basis of a “donor vs no donor” comparison that allogeneic transplant confers no overall survival advantage in patients with NPM1 mutant/FLT3-ITD− AML, whilst finding a beneficial effect of transplantation in FLT3-ITD+ patients.19 Moreover, recent studies suggest that AML characterized by biallelic CEBPA mutations with unmutated FLT3 represent a group with relatively favorable prognosis, who would appear unlikely to benefit from allogeneic transplant.20–22 In contrast, adverse prognosis associated with high risk of disease relapse has been observed in patients with AML with homozygous FLT3-ITD mutations, MLL-PTD, EVI-1 expression or with coexisting FLT3-ITD and WT1 mutations. These data may inform revised hierarchical cytogenetic and molecular risk groups and facilitate development of consensus in reporting data from clinical trials investigating treatment strategies in younger adults with AML (Table 2, Figure 2 ).
Despite these refinements, risk-stratification schemes based upon pre-treatment factors still distinguish relatively large groups of patients with broadly differing risk of relapse. Studies using flow cytometry or RQ-PCR have highlighted the potential of these techniques, which measure treatment response to achieve greater refinement, distinguishing further subgroups of patients at differing risk of relapse.37 Integration of panels of pre-treatment prognostic factors with MRD data based upon validated informative thresholds should allow development of more reliable risk-adapted treatment approaches to AML. Given that a leukemia-specific molecular target can be defined in at least 60% of patients with AML and following the introduction of flow cytometry with up to 10 colors, the longitudinal tracking of MRD status to pinpoint those patients who would be likely to relapse without additional therapy is becoming an increasingly realistic possibility. This strategy has been pioneered in APL and may allow a more individualized treatment approach in other disease subsets, potentially identifying those patients likely to benefit from further or alternative treatments including allogeneic transplant in first CR. Conversely, in older adults the focus is upon identifying patient- and leukemia-specific factors that identify patients who may be most appropriately treated using intensive or non-intensive treatment approaches.
Development of risk-directed treatment strategies is an evolving process. Further advances may arise from whole genome sequencing, with early studies suggesting that AML pathogenesis may be contingent upon cooperation between multiple mutations, with a number of particular genomic alterations being infrequent in AML as a whole or even private to individual leukemias.43,44 Many mutations may prove to be too infrequent to confidently determine their prognostic significance. However, it seems likely that the relevance of such mutations to common molecular networks involved in the generation and maintenance of leukemic stem cells and the pathogenesis of AML will become apparent over the next few years. Characterization of AML has hitherto been largely based on analysis of the bulk leukemic population, which may not effectively capture the biology of the leukemic stem cell compartment and the interaction with the marrow microenvironment. Future advances in these areas may ultimately yield the next generation of independent prognostic factors, predicting response to novel targeted therapies.
Variation in cytogenetic risk group classification across clinical trial groups. References for the various classification systems can be found in Grimwade.1 The HOVON/SAKK cytogenetic classification was derived from the study by Cornelissen et al.4 The revised MRC classification system was based on an analysis of 5635 patients aged 16 to 59 years enrolled in the MRC AML10, AML12 and AML15 trials.6
| . | Original MRC . | SWOG/ECOG . | CALGB . | GIMEMA/AML10 . | German AMLCG . | HOVON/SAKK . | Refined MRC . |
|---|---|---|---|---|---|---|---|
| Unrel abn indicates unrelated abnormality; abn, abnormal. | |||||||
| Favorable | t(15;17) t(8;21) inv(16)/t(16;16) | t(15;17) t(8;21) [lacking del(9q), complex, ie, ≥ 3 unrel abn] inv(16)/t(16;16)/del(16q) | t(15;17) t(8;21) inv(16)/t(16;16) | t(15;17) t(8;21) inv(16)/t(16;16) | t(15;17) t(8;21) inv(16)/t(16;16) | t(15;17) t(8;21) alone inv/del(16) and lacking unfav abn | t(15;17) t(8;21) inv(16)/t(16;16) |
| Intermediate | Normal Other non-complex | Normal +6, +8, -Y, del(12p) | Normal Other non- complex | Normal -Y | Normal Other non- complex | Normal Other non- complex | Normal Other non- complex |
| Adverse | abn(3q) -5/del(5q) -7 complex [≥ 5 unrel abn] Excluding those with favorable changes | abn(3q),(9q),(11q),(21q) abn(17p) -5/del(5q) -7/del(7q) t(6;9) t(9;22) complex [≥3 unrel abn] | inv(3)/t(3;3) -7 t(6;9) t(6;11) t(11;19) +8 complex (≥ 3 unrel abn) Excluding those with favorable changes | Other | inv(3)/t(3;3) -5/del(5q) -7/del(7q) abn(11q23) del(12p) abn(17p) complex (≥ 3 unrel abn) | abn(3q) -5/del(5q) -7/del(7q) abn(11q23) t(6;9) t(9;22) complex (≥ 3 unrel abn) | abn(3q) [excluding t(3;5)] inv(3)/t(3;3) add(5q)/del(5q)/ -5,-7/add(7q) t(6;11) t(10;11) t(9;22) -17 abn(17p) with other changes Complex (> 3 unrel abn) Excluding those with favorable changes |
| . | Original MRC . | SWOG/ECOG . | CALGB . | GIMEMA/AML10 . | German AMLCG . | HOVON/SAKK . | Refined MRC . |
|---|---|---|---|---|---|---|---|
| Unrel abn indicates unrelated abnormality; abn, abnormal. | |||||||
| Favorable | t(15;17) t(8;21) inv(16)/t(16;16) | t(15;17) t(8;21) [lacking del(9q), complex, ie, ≥ 3 unrel abn] inv(16)/t(16;16)/del(16q) | t(15;17) t(8;21) inv(16)/t(16;16) | t(15;17) t(8;21) inv(16)/t(16;16) | t(15;17) t(8;21) inv(16)/t(16;16) | t(15;17) t(8;21) alone inv/del(16) and lacking unfav abn | t(15;17) t(8;21) inv(16)/t(16;16) |
| Intermediate | Normal Other non-complex | Normal +6, +8, -Y, del(12p) | Normal Other non- complex | Normal -Y | Normal Other non- complex | Normal Other non- complex | Normal Other non- complex |
| Adverse | abn(3q) -5/del(5q) -7 complex [≥ 5 unrel abn] Excluding those with favorable changes | abn(3q),(9q),(11q),(21q) abn(17p) -5/del(5q) -7/del(7q) t(6;9) t(9;22) complex [≥3 unrel abn] | inv(3)/t(3;3) -7 t(6;9) t(6;11) t(11;19) +8 complex (≥ 3 unrel abn) Excluding those with favorable changes | Other | inv(3)/t(3;3) -5/del(5q) -7/del(7q) abn(11q23) del(12p) abn(17p) complex (≥ 3 unrel abn) | abn(3q) -5/del(5q) -7/del(7q) abn(11q23) t(6;9) t(9;22) complex (≥ 3 unrel abn) | abn(3q) [excluding t(3;5)] inv(3)/t(3;3) add(5q)/del(5q)/ -5,-7/add(7q) t(6;11) t(10;11) t(9;22) -17 abn(17p) with other changes Complex (> 3 unrel abn) Excluding those with favorable changes |
Pre-treatment cytogenetic and molecular entities shown to predict disease outcome in multivariable analysis studies conducted in younger adults.
| . | Cytogenetic/molecular abnormality . | Comments . |
|---|---|---|
| Favorable | t(15;17)(q22;q12~21)/PML-RARA t(8;21)(q22;q22)/RUNX1-RUNX1T1 inv(16)(p13q22)/t(16;16)(p13;q22)/CBFB-MYH11 | Irrespective of additional cytogenetic abnormalities |
| NPM1 mutant/FLT3-ITD−, WT1 wild type CEBPA mutant (biallelic, FLT3-ITD−) | ||
| Intermediate | Entities not classified as favorable or adverse | |
| Adverse | abn(3q) [excluding t(3;5)(q21~25;q31~35)], inv(3)(q21q26)/t(3;3)(q21;q26)/EVI-1 expression add(5q), del(5q), −5, −7, add(7q), t(6;11)(q27;q23), t(10;11)(p11~13;q23), t(9;22)(q34;q11), −17, abn(17p) with other changes Complex (>3 unrelated abnormalities) | Excluding cases with favorable karyotype |
| FLT3-ITD | In absence of favorable karyotype. Particularly poor prognosis with high level FLT3-ITD mutant ratio or if FLT3-ITD accompanied by WT1 mutation | |
| MLL-PTD |
| . | Cytogenetic/molecular abnormality . | Comments . |
|---|---|---|
| Favorable | t(15;17)(q22;q12~21)/PML-RARA t(8;21)(q22;q22)/RUNX1-RUNX1T1 inv(16)(p13q22)/t(16;16)(p13;q22)/CBFB-MYH11 | Irrespective of additional cytogenetic abnormalities |
| NPM1 mutant/FLT3-ITD−, WT1 wild type CEBPA mutant (biallelic, FLT3-ITD−) | ||
| Intermediate | Entities not classified as favorable or adverse | |
| Adverse | abn(3q) [excluding t(3;5)(q21~25;q31~35)], inv(3)(q21q26)/t(3;3)(q21;q26)/EVI-1 expression add(5q), del(5q), −5, −7, add(7q), t(6;11)(q27;q23), t(10;11)(p11~13;q23), t(9;22)(q34;q11), −17, abn(17p) with other changes Complex (>3 unrelated abnormalities) | Excluding cases with favorable karyotype |
| FLT3-ITD | In absence of favorable karyotype. Particularly poor prognosis with high level FLT3-ITD mutant ratio or if FLT3-ITD accompanied by WT1 mutation | |
| MLL-PTD |
Impact of karyotype on outcome in younger adults with AML. Overall survival is shown for adults with AML (median age 43 years, range 16–59) treated in successive Medical Research Council/National Cancer Research Institute trials (MRC AML10, AML12, AML15). A) Outcome of cytogenetic entities specified in the 2008 WHO classification3 B) Outcome of de novo, therapy-related and other secondary AML according to cytogenetic risk group (using revised MRC classification provided in Table 1) All patients with t(15;17) received an extended course of ATRA in addition to anthracycline-based chemotherapy.
Impact of karyotype on outcome in younger adults with AML. Overall survival is shown for adults with AML (median age 43 years, range 16–59) treated in successive Medical Research Council/National Cancer Research Institute trials (MRC AML10, AML12, AML15). A) Outcome of cytogenetic entities specified in the 2008 WHO classification3 B) Outcome of de novo, therapy-related and other secondary AML according to cytogenetic risk group (using revised MRC classification provided in Table 1) All patients with t(15;17) received an extended course of ATRA in addition to anthracycline-based chemotherapy.
Frequency of prognostically relevant molecular and cytogenetic subgroups of AML arising in younger adults. Frequencies of the various entities specified in Table 2 are based on a synthesis of published data 9,16,19–25 and analysis of adults with AML entered into the MRC AML10 and 12 trials.6
Kinetics of minimal residual disease response following induction therapy are predictive of subsequent relapse risk in AML. The predictive value of MRD assessment by WT1 RQ-PCR assay was determined in a cohort of 142 patients with AML treated with conventional anthracycline- and cytarabine-based treatment. Analysis was undertaken in patients with AML with WT1 expression exceeding 2 × 104 copies/104ABL copies in pre-treatment samples, allowing the detection of at least a 2-log reduction in WT1 transcripts following induction, taking into account the background level of expression observed in normal hematopoietic tissues.38 The patient cohort included 91 cases reported previously38 combined with a further 51 cases treated in the MRC AML15 trial (samples kindly provided by John Yin and Michelle Sale, Manchester Royal Infirmary, and analyzed by Neesa Bhudia, Guy’s Hospital, London, UK). Samples were analyzed with a standardized WT1 RQ-PCR assay (Ipsogen, Marseille, France) developed within the European LeukemiaNet, as described.38
Kinetics of minimal residual disease response following induction therapy are predictive of subsequent relapse risk in AML. The predictive value of MRD assessment by WT1 RQ-PCR assay was determined in a cohort of 142 patients with AML treated with conventional anthracycline- and cytarabine-based treatment. Analysis was undertaken in patients with AML with WT1 expression exceeding 2 × 104 copies/104ABL copies in pre-treatment samples, allowing the detection of at least a 2-log reduction in WT1 transcripts following induction, taking into account the background level of expression observed in normal hematopoietic tissues.38 The patient cohort included 91 cases reported previously38 combined with a further 51 cases treated in the MRC AML15 trial (samples kindly provided by John Yin and Michelle Sale, Manchester Royal Infirmary, and analyzed by Neesa Bhudia, Guy’s Hospital, London, UK). Samples were analyzed with a standardized WT1 RQ-PCR assay (Ipsogen, Marseille, France) developed within the European LeukemiaNet, as described.38
Disclosures Conflict-of-interest: DG is on an advisory committee for Ipsogen (Marseille, France). RKH declares no competing financial interests. Off-label drug use: None disclosed.
Acknowledgments
We are grateful to the UK MRC/NCRI AML Working Party for access to the MRC/NCRI AML trials database. We are indebted to Alan Burnett, Rosemary Gale, Charles Craddock, Torsten Haferlach and Francesco Lo Coco for critical review of the manuscript and providing helpful suggestions for improvement. DG gratefully acknowledges Leukaemia Research of Great Britain for support for minimal residual studies in the UK NCRI AML trials. DG and RKH are also grateful for support from the MRD (WP12) and AML (WP5) workpackages of the European LeukemiaNet.
References
Author notes
Department of Medical & Molecular Genetics, King’s College London School of Medicine, London, United Kingdom
Department of Haematology, School of Medicine, Cardiff University, Cardiff, United Kingdom