Abstract
Fanconi anemia (FA) is the most frequent inherited cause of BM failure (BMF). Fifteen FANC genes have been identified to date, the most prevalent being FANCA, FANCC, FANCG, and FANCD2. In addition to classical presentations with progressive BMF during childhood and a positive chromosome breakage test in the blood, atypical clinical and/or biological situations can be seen in which a FA diagnosis has to be confirmed or eliminated. For this, a range of biological tools have been developed, including analysis of skin fibroblasts. FA patients experience a strong selective pressure in the BM that predisposes to clonal evolution and to the emergence in their teens or young adulthood of myelodysplasia syndrome (MDS) and/or acute myeloid leukemia (AML) with a specific pattern of somatic chromosomal lesions. The cellular mechanisms underlying (1) the hematopoietic defect which leads to progressive BMF and (2) somatic clonal evolutions in this background, are still largely elusive. Elucidation of these mechanisms at the molecular and cellular levels should be useful to understand the physiopathology of the disease and to adapt the follow-up and treatment of FA patients. This may also ultimately benefit older, non-FA patients with aplastic anemia, MDS/AML for whom FA represents a model genetic condition.
Introduction
Fanconi anemia (FA) is the most frequent inherited cause of BM failure (BMF).1 The FA genes (genes that have been to be found mutated in FA patients) are called FANC, the most frequent being FANCA, FANCC, FANCG, and FANCD2.2 Except for the very rare FANCB, which is located on the X chromosome,3 all other FANC genes are autosomic and the disease is recessive. There are typically several clinical stages in FA that are related to age.1,4,5 At birth and early childhood, only physical signs are present and range from discreet to extensive. Many patients then experience BMF between 5 and 15 years of age, and the diagnosis of FA is often made at this stage. Later on, during their teens or young adulthood, the risk of acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS)/AML becomes very high. Still later, in adult patients, a range of solid cancers can be seen, especially mouth cancer. Throughout life, the hematopoietic situation can change spontaneously by genetic reversion or other clonal evolution, including progression to MDS or AML. Hematopoietic stem cell transplantation (HSCT) is currently the best treatment to cure severe aplastic anemia or MDS/AML.6 However, HSCT has to be carefully considered based upon clinical and biological criteria including age, severity of the cytopenia, significant BM dysplasia, excess of blast cells, cytogenetic abnormalities, and immunological compatibility with the donor.
The FA/BRCA biological pathway
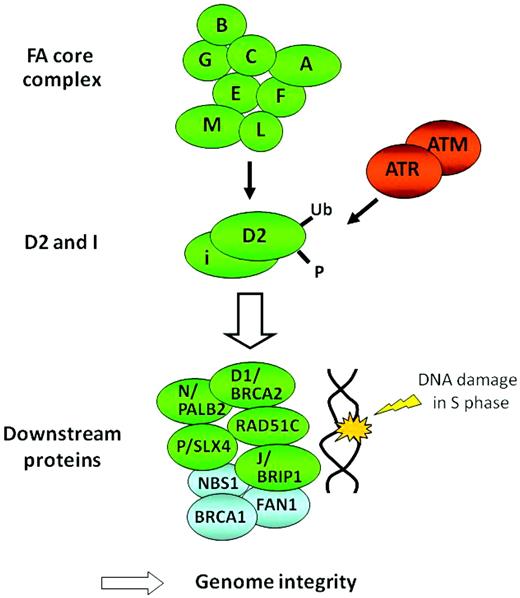
To date, 15 FA genes have been identified (FANCA, FANCB, FANCC, FANCD1/BRCA2, FANCD2, FANCE, FANCF, FANCG, FANCI, FANCJ/BRIP1, FANCL, FANCM, FANCN/PALB2, FANCO/RAD51C, and FANCP/SLX4).2,7 It is noteworthy that FANCD1 is identical to the BRCA2 gene, one of the genes predisposing to hereditary breast and ovarian cancer.8 Products of the FA genes function in a common DNA repair signaling pathway, the FA/BRCA pathway (Figure 1), which closely cooperates with other DNA repair proteins for resolving DNA interstrand cross-links (ICLs) during replication (reviewed in Moldovan et al7 and in Kee et al9 ). A central event in the pathway is the mono-ubiquitination of FANCD2 and FANCI upon DNA damage, which is mediated by a group of upstream FA proteins (FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL, and FANCM) that are assembled into a large nuclear E3 ubiquitin ligase complex called the FA core complex. The mono-ubiquitinated FANCD2 and FANCI heterodimer functionally interacts with downstream FA proteins such as FANCD1/BRCA2, FANCN/PALB2, FANCJ/BRIP1, FANCP/SLX4, RAD51C, and their associated protein, BRCA1. FAN1 (Fanconi-associated nuclease 1), a recently identified FA-associated protein, provides a nuclease activity during the ICL repair.10–13 FA proteins could also have other functions or participate in pathways other than DNA repair in response to cellular stress.14–17
FA diagnosis
FA patients often, but not always, present with a combination of various congenital abnormalities such as short stature; Fanconi facies and microphthalmia; thumb and radius deformities; skin hyperpigmentation such as “café-au-lait spots”; and cardiac, renal, genitourinary, and/or other malformations (reviewed in Shimamura and Alter1 ). Physical abnormalities can be subtle or absent. Many FA patients develop BMF some time during the course of the disease, usually during their first and second decades of life,4,5 and for the majority of patients, the suspicion of FA will only be made after the onset of pancytopenia. In some patients, the underlying diagnosis of FA is not known until MDS/AML occurs. Increases of fetal hemoglobin, serum alpha-fetoprotein, and macrocytosis are commonly noted in FA, but their absence does not rule out the disease (although not specific for FA, they may help to distinguish an inherited from an acquired BMF).1 A significant, and probably underestimated, number of patients do not develop overt BMF but still have an increased risk of malignancies.
It is crucial for family counseling and treatment centers to identify patients with FA. Patients with BMF who happen to have underlying undiagnosed FA will not respond to the immunosuppression therapy used to treat patients with idiopathic aplastic anemia. Moreover, due to a hypersensitivity to chemotherapy agents, patients with FA will often die of toxicity if given conventional conditioning for HSCT, and therefore less myeloablative regimens are used in this population.6 In addition, to being at a higher risk for developing malignancies, patients with FA need appropriate cancer surveillance throughout life.18,19 Therefore, the consensus is that FA has to be screened for in all children or young adults with hypoplastic or aplastic anemia or cytopenias. It is also safe to screen children and young adults with MDS or AML with physical signs and/or FA-associated chromosomal abnormalities in the BM, such as 1q+ or 3q+ (see below).
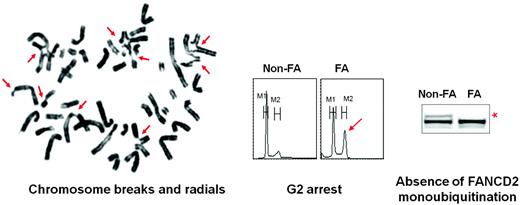
Due to the many genes and mutations associated with FA, a single genetic test cannot be used as a first approach for FA diagnosis in unselected BMF patients. The biological diagnosis of FA is primarily based on the exquisite sensitivity of FA cells to DNA ICL chemicals such as diepoxybutane or mitomycin C. The chromosomal breakage test with these agents is the technique of reference for diagnosing FA.1,20 In the majority of cases, a precise diagnosis can be made with careful history, physical examination, and a positive chromosomal breakage blood test (Figure 2). Other blood tests include cell-cycle analysis21 and evaluation of FANCD2 mono-ubiquitination, which can positively diagnose FA core patients.22 However, all of these tests can be ambiguous or even falsely negative in patients who develop hematopoietic reversion and somatic mosaicism. Hematopoietic reversion occurs when, after a spontaneous genetic event in a hematopoietic stem cell (ie, a reverse point mutation or intragenic recombination), one FA allele is corrected, with a consequent recovery of a normal or subnormal protein activity and cellular phenotype.23,24 Because there has been no evidence that this same phenomenon could happen in primary skin fibroblasts, these cells have been used to overcome misleading results in blood due to somatic mosaicism.25–28 Primary fibroblast cells can be tested for their ICL sensitivity by chromosome breaks (although less conveniently than in blood cells), growth curves, FANCD2 test, or flow-derived tests.26–29 Moreover, fibroblast DNA is useful for subsequent screening for the FANC mutation and also as a constitutional reference when analyzing somatic molecular and chromosomal abnormalities in the BM.30 Once the FA diagnosis is established at the cellular level, FANC gene mutations can be screened. The most frequent gene (FANCA) is analyzed first, then FANCG, FANCC, and so on. Alternatively, or in difficult cases, retroviral complementation can be used to ascertain the FA group before sequencing. The FANCD2 mono-ubiquitination test in fibroblasts is useful to orientate the search toward a FA core, FANCD2 or downstream gene. FA mutations are being referred to in the Fanconi Anemia Mutation Database (www.rockefeller.edu/fanconi). For FANCD2 and genes from the downstream groups, including FANCD1/BRCA2, at least one of the mutations is hypomorphic because the complete absence of protein is lethal.
FA biological diagnosis is based on the hypersensitivity of FA cells to ICL agents.
FA biological diagnosis is based on the hypersensitivity of FA cells to ICL agents.
There are few clear genetic-phenotype correlations in the classical FA core patients, although “hypomorphic” mutations might be associated with milder phenotypes and could allow time to develop to clonal evolution and late-onset solid cancer.31–33 In contrast, FANCD2 patients usually experience a more severe phenotype,34 and FANCD1/BRCA2 patients develop an early and rapidly lethal cancer-prone syndrome.35–37
Clonal evolution, MDS, and AML in FA patients
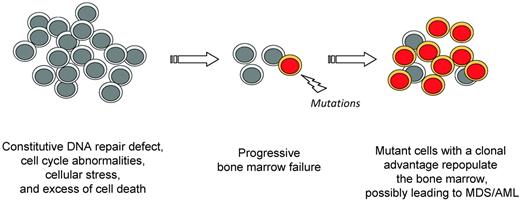
FA patients have a cumulative hematopoietic dysfunction likely related to an excess of genetic instability, cellular stress, p53 activation and cell death, which is proved to be intrinsic by the excellent efficiency of allogeneic HSCT in this population. In such a background of deficient hematopoiesis, FA BM cells experience a strong selective pressure toward clonal evolution, probably favored by the constitutive chromosomal instability (Figure 3).38–40 Myelodysplasia and AML cells have experienced clonal genetic evolution. Although a case of an AML cell line (from the D1/BRCA2 group) with genetic reversion has been reported,41 reversion does not seem frequent in the MDS and AML of FA patients,30 suggesting that genetic and/or epigenetic evolutions other than FA reversion happen. Acquired attenuation of the FA-prototypical G2 checkpoint and resistance to TNFα have been described in humans and mice, respectively.42,43 It is likely that these and other cellular phenotypes rescue FA cells and confer a selective advantage, but also predispose patients to develop malignancies. Therefore, a thorough characterization of the molecular and cellular features associated with the BM progression in FA should lead to an understanding the step-wise mechanisms of transformation, and to better prevention or treatment in these patients.
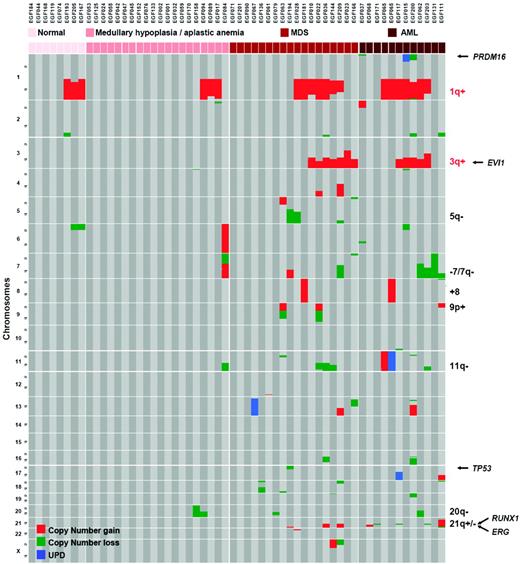
Clinically, FA patients experience MDS and AML with the highest frequency during their teens or young adulthood.1,44 These diseases are often, but not always, preceded by an hypoplastic or aplastic phase. MDS in FA often presents as refractory cytopenia with multilineage dysplasia (according to the WHO 2008 classification), with or without excess of blasts.30,45 A certain level of dyserythropoiesis is almost constant in FA, and a mild dyserythropoiesis is not considered as an MDS criteria in this population. Acute leukemia can be diagnosed primarily (in approximately 30% of the AML in FA patients) or after a MDS phase with an increasing fraction of blast cells in the BM. Karyotypic abnormalities are frequently found, and translocations of chromosome 1q, monosomy 7, and gains of 3q have been reported.30,44–48 Using array-based high-density chromosomal profiling and oncogene sequencing, we recently analyzed BM samples from a large series of FA patients at various stages of the disease (aplastic, MDS, and AML).30 We found a highly recurrent pattern of somatic abnormalities that were related to unbalanced chromosomal translocations and led to partial chromosomal arm duplications or losses (Figure 4). In contrast to what is seen in non-FA MDS/AML patients,49 somatic copy-neutral loss of heterozygosity (uniparental disomy) were rarely found in FA, which is consistent with a constitutive defect of homologous recombination repair in FA. The most frequent lesion was partial duplication of chromosome 1q (1q+, 44.8% of 29 MDS/AML patients), after by 3q+ (41.3%), RUNX1/21q− (20.7%), monosomy 7/7q− (17.2%), and 11q− (13.8%). Mutations of MDS/AML oncogenes and tumor-suppressor genes were rarely found (isolated FLT3-ITD, NRAS, MLL-PTD, but no TP53, CBL, TET2, CEBPα, NPM1, and FLT3-TKD mutations in this series). It appears that the oncogenesis in FA shares common pathways with non-FA patients (monosomy 7/7q−, 5q−, RUNX1/21q− lesions, PRDM16 translocations, MLL-PTD), but also has additional FA-specific chromosomal lesions, especially 1q+ and 3q+. The molecular targets in these 2 chromosomal lesions are not known, although the EVI1 oncogene at 3q26 is a candidate. Interestingly, whereas the lesions 7q−, 3q+, and RUNX1 abnormalities were found at the MDS and AML stages only, translocation/duplications 1q+ can be seen at all stages in the BM, including “normal” or hypoplastic BM without apparent transformation signs (Figure 4), suggesting that 1q+ could rescue the FA cells without necessarily transforming them into MDS/AML.30
A recurrent profile of acquired chromosomal abnormalities in BM samples of FA patients. Samples are grouped by BM stages (normal, aplasia, MDS, and AML). The 1q+ and 3q+ lesions are specific from FA compared with MDS/AML of non-FA patients. Other abnormalities, including 7q− and RUNX1 mutations, can be found in non-FA patients. 1q+ is found at all stages of the BM progression, whereas most other lesions are found at the most advanced MDS/AML stages. (Used with permission from Quentin et al.30 )
A recurrent profile of acquired chromosomal abnormalities in BM samples of FA patients. Samples are grouped by BM stages (normal, aplasia, MDS, and AML). The 1q+ and 3q+ lesions are specific from FA compared with MDS/AML of non-FA patients. Other abnormalities, including 7q− and RUNX1 mutations, can be found in non-FA patients. 1q+ is found at all stages of the BM progression, whereas most other lesions are found at the most advanced MDS/AML stages. (Used with permission from Quentin et al.30 )
Because MDS/AML is a frequent and severe occurrence in FA, it is necessary to follow up on patients with regular BM aspirate tests with expert morphological and karyotype analyses to detect transformation before the onset of overt MDS/AML.1,20 In our study, conventional karyotype analysis appeared to be more sensitive than comparative genomic hybridization/single nucleotide polymorphism arrays for the early detection of discrete 1q+ subclones due to the possibility of observing individual cells and probably a clonal advantage in culture. Systematic interphasic FISH screening using probes for chromosome 7q, 3q, and break-apart RUNX1 might increase the sensitivity of detection of transformed cells. MDS/AML cases can have a normal karyotype but cryptic chromosomal or genomic abnormalities detected by FISH and/or array analysis.30 Therefore, when MDS or AML is diagnosed with a BM smear in FA patients, we now implement the karyotype analysis using high-density DNA arrays and RUNX1 FISH. This approach is useful in detecting cryptic lesions and in characterizing the abnormal karyotypes precisely, frequently highlighting meaningful chromosomal/molecular lesions.30 The predictive value of the various chromosomal/genomic abnormalities in patients with or without MDS/AML will have to be carefully evaluated on the long term in large cohorts of FA patients with respect to the therapeutic options and clinical benefits. For example, a sole clonal abnormality such as 1q+ can be present in a “normal” or non-MDS hypoplastic BM, and may not necessarily predict a progression into AML in the following years.
Conclusion
FA is a genetic condition that strongly predisposes patients to aplasia, MDS, and AML. Follow-up of FA patients requires a specialized multidisciplinary clinical and biological expertise. To optimize treatment, it is of critical importance to understand the step-wise molecular and cellular mechanisms of the BMF and clonal evolution. This knowledge may also ultimately benefit older, non-FA patients with aplastic anemia or MDS/AML, for whom close physiopathological cellular mechanisms are likely involved, including genomic instability, cytopenia, and clonal evolution.
Acknowledgments
Thanks to the patients and families, the French Association AFMF, the physicians who take care of the patients, the Centre de Référence Aplasies médullaires constitutionnelles Saint-Louis, Robert Debré, the Réseau INCa Maladies cassantes de, l' ADN, and to the members of the team Génome et Cancer at Saint-Louis Hospital, Paris, France, especially Raphael Ceccaldi.
Disclosures
Conflict-of-interest disclosure: The author declares no competing financial interests. Off-label drug use: None disclosed.
Correspondence
Jean Soulier, Hôpital Saint-Louis, 1 Avenue Claude-Vellefaux, 75010 Paris, France; Phone: +33-14249-9891; Fax: +33-14249-4027; e-mail: jean.soulier@sls.aphp.fr.