Abstract
Childhood acute lymphoblastic leukemia (ALL) provides an outstanding model for pharmacogenomic research: it is a drug-responsive disseminated cancer that is cured with medications alone in ∼ 85% of patients, but relapse remains unacceptably high for some subgroups. Inherited genomic variation contributes to the risk of relapse and to the risk of short- and long-term serious adverse effects of therapy. Our goal is to identify the inherited genomic variants that contribute to interindividual differences in response in patients with ALL. We discuss results of whole-genome interrogations of germline DNA in ALL.
Introduction
Pharmacogenomics is the study of how genetic variation among individuals contributes to interindividual differences in efficacy and toxicity of drugs. It began with candidate gene studies and, as technology has progressed, has moved to analysis of variants across the entire genome. Genome-wide association studies (GWAS) analyze the association of individual genetic variants across the entire genome with a phenotype of interest. GWAS generally focuses on common variants that are genotyped using commercially available high-throughput arrays testing more than 500 000 single nucleotide polymorphisms (SNPs). Because of the possibility of a high rate of false positives when doing so many association tests, the P value threshold for genome-wide significance is very low, generally P < 5 × 10−7. As for identification of any prognostic features, variations in therapy may affect which genetic features are discovered, so studies of pharmacogenetic features must be conducted against the background of multiple different treatment settings. Clinical trials afford the best platforms for pharmacogenetic discovery, and nongenetic features should be considered as covariates in GWAS.
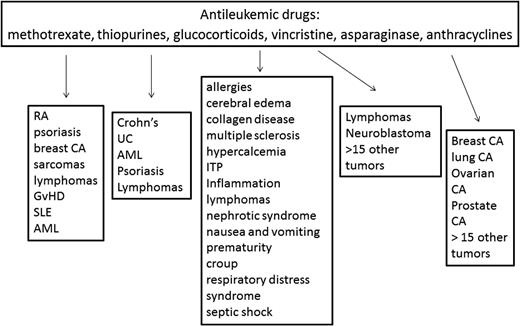
Childhood acute lymphoblastic leukemia (ALL) represents an outstanding platform for the conduct of pharmacogenomic research for several reasons. First, most children with ALL are treated on standardized treatment protocols, so their treatment is relatively uniform, many important nongenetic covariates are captured, and their antileukemic and adverse effect outcomes are recorded. Second, clinicians are accustomed to adjusting the intensity and content of therapy based on the acquired genomic characteristics of the ALL blasts, so the concept of adjusting therapy based on genomics is not a foreign one. Third, the disease is cured in the majority of patients using medications alone. Fourth, the medications have narrow therapeutic indices, meaning that understanding the basis for both antileukemic response and unacceptable adverse events is desired. Finally, many of the medications used to treat childhood ALL (glucocorticoids, vincristine, anthracyclines, thiopurines, and methotrexate [MTX]) are used to treat many other cancers and nonmalignant conditions (Figure 1), so knowledge generated on the pharmacogenomics of these agents can be leveraged to inform the use of these agents for other diseases.
Medications commonly used to treat childhood ALL. The medications commonly used to treat childhood ALL are also used to treat many other disorders. Genetic variants associated with drug effects, particularly adverse events, may have relevance for these other indications as well. RA indicates rheumatoid arthritis; SLE, systemic lupus erythematosus; AML, acute myeloid leukemia; UC, ulcerative colitis; ITP, idiopathic thrombocytopenia purpura; and CA, cancer.
Medications commonly used to treat childhood ALL. The medications commonly used to treat childhood ALL are also used to treat many other disorders. Genetic variants associated with drug effects, particularly adverse events, may have relevance for these other indications as well. RA indicates rheumatoid arthritis; SLE, systemic lupus erythematosus; AML, acute myeloid leukemia; UC, ulcerative colitis; ITP, idiopathic thrombocytopenia purpura; and CA, cancer.
There have been many candidate gene pharmacogenomic studies in ALL. Such studies have been useful in ALL, particularly those of thiopurine methyltransferase (TPMT), with its activity inherited as an essentially monogenic, codominant trait. TPMT has made its way from research subject to clinical implementation,1 in that acute myelosuppression can be prevented be adjusting doses of thiopurines based on TPMT phenotype or genotype without compromising ALL effectiveness.2 However, many candidate gene studies have resulted in associations with relatively small effect sizes or are subject to poor replication (possibly due to false discoveries and biases toward reporting positive studies). With the advent of high-throughput genome-wide arrays, several GWAS have been performed in ALL. The increasing recognition that large sample sizes are needed to improve the power of discovering real genotype/phenotype associations has also led to improved quality in pharmacogenomic studies in ALL.
Variants associated with risk of ALL and risk of relapse
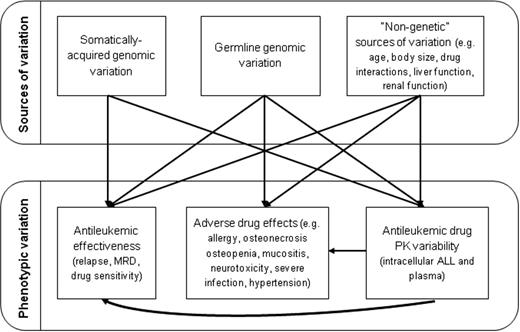
The most important pharmacogenomic phenotypic indicator of efficacy is of course relapse; minimal residual disease (MRD) is highly related to relapse and is also a useful phenotypic end point. In cancer, both the somatically acquired genomic features of the malignancy and the inherited genomic variation of the host have the potential to influence efficacy of therapy (Figure 2), and this is certainly true in ALL. In fact, much of the time, the mechanism by which somatically acquired genomic variations (eg, presence of the ETV6/RUNX1 fusion) confer either a favorable or an unfavorable prognosis with “standard” therapy remain unknown. Therefore, one of our first steps was to determine whether germline variation was associated with specific molecular and phenotypically defined ALL subtypes and if that variation could both explain the risk of specific ALL subtypes and account for some of the variation in its response to therapy.3 We found that ARID5B polymorphisms were associated with the incidence of childhood ALL, particularly hyperdiploid B-lineage ALL. These same polymorphisms are related to accumulation of MTX active polyglutamate metabolites (MTXPGs) in ALL blasts, showing that germline polymorphisms may explain the risk of specific ALL subtypes and why they respond well to therapy. ARID5B's association with risk of ALL has been replicated in multiple pediatric studies,4-10 but not in adult studies.11 The associations of ARID5B with childhood but not adult ALL are likely partly due to the abundance of hyperdiploid ALL among children, but the associations are also stronger in younger children than in older children.9 IKZF1 polymorphisms associated with ALL risk were also confirmed by several studies,4,6-10 which we found were able to distinguish T-cell cases from B-lineage ALL.3 Many inherited DNA variants differ in frequency among racial/ancestral groups. Interestingly, although discovered in Europeans, the frequency of ARID5B polymorphisms is highest in Hispanics and lowest in blacks, which mimics the frequency of childhood ALL in these racial groups. ARID5B polymorphisms also account for some of the racial disparities in outcome due to the higher frequency of variants associated with relapse in Hispanic versus white patients.12 Interestingly, in follow-up studies that included non-Europeans in a GWAS for ALL risk that adjusted for ancestry, an additional predisposing locus was identified,9 illustrating the power of including diverse populations in some GWAS.
Genomic variation in ALL. Genomic variation that is somatically acquired in the ALL blasts or that is inherited in the germline can affect interindividual variability in response, whereas adverse effects are affected by inherited variations. All phenotypes can be affected by several nongenetic features, so controlling and adjusting for these nongenetic features in GWAS of ALL is critical.
Genomic variation in ALL. Genomic variation that is somatically acquired in the ALL blasts or that is inherited in the germline can affect interindividual variability in response, whereas adverse effects are affected by inherited variations. All phenotypes can be affected by several nongenetic features, so controlling and adjusting for these nongenetic features in GWAS of ALL is critical.
Genome-wide approaches have also been used to discover germline variations associated with ALL relapse and early response to therapy (as assessed by MRD). Of the approximately 100 top SNPs associated with increased levels of MRD, approximately 20% were also associated with unfavorable systemic pharmacokinetics for the 2 agents for which data were available (MTX and etoposide).13 Given that up to 7 drugs (prednisone, vincristine, asparaginase, anthracyclines, etoposide, cytarabine, and MTX) were used during induction, and even considering that variants associated with the pharmacokinetics of etoposide might also affect vincristine, prednisone, and anthracyclines to some extent, one might assume that a total of approximately 40% to 50% of the variants exerted an effect on MRD via host pharmacokinetics. In addition, multiple SNPs associated with MRD were closely linked to the inherent susceptibility of ALL cells to drug-induced apoptosis, such as those in IL15.13 In a somewhat larger study using a genome-wide approach to assess ancestry, high Native-American ancestry was associated with a higher relapse risk, but that relapse risk could be attenuated by giving an extra phase of delayed intensification chemotherapy.14 In a multiethnic cohort of approximately 2500 newly diagnosed children with ALL, a GWAS of relapse risk was performed using multiple rounds of discovery and validation.15 Analyses accounted for treatment arm and ancestry, and we identified 134 SNPs that were associated with ALL relapse. The strongest association was for the C-allele at rs7142143 (in the PYGL gene) and a 3.6-fold higher risk of relapse than the T-allele (P = 6.7 × 10−9). We found that 14 of the 134 SNPs associated with relapse, including variants in PDE4B and ABCB1, were also associated with antileukemic medication pharmacokinetics, thereby suggesting a mechanism by which some germline SNPs may affect response to therapy in ALL.
Variants associated with MTX efficacy and pharmacokinetics
MTXPG accumulation differs among leukemic subtypes, with higher accumulation often corresponding to better subtype response. We applied a genome-wide approach to identify genomic determinants of MTXPG accumulation in primary ALL blasts and normal lymphoid cell lines using gene expression, somatic copy number variation, and inherited SNP genotypes.16 We found that all 3 types of genomic variation in 7 genes (FHOD3, IMPA2, ME2, RASSF4, SLC39A6, SMAD2, and SMAD4) were associated with MTXPG in blasts. To begin to assess the relative importance of the 3 types of variation, we found that expression of the top 7 genes in primary ALL cells accounted for more variation in MTXPGs than did expression of the top 7 genes in normal lymphoid cell lines and that the top 7 inherited SNP variants in children with ALL patients accounted for approximately the same degree of variation in MTXPGs as did the top 7 SNP genotypes in HapMap cell lines. In a classification and regression tree (CART) analysis for the ALL patients, ALL subtype was the strongest predictor of MTXPG accumulation, followed by ALL cell chromosome 18 copy number and then chromosome 10 copy number (analyzed via whole-genome SNP arrays or cytogenetically). These data provide evidence that acquired genetic variation in the ALL cells themselves had a stronger influence on MTXPG accumulation than did inherited germline variation.
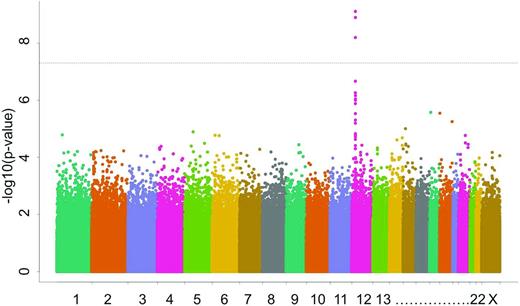
MTX effectiveness is also influenced by interindividual variation in its plasma clearance, which then results in heterogeneous systemic exposure. In a GWAS of children with ALL who received high-dose MTX in St. Jude frontline trials, multiple common polymorphisms in SLCO1B1 were associated with MTX clearance.17 In a completely separate group of children with ALL, we confirmed the importance of SLCO1B1 (Figure 3).18 MTX plasma clearance was estimated for 1279 patients with ALL treated with MTX (24-hour infusion of a 1 g/m2 dose or 4-hour infusion of a 2 g/m2 dose) on the Children's Oncology Group P9904 and P9905 protocols. Several features that have been repeatedly associated with low clearance, such as older age and female sex, were confirmed in these cohorts. The GWAS showed that MTX clearance was associated with SNPs in SLCO1B1 at a genome-wide level of significance (P = 2.1 × 10−11). Therefore, we replicated findings using different schedules of high-dose MTX from the St. Jude ALL treatment protocols. The SLCO1B1 SNP rs4149056 has now been replicated for 5 different treatment regimens of MTX.
Manhattan plot illustrating results of a GWAS with MTX clearance as the phenotype. The y-axis plots the inverse of the log of the association P value for each typed SNP (> 500 000 SNPs interrogated per patient) as it relates to MTX clearance; the x-axis sorts SNPs based on chromosomal position. The peak of P values corresponds to SNPs that localize to SLCO1B1, indicating that variation in this gene identified genetic variation associated with interpatient variability in MTX clearance among 1279 children with ALL.18
Manhattan plot illustrating results of a GWAS with MTX clearance as the phenotype. The y-axis plots the inverse of the log of the association P value for each typed SNP (> 500 000 SNPs interrogated per patient) as it relates to MTX clearance; the x-axis sorts SNPs based on chromosomal position. The peak of P values corresponds to SNPs that localize to SLCO1B1, indicating that variation in this gene identified genetic variation associated with interpatient variability in MTX clearance among 1279 children with ALL.18
We further evaluated the influence of rare versus common variants on MTX clearance in analyses that included all clinical covariates on clearance. After sequencing SLCO1B1 in 699 children, we identified 93 SNPs and, of these, 15 were nonsynonymous (NS).19 Three of these 15 NS SNPs were common, with an allele frequency > 5%, one had a low minor allele frequency (MAF; 1%-5%), and 11 had an MAF of < 1%. We found that, regardless of their MAF, NS SNPs that were predicted to be functionally damaging by computational algorithms were more frequent among patients in the lowest decile for MTX clearance than among those with the highest clearance. By expressing variants in mammalian cell lines, we verified that 4 SLCO1B1 haplotypes that were associated with reduced MTX clearance in patients indeed had reduced MTX transport capacity in vitro. Quantitatively, after adjusting for other genetic and nongenetic covariates, SLCO1B1 variants accounted for 10.7% of the population variability in clearance, a percentage that is relatively high compared with the percentage variability accounted for by genetics for some other inherited traits such as serum lipids.20 Of that 10.7% variability, rare damaging NS variants constituted 17.8% of SLCO1B1's effects on MTX clearance (1.9% of total variation). Rare variants had larger effect sizes than common NS variants, as is true for some nonpharmacogenetic traits.21 These findings support the notion that both rare and common inherited genomic variants have an important effect on pharmacogenetic phenotypes in ALL.
Variants associated with asparaginase efficacy and allergy
Asparaginase is an important drug for ALL, and overall response has been related to both the intrinsic sensitivity of the ALL blasts to the drug and to the total number of asparaginase doses delivered during continuation therapy.22 There is evidence that ALL subtypes differ in their responsiveness to asparaginase.23 We used a genome-wide approach to identify genetic variants that affect asparaginase sensitivity using normal lymphoblastoid cell lines from 87 individuals of European ancestry and primary diagnostic ALL blasts from 42 children with ALL of European ancestry.24 We assessed in vitro sensitivity by estimating the half-maximal inhibitory concentration (IC50) after exposure to native E coli asparaginase. At a threshold of P < .001, we found 329 SNPs representing 94 genes associated with asparaginase IC50. We then combined the germline SNP data with genes for which expression was associated with IC50 at the P < .05 level (1706 genes). Testing for pathways overrepresented by the 94 top-ranked genes (329 SNPs), we found that the top ranked pathway was that of aspartate metabolism, which may be linked directly to the mechanism of action of asparaginase. The 2 most highly ranked genes (ADSL and DARS) in this pathway encompassed 7 SNPs. Overall, approximately one-third of the variability in asparaginase IC50 among the lymphoid cell lines could be accounted for by these 7 SNPs. Moreover, we found that more sensitive ALL subtypes (hyperdiploid and TEL-AML1) had lower ADSL expression than more resistant subtypes (T-ALL), which is consistent with higher ADSL expression associating with resistance. Therefore, we found that an agnostic genome-wide approach yielded insights that both inherited and acquired genomic interindividual variation in the aspartate metabolic pathway contribute to asparaginase resistance in ALL.
One of the most common adverse effects of asparaginase is allergy, with up to 40% of patients developing hypersensitivity to the most commonly used preparation (E coli L-asparaginase). Allergy to the drug is problematic because it often results in lower serum asparaginase concentrations and thus less-than-optimal asparagine depletion. We performed a GWAS to determine whether there were inherited variations associated with allergy to Elspar (native E coli asparaginase) in 485 children with ALL treated in a frontline St Jude trial.25 Accounting for age, sex, treatment arm, race, and the number of episodes of asparaginase allergy, we assigned children into discovery (n = 322) and validation (n = 163) cohorts. Of the top-ranked 100 SNPs associated with allergy in the discovery cohort, chromosome 5 was overrepresented, with 10 SNPs annotated to genes. Among these 10 SNPs, we replicated the association of one SNP (rs4958381), in GRIA1 on chromosome 5q33, in the validation cohort. GRIA1 had an additional 4 SNPs that were associated with asparaginase allergy (P < .05) in both cohorts. SNPs on 5q33 have previously been associated with asthma and atopy in non-ALL settings. These data contribute to the growing body of evidence that there is an inherited component to predisposition to drug allergy. Whether different variants will be important for allergy to alternative forms of asparaginase and in additional clinical settings is under investigation.
Variants associated with osteonecrosis
Osteonecrosis is caused by glucocorticoids, but additional treatment- and host-related factors also play a role. To detect both symptomatic and asymptomatic osteonecrosis, we prospectively screened children (n = 364) with MRI of hips and knees at standard times in ALL therapy. We found that the cumulative incidence of any (grade 1-4) or of symptomatic (grade 2-4) osteonecrosis was 71.8% and 17.6%, respectively.26 The covariates that we evaluated included age, race, sex, ALL treatment arm, body mass, serum lipids, albumin and cortisol levels, dexamethasone pharmacokinetics, and genome-wide germline genetic polymorphisms. As expected, age > 10 years (odds ratio: 4.85; P = .00001) was a strong risk factor for osteonecrosis. We also found that the more intensive treatment arm, which included more asparaginase and higher doses of dexamethasone, was a risk factor and both were included as covariates in the GWAS. Lower serum albumin (P = .05) and higher serum cholesterol (P = .02) during therapy were associated with symptomatic osteonecrosis and severe (grade 3-4) osteonecrosis was linked to poor dexamethasone clearance (P = .0005), measured during continuation therapy. After adjusting for all relevant clinical covariates, we found that polymorphisms of ACP1 (odds ratio: 5.6) were associated with osteonecrosis. ACP1 is involved in the regulation of lipid levels. The ACP1 SNPs that were associated with risk of osteonecrosis were also associated with lower albumin and higher cholesterol. The features of older age (> 10 years of age), lower albumin, higher lipid levels, and dexamethasone exposure were associated with osteonecrosis and may be linked by common inherited genomic variation, suggesting a pleiotropic effect of the inherited variation. Moreover, several of these features (lower albumin, higher lipids, and higher dexamethasone plasma levels) may reflect higher asparaginase exposure and its possible contribution to osteonecrosis. Asparaginase has been shown to increase the frequency of dexamethasone-induced osteonecrosis in our murine model of the disorder.27 Additional studies in larger cohorts and in cohorts of differing age groups are ongoing.
Clinical implementation of pharmacogenomics
We and others incorporate genetic testing of TPMT before initiation of thiopurines1 to minimize acute myelosuppression. This is the only antileukemic agent for which the evidence is sufficient to warrant clinical use of germline genetic variation at this time.
Conclusions
Although GWAS have been effective for the discovery of multiple variants affecting de novo disease risk,28 the number of adequately powered GWAS for pharmacogenomics is far fewer.29 Given that the strongest determinant of treatment outcomes is generally related to the treatment delivered and that most treatment arms are relatively small in sample size and are rarely precisely replicated, it is not surprising that the conduct of adequately powered pharmacogenomic studies is challenging. ALL offers some of the largest cohorts of patients who receive uniform chemotherapy via clinical trials, so is well suited to continued genome-wide pharmacogenomic studies. We are continuing to perform GWAS in additional frontline ALL trials with the aim of determining whether findings replicate and increasing sample sizes to be able to study more rare clinical phenotypes.
Acknowledgments
This work was supported by the National Institutes of Health (grants U01 GM092666, CA142665, CA21765, CA36401, CA98543, and CA156449), the Leukemia & Lymphoma Society (grant 6168-12), and the American Lebanese Syrian Associated Charities.
Disclosures
Conflict-of-interest disclosure: M.V.R. holds patents with or receives royalties from Prometheus and Specialty Labs. L.B.R. declares no competing financial interests. Off-label drug use: None disclosed.
Correspondence
Mary Relling, PharmD, Pharmaceutical Sciences, MS313, Room I5112, St Jude Children's Research Hospital, 262 Danny Thomas Place, Memphis, TN 38105; Phone: 901-595-2348; Fax: 901-525-8869; e-mail: mary.relling@stjude.org.