Abstract
The discovery of mutations activating JAK-STAT signaling in the majority of patients with myeloproliferative neoplasms (MPNs) led to identification of tyrosine kinase activation as a predominant mechanism driving MPN pathogenesis. Despite this, the existence of additional genetic events that modify the MPN phenotype, predate JAK2 mutations, and/or contribute to leukemic transformation of MPNs has been suggested. Recently, mutations in several epigenetic modifiers have been described in patients with MPNs, including mutations in ASXL1, DNMT3A, EZH2, IDH1, IDH2, and TET2. Moreover, the mutant JAK2 itself has been shown recently to affect histone posttranslational modifications directly. Here we review the biological and clinical implications of epigenetic alterations in the pathogenesis of MPNs.
Somatic mutations activating JAK-STAT signaling
The classic BCR-ABL–negative myeloproliferative neoplasms (MPNs) are clonal disorders of hematopoiesis characterized by the production of mature-appearing cells within the bloodstream. In 2005 to 2007, a series of studies found that a very high frequency of activating mutations in the JAK-STAT pathway is present in MPN patients. This includes JAK2V617F mutations in 90% to 95% of patients with polycythemia vera (PV), 50% to 60% of patients with essential thrombocytosis (ET), and 50% to 60% of patients with primary myelofibrosis (PMF)1-4 ; JAK2 exon 12 mutations in JAK2V617F-negative PV patients5 ; and mutations activating the thrombopoietin receptor MPL in 3% to 5% of patients with ET and 8% to 10% of patients with PMF.6 More recently, mutations in LNK,7 a negative regulator of JAK2 signaling, have also been identified in 3% to 6% of patients with ET and a similar percentage of patients with PMF.
Evidence for alterations outside of JAK-STAT mutations in MPN pathogenesis
In addition to these genetic data indicating a disease-defining high frequency of JAK-STAT pathway activating mutations in MPN patients, functional studies using in vitro and in vivo systems have shown repeatedly that activating mutations in JAK2 and MPL appear to be sufficient for hematopoietic transformation and critical phenotypic aspects of MPNs.
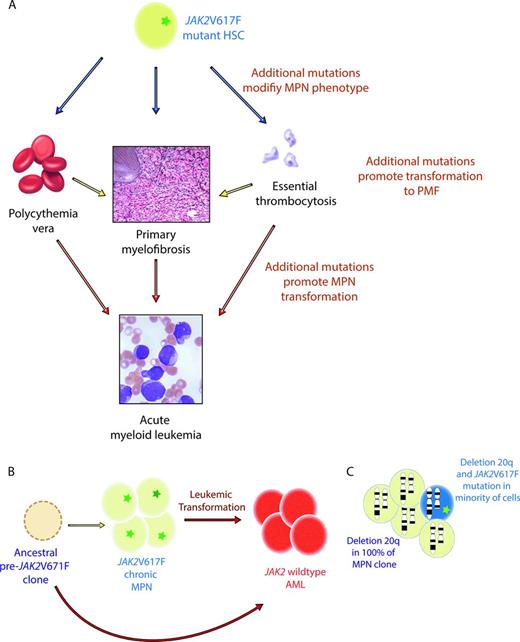
Despite the clear link between JAK-STAT pathway mutations and MPN pathogenesis, multiple pieces of evidence suggest that genetic events outside of JAK-STAT–activating mutations are likely in MPN patients (Figure 1, Table 1). First is the fact that a significant proportion of patients with ET and PMF have no identifiable JAK2, MPL, or LNK mutations. Second is the conundrum of how a single mutation in JAK2, which appears to be sufficient for MPN pathogenesis, could result in the development of 3 phenotypically variable diseases. One attractive hypothesis to answer this question was that additional acquired or inherited genetic modifiers outside of JAK2 could be present and modify the MPN phenotype induced by the JAK2V617F mutation. Moreover, clonal analysis of patients with JAK2/MPL mutations have consistently demonstrated the presence of occasional patients with JAK2 wild-type erythropoietin-independent erythroid colony formation—clear evidence that an additional aberration responsible for erythropoietin-independent growth must be present.8 Third, clonality analyses of patients with a cytogenetic abnormality in conjunction with the JAK2V617F mutation also revealed that patients in whom cytogenetically abnormal clones with and without the JAK2V617F mutation could be identified.9 Finally, since the discovery of the JAK2V617F mutation in 2005, several reports have shown consistently that leukemic blasts of acute myeloid leukemia (AML) derived from a JAK2V617F MPN are frequently JAK2 wild-type.10,11 This suggests that the MPN and AML clones may arise from 2 distinct progenitor cells or that an ancestral clone bearing an abnormality preceding the JAK2V617F mutation could be present, giving rise to both MPN and AML.
Evidence for somatic mutations in genes other than those activating JAK-STAT signaling in MPN patients. (A) Given that JAK2V617F mutations are common to 3 MPN disorders with distinct phenotypes (PV, ET, and PMF), it has been postulated that additional somatic or germline variants might contribute to the MPNs produced. In addition, mutations in genes outside of JAK2V617F are thought to play a role in transformation of PV and ET to PMF and in the transformation of chronic MPNs to AML. (B) Since the discovery of the JAK2V617F mutation, it was noted that patients with JAK2 mutant chronic MPN may undergo transformation to a JAK2 wild-type AML. This observation suggests that the JAK2V617F mutation may not be required for leukemic transformation or that a clone ancestral to the JAK2V617F mutant cell was subject to leukemic transformation. (C) Finally, it has also been observed that occasional patients with the JAK2V617F mutation in addition to a cytogenetic alteration (illustrated here by deletion 20q) may have the JAK2 mutation in only a portion of the MPN clones, whereas the majority of the MPN cells bear both the cytogenetic alteration and the JAK2 mutation. This finding again suggests that the JAK2V617F mutation may not be the initiating genetic event in MPN pathogenesis.
Evidence for somatic mutations in genes other than those activating JAK-STAT signaling in MPN patients. (A) Given that JAK2V617F mutations are common to 3 MPN disorders with distinct phenotypes (PV, ET, and PMF), it has been postulated that additional somatic or germline variants might contribute to the MPNs produced. In addition, mutations in genes outside of JAK2V617F are thought to play a role in transformation of PV and ET to PMF and in the transformation of chronic MPNs to AML. (B) Since the discovery of the JAK2V617F mutation, it was noted that patients with JAK2 mutant chronic MPN may undergo transformation to a JAK2 wild-type AML. This observation suggests that the JAK2V617F mutation may not be required for leukemic transformation or that a clone ancestral to the JAK2V617F mutant cell was subject to leukemic transformation. (C) Finally, it has also been observed that occasional patients with the JAK2V617F mutation in addition to a cytogenetic alteration (illustrated here by deletion 20q) may have the JAK2 mutation in only a portion of the MPN clones, whereas the majority of the MPN cells bear both the cytogenetic alteration and the JAK2 mutation. This finding again suggests that the JAK2V617F mutation may not be the initiating genetic event in MPN pathogenesis.
Mutations in epigenetic modifiers in MPN patients

5-mC indicates 5-methylcytosine; and 5-hmC, 5-hydroxymethylacytosine.
Given the above evidence for potential somatic genetic alterations, germline variants, and epigenetic events that might contribute to MPN development, influence the MPN disease phenotype, and/or promote leukemic transformation, several candidate gene-resequencing efforts, array-based gene discovery technologies (including single nucleotide polymorphism arrays and comparative genomic hybridization arrays), and exome/whole genome sequencing studies have uncovered a series of somatic mutations in epigenetic modifiers in MPN patients. This includes mutations in ASXL1, DNMT3A, EZH2, IDH1/2, and TET212-16 (Figure 2).
Timeline of gene discovery efforts in patients with MPNs. The MPNs were initially grouped together based on prescient clinical insights by William Dameshek in 1951.54 The earliest insights into the genetic causes for the MPNs were then made in 1976 to 1981, when a series of studies by Philip Fialkow et al demonstrated that all 3 classic MPNs represented clonal disorders derived from a genetically aberrant HSC.55-57 This was followed by the discovery of activating mutations in JAK2 and the thrombopoietin receptor MPL in the majority of patients with PV, ET, and PMF in 2005 to 2007. More recently, a series of mutations in genes with the primary known function of epigenetic regulation of transcription have been identified in MPN patients. This includes mutations in TET2, ASXL1, EZH2, and DNMT3A in MPN patients. Finally mutations in the spliceosomal proteins of unclear function have also been found to occur in MPN patients.
Timeline of gene discovery efforts in patients with MPNs. The MPNs were initially grouped together based on prescient clinical insights by William Dameshek in 1951.54 The earliest insights into the genetic causes for the MPNs were then made in 1976 to 1981, when a series of studies by Philip Fialkow et al demonstrated that all 3 classic MPNs represented clonal disorders derived from a genetically aberrant HSC.55-57 This was followed by the discovery of activating mutations in JAK2 and the thrombopoietin receptor MPL in the majority of patients with PV, ET, and PMF in 2005 to 2007. More recently, a series of mutations in genes with the primary known function of epigenetic regulation of transcription have been identified in MPN patients. This includes mutations in TET2, ASXL1, EZH2, and DNMT3A in MPN patients. Finally mutations in the spliceosomal proteins of unclear function have also been found to occur in MPN patients.
Mutations affecting histone posttranslational modifications in MPN pathogenesis
Outside of JAK2, mutations in genes encoding the core members of the polycomb repressive complex 2 (PRC2) and in the polycomb-associated protein ASXL1 represent the most frequently described mutations that regulate histone modifications directly in MPN patients.
Using Agilent 244K comparative genomic hybridization arrays, Gelsi-Boyer et al discovered mutations in ASXL1 initially through the identification of a deletion in an MDS patient at the ASXL1 locus.15 Further sequencing of ASXL1 by this group and others led to reports of ASXL1 mutations in 2% to 5% of patients with PV, 5% to 8% of patients with ET, and 7% to 17% of patients with MF. ASXL1 is 1 of 3 mammalian homologs of the additional sex combs gene in Drosophila. The genes are named for the fact that deletion in Drosophila leads to homeotic transformations. This occurs because ASXL appears to regulate the expression of both polycomb group and trithorax group proteins in Drosophila. The known functions of ASXL1 that have been identified in mammalian hematopoietic cells thus far include physical association with the PRC2 complex17 and physical interaction with the H2AK119 deubiquitinase enzyme BAP1.18 ASXL1 appears to be a critical factor for the function of both BAP1 and PRC2 function in myeloid HSCs. Conditional deletion of Asxl1 alone in vivo results in a phenotype most resembling BM failure with morphologic dysplasia.19 Further work to examine the combined phenotype of Asxl1 loss with Jak2 activation may be particularly enlightening given the important prognostic importance of ASXL1 mutations in PMF (described below).20
After mutations in ASXL1, EZH2, the catalytic member of the PRC2 complex, was found to be mutated in 2010 in patients with classic MPNs, MDS, and MPN/MDS overlap disorders. Mutations in EZH2 occur in ∼ 10% to 12% of patients with MDS/MPN, 5% to 15% of patients with MDS, and 5% to 13% of patients with classic MPNs.21-23 Moreover, mutations in EZH2 predominate in MPN patients with PMF or post-PV/ET MF and are far less frequent in ET and PV, possibly indicating a role for EZH2 mutations in fibrotic transformation.21
Mutations in EZH2 in myeloid malignancy patients appear to be loss-of-function mutations.23 Although the in vivo effects of Ezh2 loss in the hematopoietic system have been described, the myeloid effects of Ezh2 loss in vivo are not well understood.24 Mice with heterozygous deletion of Eed, a noncatalytic core PRC2 member, have been created and display severe myeloproliferation by 7 months of age, suggesting a pathogenetic role of PRC2 loss in myeloid malignancy.25
After the description of EZH2 mutations in MPN patients, several groups have performed candidate gene-sequencing studies of additional PRC2 members in patients with myeloid malignancies. In addition to somatic loss-of-function mutations in EZH2, rare additional deletions and putative loss-of-function mutations have been identified in the other core PRC2 members in patients with MDS, including SUZ12 and EED mutations (all at < 5% frequency).26,27
Mutations affecting DNA cytosine modifications in MPN pathogenesis
The discovery of TET2 mutations in patients with myeloid malignancies in 2009 by Delhommeau et al and by Langemeijer et al was a landmark finding that gave rise to studies on the function of the TET family of proteins in transcriptional regulation and stem cell biology and the importance of TET2 mutations in the clinical management of patients with myeloid malignancies.14,16 Initial studies in patients with MPN suggested the occurrence of TET2-mutant/JAK2-mutant and TET2-mutant/JAK2-wild-type clones, but not TET2-wild-type/JAK2-mutant clones, suggesting that TET2 mutations occur as a “pre-JAK2” event.14 However, subsequent studies have noted the post-JAK2V617F acquisition of TET2 mutations, refuting a paradigm that mutations in TET2 represent the earliest genetic aberration in MPN.28 Ongoing work examining the cooperativity of Jak2V617F mutations and Tet2 loss in vivo will hopefully address the potential importance of the order of JAK2/TET2 mutation acquisition on disease phenotype and HSC self-renewal in MPN.29
Our current understanding of TET2 mutations in MPN patients is that these mutations are loss-of-function mutations that result in decreased 5-hydroxymethylcytosine,30 exist in chronic-phase MPN patients, and may predate the JAK2V617F mutation, but at the same time are more frequent at leukemic transformation of MPNs.28 There is currently no evidence that TET2 mutations affect survival in PMF patients.31 However, large studies with adequate clinical correlates are still needed to clarify the clinical correlates of TET2 mutations in PV and ET patients.
Perhaps more advanced than the clinical effects of TET2 mutations in MPN patients has been the in vivo evidence that TET2 loss results in increased HSC self-renewal and myeloproliferation. Currently, 5 different Tet2 knockout mouse models have been created and characterized.32-35 In all of these models, Tet2 loss leads to a progressive enlargement of the HSC compartment and eventual myeloproliferation in vivo, including splenomegaly, monocytosis, and extramedullary hematopoiesis.33 In addition, Tet2+/− mice also display increased stem cell self-renewal and extramedullary hematopoiesis, suggesting that Tet2 haploinsufficiency contributes to hematopoietic transformation in vivo. To date, there is no evidence that Tet2 loss in vivo results in the development of MF in mice.33 Three of the 4 Tet2 knockout mouse studies have revealed a clear linkage between loss of Tet2 and decreased 5-hydroxymethylcytosine in vivo.32,34,35 However, genetic targets of TET2 loss in the hematopoietic system are not yet well understood.
In addition to TET2 mutations, DNMT3A mutations have also been described in MPN.36,37 Although these mutations are more frequent in AML and post-MPN AML, they also occur in chronic-phase MPN, including in 10% to 15% of PMF patients and 5% to 7% of PV patients. Until recently, data from murine studies had suggested that Dnmt3a may be dispensable in HSCs.38 However, careful characterization of the HSCs from Dnmt3a knockout mice in serial transplantation experiments by Challen et al has revealed that Dnmt3a loss results in a striking expansion of HSCs.39 Despite the dramatic effects of Dnmt3a loss on HSC number and frequency with serial transplantation, the effects of Dnmt3a loss on DNA methylation and gene expression are perplexing. From in vivo studies of genome-wide methylation status using HPLC-MS and bisulfite sequencing of purified HSC populations, there appears to be very little correlation between Dnmt3a loss and DNA methylation/gene expression at specific loci.39 Nonetheless, several genes that should be repressed for normal HSC differentiation were found to be consistently up-regulated and hypomethylated with Dnmt3a loss, including Runx1 and Gata3.39 Although the loss of Dnmt3a in vivo still appears from this work to be insufficient for transformation or disease phenotype, future work to address the combined effect of DNMT3A loss with activating alterations in JAK2 may be very enlightening.
Mutations affecting modifications of DNA and histones in MPN patients
Gain-of-function mutations in the genes encoding isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2) are now well established genetic events in patients with glioblastoma multiforme and AML. Rare IDH1/2 mutations were subsequently identified in smaller fraction of patients with MDS and MPNs.40 The existing mutational data in MPN patients clearly indicate that IDH1/2 mutations cluster in patients with blast-phase MPNs and MF but are rare in patients with chronic-phase MPNs. From a large study of 1473 MPN patients, only 38 IDH mutant patients were seen, with IDH1/2 mutations being found in 1.9% of PV patients, 0.8% of ET patients, 4.2% of PMF patients, but in 21.6% of patients with blast-phase MPN. Moreover, in blast-phase MPN, the presence of an IDH mutation predicted worse survival. In a more recent study of IDH1/2 mutations in a cohort of 301 patients with PMF, it was shown that IDH1/2 mutations are associated with decreased overall survival and leukemia-free survival in PMF.41
It is now established that IDH1/2 mutations are gain-of-function mutations that result in neomorphic enzymatic activity and the production of 2-hydroxyglutarate by the mutant enzymes42 and are transforming in classic in vitro experiments.43 It has been shown by at least 2 groups that 2-hydroxyglutarate may compete with alpha-ketoglutarate (aKG) and thereby inhibit the function of aKG-dependent enzymes.44 This includes impairment of the function of TET2, a known aKG-dependent enzyme, as well as the Jumonji C-terminal domain family of histone demethylases, another group of aKG-dependent enzymes. These findings suggest that aberrant hypermethylation of histone lysine tails and DNA cytosine residues may be important in the pathophysiology of IDH1/2-mutant disease. Additional work to clarify the targets of aberrant DNA and histone lysine methylation in malignant HSCs with IDH1/2 mutations is needed. Recently, initial inhibitors of mutant selective IDH145 and IDH2R140Q inhibitors46 have been described and the effects of mutant IDH2 inhibition have been demonstrated in vitro in AML cell lines and patient samples.46
Involvement of JAK2 in epigenetic regulation
The transforming effects of activating mutations in JAK2 have been mostly ascribed to constitutive activation of downstream mitogenic pathways such as the STAT family of transcription factors, MAPK, and AKT. Surprisingly, in 2009, Dawson et al made the intriguing observation that JAK2 is also found within the nucleus of both normal and malignant HSCs.47 At least one functional consequence of this finding is that JAK2 phosphorylates histone H3 among all core histones. This was confirmed through evidence of decreased H3 phosphorylation in the presence of at least 2 different JAK2 inhibitors and phosphorylation of H3Y41 only after JAK2 transfection in JAK2-null g2A cells. The investigators then demonstrated that phosphorylation of H3Y41 results in displacement of HP1a and, subsequently, overexpression of LMO2, an oncogene with a known role in leukemogenesis. The findings of that study provide evidence that a kinase thought to be restricted to the cytoplasm may regulate gene expression directly by affecting chromatin structure. Evidence for the nuclear localization of JAK2 has since been confirmed by several additional groups and JAK1 has also been discovered to be present in the nucleus.48,49
Identification of JAK2 within the nucleus led to the question of possible additional nuclear substrates of JAK2 phosphorylation other than histone H3Y41. Previously, study of the physical interactions of JAK2 with other proteins had focused on the association of JAK2 with cytoplasmic domains of type II cytokine receptors to mediating signals that are triggered by hematopoietic growth factors and activate the STAT5/BCL-XL, PI3K/AKT, and ERK/MAPK pathways. However, as mentioned above, activation of these mitogenic pathways may not completely account for the MPN phenotype.
The type II arginine methyltransferase PRMT5 was first identified as JAK-binding protein 1 (JBP1) in a yeast 2-hybrid assay. It mediates the symmetrical dimethylation of arginine residues within histones H2A, H3, and H4 and methylates other cellular proteins as well, such as p53, SPT5, and MBD2. Through investigation of the in vivo interaction between PRMT5 and the oncogenic mutant JAK2 kinases (JAK2V617F and JAK2K539L), Liu et al found that mutant JAK2 protein bound PRMT5 more strongly than did wild-type JAK2.48 These oncogenic kinases also acquired the ability to phosphorylate PRMT5, greatly impairing its ability to methylate its histone substrates and representing a specific gain of function that allows them to regulate chromatin modifications. The investigators also readily detected PRMT5 phosphorylation in JAK2V617F-positive patient samples and when PRMT5 was depleted in human CD34+ cells using shRNA, increased colony formation and erythroid differentiation resulted. These results indicate that phosphorylation of PRMT5 contributes to the mutant JAK2-induced myeloproliferative phenotype.
Clinical importance of epigenetic alterations in MPN patients
Great progress has been made recently in our understanding of the clinical importance of mutations in genes other than JAK2 in patients with MPNs. Although studies of individual genetic alterations had been reported in various single-center cohorts previously, the first comprehensive mutational analysis in a large multicenter MPN patient cohort was described recently by Vannucchi et al20 In that study, mutational analysis for 10 genes was performed in a test and validation cohort of > 800 PMF patients with a median follow-up of 3.7 years. This landmark study revealed that overall and leukemia-free survival were predicted by ASXL1, SRSF2, or EZH2 mutations and ASXL1, SRSF2, and IDH1/2 mutations, respectively. When the analysis was subjected to multivariate analysis including the Dynamic International Prognostic Scoring System-Plus (DIPSS-Plus) prognostic score, only ASXL1 mutations retained their prognostic significance. These data indicate the utility of additional mutational analyses for ASXL1 (at a minimum) in routine clinical practice and the importance of epigenetic dysregulation in the disease course of PMF patients.
In addition to the utility of epigenomic alterations in aiding in the prognostication of MPN patients, epigenomic alterations may aid in the discrimination of particular clinical subsets of MPN patients. Recently, Nischal et al reported that PV and ET are characterized by aberrant promoter hypermethylation, whereas PMF is an epigenetically distinct subgroup characterized by both aberrant hypermethylation and hypomethylation. In addition, they found that within the PMF subgroup, cases with ASXL1 disruptions formed an epigenetically distinct subgroup with relatively increased methylation and MPNs with TET2 mutations showed decreased levels of hydroxymethylation and distinct set of hypermethylated genes. Furthermore, they suggest that numerous significantly and uniformly hypermethylated loci in PV, EF, and PMF may be targeted by epigenetic modifiers in future clinical trials.50 The use of epigenetic-targeted therapeutics in MPNs is an area of active clinical investigation, which thus far has consisted of the study of the use of histone deacetylase inhibitors in MPN patients. DeAngelo et al presented the results of a phase 2 trial of panobinostat, an orally available histone deacetylase inhibitor, in MPN patients including both PV and PMF.51 In addition, Mascarenhas et al reported that panobinostat is a well-tolerated, clinically active treatment for PMF patients.52 Moreover, in a phase 2 study of 20 patients with PMF treated with low-dose decitabine, a DNA methyltransferase I inhibitor, a 37% response rate was seen.53
Conclusion
Mutations in epigenetic modifiers discovered in MPN patients have provided important insights into the pathogenesis of MPNs and cancer biology in general. For example, mutations in TET2 have been discovered recently to be important drivers of myeloid leukemic transformation with importance in the prognostication of AML. Moreover, mutations in ASXL1 appear to be novel biomarkers of adverse disease outcome in PMF patients. Despite this, several unresolved questions regarding the biological and clinical importance of many of these alterations still exist. For example, the prognostic importance of many epigenetic mutations has not been well described in PV and ET patients, including mutations in TET2, ASXL1, and DNMT3A. Given the relative rarity of these mutations in many chronic-phase MPN patients, larger sequencing studies with comprehensive mutational data and pristine clinical annotation are greatly needed. Moreover, further functional studies to understand the effects of these alterations in combination with JAK2 and MPL mutations are needed for a deeper understanding of the biological contribution of these alterations to MPN disease phenotype and outcome. Lastly, in vitro and in vivo studies will be needed to address the effect, if any, of epigenetic alterations on the response to therapeutics administered to MPN patients, such as hydroxyurea, IFN-α, and JAK2-targeted therapy.
Disclosures
Conflict-of-interest disclosure: The authors declare no competing financial interests. Off-label drug use: None disclosed.
Correspondence
Omar Abdel-Wahab, Human Oncology and Pathogenesis Program and Leukemia Service, Memorial Sloan-Kettering Cancer Center, 1275 York Ave, New York, NY 10065; Phone: 646-888-2796; Fax: 646-422-0890; e-mail: abdelwao@mskcc.org.