Abstract
Despite the high cure rate in classical Hodgkin lymphoma (CHL), more accurate tailoring of upfront treatment is required to maximize cure while avoiding unnecessary short- and long-term treatment side effects. To this end, the unique tumor microenvironment of CHL has been searched extensively for prognostic biomarkers. Beyond targeted immunohistochemistry (IHC) studies, gene expression profiling (GEP) of diagnostic whole tissue biopsies has allowed a de novo approach to biomarker discovery. Among numerous candidate biomarkers, an association between the number of tumor-associated macrophages in the microenvironment and outcomes after ABVD (doxorubicin + bleomycin + vinblastine + dacarbazine) chemotherapy emerged, and multiple subsequent studies have validated this biological relationship using IHC. These studies have also defined key aspects for macrophage interrogation, including the characteristics of the CD68 and CD163 antibodies, appropriate scoring methodologies, and the identification of specific patient populations in which macrophage IHC may not be prognostic. The GEP studies also led to the development of gene expression-based prognostic models for advanced-stage CHL, with new technologies allowing reliable gene expression quantitation using RNA from routinely produced formalin-fixed paraffin-embedded biopsies. The bridge to predictive biomarkers that can be used reliably to inform upfront treatment selection requires further studies to demonstrate that these biomarkers can identify robustly, at diagnosis, patients at high risk of treatment failure with ABVD and that this risk may be overcome using alternative treatments.
Learning Objective
To understand the biological and prognostic significance of the microenvironment of CHL
Introduction
The treatment of classical Hodgkin lymphoma (CHL) has been hailed as a success story for multiagent chemotherapy (with or without radiotherapy), with ∼80% of patients eventually being cured by this approach.1 However, with current strategies to select upfront treatment being largely based on tumor burden, dividing patients into limited or advanced stage, a significant proportion of patients may be either overtreated, experiencing unnecessary short- and long-term sequelae, or undertreated, experiencing disease relapse. To tailor management more accurately to individual patients, tools are required that provide risk stratification (prognostic biomarkers) and, ideally, inform decisions by predicting response to alternative treatment regimens (predictive biomarkers).
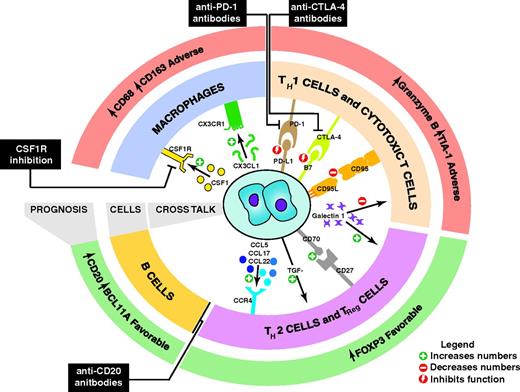
Over the past decades, an increasing array of potential biomarkers have been described, ranging from tests allowing risk stratification at diagnosis through to those that monitor treatment response, guiding dynamic management decisions. Sources of biomarkers include the ongoing elucidation of the biology of the malignant Hodgkin Reed-Sternberg (HRS) cells, the extensive collection of nonmalignant cells that make up the tumor microenvironment (TME) and the molecular interactions between these elements (Figure 1).
Biology and clinical significance of the TME in CHL. This schematic shows a selection of the interactions between the HRS cells and cells in the microenvironment. The prognostic significance of components of the microenvironment as detected by IHC is shown. Increased galectin 1 in the microenvironment is also associated with adverse prognosis. Therapeutic strategies that target the cross-talk between the HRS cells and the TME are shown in black boxes.
Biology and clinical significance of the TME in CHL. This schematic shows a selection of the interactions between the HRS cells and cells in the microenvironment. The prognostic significance of components of the microenvironment as detected by IHC is shown. Increased galectin 1 in the microenvironment is also associated with adverse prognosis. Therapeutic strategies that target the cross-talk between the HRS cells and the TME are shown in black boxes.
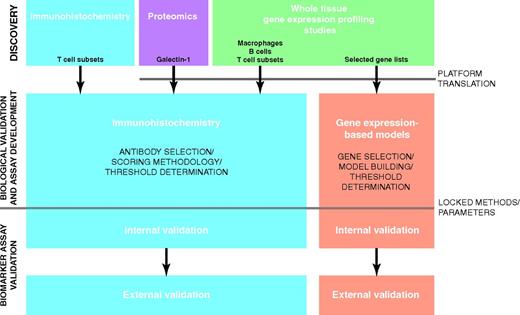
Here, we focus on the biological and clinical importance of tumor-associated macrophages and gene expression-based prognostic models, both of which have emanated from gene expression profiling (GEP) discovery studies using pretreatment whole tissue biopsies. Specifically, we highlight the road to developing clinically relevant biomarkers and the challenges therein (Figure 2).
Roadmap from discovery to clinical implementation for candidate biomarkers in the TME in CHL. This diagram shows the path from discovery of candidate biomarkers through to assays suitable for broad clinical implementation. Examples of candidates emanating from targeted discovery approaches and the de novo approaches are shown. Although not absolutely required, several studies have incorporated an internal validation, which increases confidence that the biomarker assay will validate in other cohorts and laboratories: the external validation. Increasingly, biomarker assay development and validation are being performed using material from clinical trials either retrospectively in prospectively collected biopsies or in a prospective fashion.
Roadmap from discovery to clinical implementation for candidate biomarkers in the TME in CHL. This diagram shows the path from discovery of candidate biomarkers through to assays suitable for broad clinical implementation. Examples of candidates emanating from targeted discovery approaches and the de novo approaches are shown. Although not absolutely required, several studies have incorporated an internal validation, which increases confidence that the biomarker assay will validate in other cohorts and laboratories: the external validation. Increasingly, biomarker assay development and validation are being performed using material from clinical trials either retrospectively in prospectively collected biopsies or in a prospective fashion.
TME
The TME of CHL has been described extensively using morphology, flow cytometry, and immunohistochemistry (IHC) to survey the composition and spatial arrangement of the nonmalignant cells. Studies specifically examining components of the TME, isolated or enriched using techniques such as flow cytometry, are providing texture to this cellular survey.2 Typically, the malignant HRS cells make up <5% of the tumor, with the TME dominated by T cells, along with variable numbers of macrophages, B cells, eosinophils, plasma cells, neutrophils, and fibroblasts.3 Four broad microenvironment patterns are observed and form the basis of the subtypes in the WHO classification, namely nodular sclerosis (NS), mixed cellularity (MC), and lymphocyte-rich and lymphocyte-depleted,3 with NS and MC collectively making up >90% of total cases.
Determinants of the TME composition
The mechanisms underlying the differences in TME observed between and within the CHL subtypes are not fully understood. Evidence is accumulating that, despite the similar morphology and immunophenotype of the HRS cells, the malignant cells in the NS and MC subtypes can be distinguished at the level of gene expression4 and have distinct mutational landscapes.5 Another example in which the phenotype of the HRS cells affects the TME is the impact of latent EBV infection of the HRS cells, which is seen uncommonly in NS but frequently in MC. EBV+ disease is associated with a whole tissue gene expression profile consistent with a Th1 response and overexpression of macrophages genes.6 Finally, colony stimulating factor 1 receptor (CSF1R) overexpression in HRS cells has been associated with increased macrophages.7 With the TME reflecting the interplay between the HRS cells and the host immune system, differences in host immunity, including polymorphisms within immune response genes and the effects of age, should not be overlooked.8-10 Ultimately, a full understanding of these relationships will require comprehensive integration of genetic aberrations and transcription patterns in the HRS cells, the composition of the microenvironment, and determinants of host immune response.
TME–outcome associations
The association between elements of the TME in diagnostic biopsies and treatment outcomes in CHL has been identified using 2 broad approaches: the de novo discovery approach allowed by techniques such as proteomics and whole tissue GEP and the targeted hypothesis-driven approach, exemplified by many IHC studies (Figure 2). In aggregate, these approaches have revealed numerous factors in the TME associated with treatment outcomes, as reviewed recently11,12 (Figure 1).
Precisely what underlies the relationship between elements in the TME and treatment outcomes is unknown. Conceptually, the association between cells in the TME and outcome may indicate that the presence of these cells affects the efficacy of standard treatment directly, making the additional targeting of these cells and their interactions with the HRS cells attractive. Alternatively, the TME composition may represent an epiphenomenon or a surrogate for genetic aberrations harbored by the HRS cells that impart resistance to chemotherapy. By extension, the TME-outcome correlates may thus be only limited to certain CHL subtypes and/or EBV+ or EBV− disease.
GEP in whole tissue: a discovery tool
The ability to measure gene expression changes on a transcriptome-wide scale presented an opportunity to discover gene expression signatures associated with treatment outcomes in CHL.6,13-15 In contrast to other cancers and lymphomas, in which the proportion of malignant cells is typically high and the gene expression profile is dominated by gene expression of the malignant cells, the HRS cells in CHL tumors are vastly outnumbered by nonmalignant cell subsets. With the resulting gene expression representing a summation of the expression profiles of the individual, predominantly non-neoplastic cells, the profiles in CHL reflect the variable cellular composition of the TME.
The pioneering study by Devilard et al,13 published in 2002, used microarrays to measure the expression level of ∼1000 selected genes in 21 diagnostic and/or relapse biopsies from 20 patients. Although the small sample size and heterogeneity of the biopsies make the results of this study difficult to interpret, it provided early evidence that GEP contained signals from the TME that could be used to predict response to treatment. In the decade after this study, 3 further studies were performed,6,14,15 focusing on pretreatment biopsies and outcomes after upfront treatment. Sanchez-Aguilera et al14 and Steidl et al15 took an approach of outcome extremes, enriching their respective cohorts with biopsies from patients who had experienced treatment failure and using t-test-based methods to identify the genes that were differentially expressed between the 2 groups. Conversely, Chetaille et al6 used a cohort more representative of the outcomes typically observed in clinical practice and used univariate Cox analyses on time-to-event data to identify genes of interest.
At an individual gene level, there is very little consistency among these 3 studies, likely related to differences in cohort selection, the modest sample sizes, differences in microarray platforms, and the statistical strategies used. However, when these genes were organized into signatures representing biological processes and cell types, more commonality was seen. An overexpression of macrophage genes was found by both Sanchez-Aguilera et al14 and Steidl et al.15 Sanchez-Aguilera et al14 identified overexpression of genes associated with macrophages (including ALDH1A1, LYZ, and STAT1) as being associated with poor outcome. They validated these findings by performing IHC for STAT1 and ALDH1A1 on a tissue microarray (TMA) comprising biopsies from an independent cohort of 235 patients with CHL. STAT1 and ALDH1A1 staining was mainly restricted to macrophages and an increased number of stained cells (the upper quartile) was associated with inferior disease-specific survival (DSS). Steidl et al15 leveraged the concept that the GEP mainly reflected the absolute numbers of cell types present by comparing the GEP results with gene expression signatures characteristic of a broad array of cell types. Using this “molecular microscope” approach, they observed overexpression of macrophage genes in biopsies from patients who eventually experienced treatment failure.
As signatures of tumor-associated macrophages independently emerged from 2 of these discovery studies and the importance of macrophage polarization was well referenced for the pathogenesis and treatment response of solid cancers and other lymphomas,16,17 several follow-up studies prioritized the biological validation of tumor-associated macrophages in CHL.
Macrophages
The findings of GEP studies that a monocyte/macrophage signature was associated with outcome in the modern treatment era represented a reestablishment of a relationship that was first observed 30 years previously. In 1985, Ree and Kadin described patterns of peanut-agglutinin-binding cells in CHL.18 Staining was seen in both malignant cells and in macrophage histiocytes, with increased numbers of macrophages portending poor prognosis in a small group of MC and nodular-lymphocyte-predominant cases.
Steidl et al used IHC with an anti-CD68 antibody on a TMA containing the diagnostic biopsies of an independent cohort of 166 patients to validate the macrophage and monocyte signatures they had observed to be overexpressed in the biopsies of patients who experienced treatment failure.15 Increased CD68+ cells were significantly associated with reduced progression-free survival and DSS after ABVD (doxorubicin + bleomycin + vinblastine + dacarbazine) chemotherapy (with or without radiotherapy). Importantly, the prognostic impact of ≥5% CD68+ cells on DSS was independent of other recognized adverse clinical and laboratory parameters.
Biological validation
Many,19-31 but not all,24,32-34 IHC studies that followed have confirmed that increased numbers of tumor-associated macrophages are associated with inferior outcomes after upfront treatment (Table 1). These studies consolidate the biological validity of the relationship, extending the finding beyond the initial description into independent patient cohorts using a range of different antibodies and scoring methodologies to investigate outcome correlations across a range of end points, including overall survival (OS). Because these studies were almost universally conducted using biopsies from patients treated with ABVD (or equivalent) chemotherapy with or without radiotherapy, it remains unclear whether the biological association is maintained when alternative upfront treatment regimens, such as escalated BEACOPP (bleomycin + etoposide + doxorubicin + cyclophosphamide + vincristine + procarbazine + prednisone), are used.
Published studies examining the association between macrophages and outcome in adult classical Hodgkin lymphoma
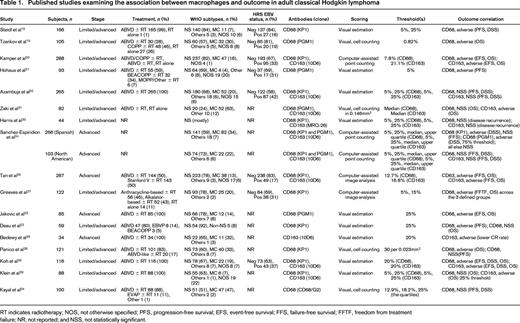
RT indicates radiotherapy; NOS, not otherwise specified; PFS, progression-free survival; EFS, event-free survival; FFS, failure-free survival; FFTF, freedom from treatment failure; NR, not reported; and NSS, not statistically significant.
Developing macrophages as a biomarker
Further studies are required to establish IHC for macrophages as a biomarker that can be used prospectively to risk-stratify patients (Figure 2). Specifically, an assay needs to be defined including a suitable antibody, staining methods, scoring methodologies, and thresholds that identify groups at different risk. Ultimately, to demonstrate the robustness and portability of the assay, it then needs to be externally validated in another laboratory in cohorts of patients independently from those in which the assay was trained.
Antibody characteristics.
The IHC studies published to date used a variety of different antibodies to stain for macrophages and, in doing so, have identified their performance characteristics. Azambuja et al35 reported that using antibodies to CD68 (the KP1 clone as used by Steidl et al15 ) resulted in low interobserver agreement with the visual estimation method—a finding confirmed by Klein et al.29 CD68 belongs to the family of scavenger receptors and represents a marker of lysosomes/endosomes. Accordingly, antibodies against CD68 are known to recognize not only macrophages, but also other cells potentially found in the TME, such as myeloid cells, fibroblasts, and some T cells.36 Related issues include the granular staining pattern, concentrated near the nucleus, which can be weak in the cytoplasmic projections, and the tendency toward high background staining; these issues may be dependent on the antibody clone used. Interobserver agreement is improved using antibodies to CD163,29,32 which are more specific for macrophages than either the KP1 or PGM1 clone for CD68.37 In addition, CD68 and CD163 may stain different cells within macrophage populations, with antibodies to CD163 reportedly staining alternatively activated (M2) macrophages, whereas antibodies to CD68 stain all macrophage populations. When antibodies to CD68 and CD163 have both been used, some studies have noted discrepancies between their associations with outcome. Sanchez-Espiridion et al24 observed an association between increased CD68 (KP1 clone)-stained cells and DSS in a Spanish cohort, but no association was seen using an antibody to CD163. In contrast, Zaki et al21 and Klein et al29 only saw an association between CD163 and outcome.
Scoring methods and thresholds.
Beyond the visual estimation method, several scoring methodologies have been used, ranging from cell counting19,21,34 to computer-assisted methods of point counting20,24 and computer-assisted image analysis.25,27 Although it is likely that these methods improve interobserver and interlaboratory agreement, this improvement comes at the expense of either being labor intensive or requiring specialized equipment. Few studies have retained the somewhat arbitrarily selected thresholds of <5%, 5%–25%, and >25% originally reported.15 This has led to a very wide range of suggested thresholds for separating patients into low- and high-risk populations, ranging from 0.82%19 to 25%. Although these thresholds are likely to be dependent on scoring methodology, few studies have validated these optimized thresholds.25,27
A recent study conducted using TMAs comprising biopsies from 287 participants in the Intergroup E2496 trial moved some way toward the goal of establishing macrophage IHC as a prognostic biomarker.25 This multicenter, phase 3, randomized controlled trial compared outcomes in patients treated with ABVD or Stanford V regimens. An independent training/validation cohort design was used to establish and then test thresholds for CD68 and CD163 IHC scored using an objective computer-assisted image analysis platform. Future studies will need to address the reproducibility of assay methods in tissue sections and between laboratories.
Macrophages and outcomes in specific populations
The population used by Steidl et al15 to establish the relationship between macrophages and outcomes was dominated by the NS subtype, with only 7% of the cases being the MC subtype. In total, 16% of the cases were EBV+ (Christian Steidl, BC Cancer Agency, Vancouver, Canada, April 2014, personal communication of unpublished data). The subsequent IHC studies have also treated CHL as a single disease, pooling patients with the 4 subtypes and cases that are EBV+ and EBV−. Although the prognostic significance of macrophages in adult patients with the NS subtype is well supported, the relationship between macrophages and outcomes in other patient populations is less certain.
EBV and MC subtype.
In the GEP study by Chetaille et al,6 overexpression of macrophage genes, including CD68 and CD163, were a prominent part of the signature that distinguished biopsies with EBV+ HRS cells from those that were EBV−. In all studies that have examined the relationship, EBV+ cases have a significantly higher number of CD68+20,25,31 and CD163+20,25 cells in the TME. When the prognostic significance of macrophages was examined in 49 cases of EBV+ advanced-stage CHL, there was no association found between CD68 IHC and failure-free survival or OS, although increased CD163 was associated with inferior outcomes.25 In a larger cohort of EBV+ cases, including limited and advanced stage, neither CD68 nor CD163 was associated with outcomes.35 This strongly suggests that the association between macrophages and outcomes is limited to EBV− cases. Given the strong association between EBV and the MC subtype, it is not surprising that biopsies of the MC subtype consistently show higher numbers of macrophages, a finding dating back to Ree and Kadin's study and confirmed using antibodies to both CD68 and CD163.20,25 Given that the MC subtype is not, in and of itself, associated with poor outcome, this raises doubt as to whether macrophages have prognostic power in this subtype.
Pediatric CHL.
Two studies, by Barros et al38 and Gupta et al,39 have investigated the relationship between macrophages and outcome in pediatric CHL, in 100 and 96 patients, respectively. Neither study found any significant relationship between CD68 IHC and outcomes, although Barros et al38 found that CD163 IHC was associated with progression-free survival in EBV− but not EBV+ cases. Gupta et al39 did not observe an association between CD163 and outcomes, but did not determine the EBV status of the HRS cells in their study.
CHL at relapse/progression.
Steidl et al15 reported that having ≥5% CD68+ cells in the diagnostic biopsy was associated with failure of autologous hematopoietic stem cell transplantation (ASCT) at relapse. Only one study, Casulo et al,40 has examined the prognostic significance of macrophages in the biopsy obtained at relapse in CHL. In a cohort of 81 patients with relapsed or refractory CHL undergoing salvage treatment before ASCT, increased CD68+ macrophages were associated with inferior OS in a univariate analysis. When they considered the 70 patients who received ASCT, those that had >30% CD68+ macrophages had inferior event-free survival and OS.
Gene expression-based models
A fundamentally different approach from that pursued with the macrophage studies is seen with the development of prognostic gene expression-based models. In these studies, the investigators used the combined prognostic power of individual genes and signatures with the express purpose of producing multigene biomarkers for risk stratification. This requires the transition of gene expression signatures derived from previous studies to technology platforms suitable for the reliable quantitation of gene expression in the highly fragmented RNA extracted from routinely produced formalin-fixed paraffin-embedded tissue (FFPET) (Figure 2).
Quantitative RT-PCR-based assays
The 13 gene assay described in Sanchez Espiridion et al41 in 2010 represents a distillation and translation of the findings from their GEP study14 into an assay suitable for FFPET. By reanalyzing their gene expression data and using bioinformatics tools, they identified 56 genes that were representative of pathways from both the malignant cells and the TME associated with poor outcomes.42 IHC was used to validate that, at a protein level, expression of a selection of these genes was enriched in either the HRS cells or the TME. The prognostic ability of these genes, quantitated using an RT-PCR assay based on Taqman low-density assay technology, was tested in 56 FFPET biopsies. Thirty of these genes were then carried forward to build and test a prognostic model using an independent training/validation approach in a cohort of 282 adult patients with advanced-stage CHL treated with ABVD or ABVD-like chemotherapy.41 In the training cohort of 183 patients, the final model of 11 genes (and 2 reference genes) was trained against the end point of unfavorable outcome (ie, failure to attain complete response or relapse within 12 months) and a threshold for separating patients into low-risk and high-risk was determined. These genes were selected to represent the pathways of apoptosis and cell cycle, purportedly in the HRS cells, and macrophage activation from the TME with the addition of IRF4. In the independent validation cohort of 79 patients, the assay's high-risk group had significantly inferior FFS but not OS. They report that the difference in FFS was significant in both NS and MC subtypes.
NanoString-based assays
Scott et al used an approach that integrates and harnesses the prognostic power of a multitude of previously described biomarkers, including cellular components of the TME discovered by IHC and individual genes and signatures identified through GEP.43 In total, 259 genes were examined using multiplex digital GEP on the NanoString platform. A model with 23 genes (with 3 reference genes) was built using penalized Cox regression on the data from 290 patients with advanced-stage CHL from the intergroup phase 3 randomized controlled trial E2496 comparing ABVD and Stanford V regimens. The model was trained on an end point of OS because this represents the outcome of the “package” of upfront ABVD and ASCT at relapse (for younger patients). The model and established threshold were then tested in a population-based cohort of advanced-stage CHL patients uniformly treated with ABVD enriched for treatment failure. A weighted analysis approach was used to correct for the enrichment, allowing unbiased estimates of the model's performance. The assay identified a high-risk group with significantly worse FFS and OS in the validation cohort independently of the International Prognostic Factors Project score, age >45 years, and CD68 IHC. The relatively low numbers of patients with the MC subtype and/or EBV+ CHL resulted in insufficient statistical power to determine whether the assay was prognostic in those specific groups.
Concluding remarks
In the years since the discovery of the relationship between tumor-associated macrophages and outcomes after upfront ABVD, a body of literature has accumulated validating and giving texture to this biological relationship. The precise biology underlying the association remains unclear and forthcoming studies are anticipated to determine whether macrophages represent direct participants in the chemoresistant phenotype or if they are a surrogate for genetic aberrations within HRS cells. The former scenario would spark further studies to target not only macrophages, but also by extension other elements in the TME therapeutically.
The initial promise that macrophage IHC would guide upfront treatment selection is yet to be realized. It is unclear whether it will be this biomarker or the potentially more robust gene expression-based assays that will penetrate clinical practice. Ultimately, for these biomarkers to guide upfront treatment choices, clinicians will need to be shown that alternative treatments overcome the poor prognosis in identified high-risk groups. In an era when treatment regimens continue to evolve, the further development of such predictive biomarkers will have to be achieved in parallel with clinical trials. A future may be envisaged when these predictive biomarkers, determined at diagnosis, are integrated with biomarkers of dynamic treatment response, tailoring the entire treatment package to produce optimal patient care.
Disclosures
Conflict-of-interest disclosure: The authors declare no competing financial interests. Off-label drug use: None disclosed.
Correspondence
David W. Scott, 675 W 10th Street, Vancouver, British Columbia V5Z 1L3, Canada. Phone: (604)675-8000; Fax: (604)675-8183; e-mail: dscott8@bccancer.bc.ca.