Abstract
Philadelphia chromosome–like acute lymphoblastic leukemia (Ph-like ALL) is a newly identified high-risk (HR) B-lineage ALL subtype, accounting for ∼15% of children with National Cancer Institute–defined HR B-ALL. It occurs more frequently in adolescents and adults, having been reported in as much as 27% of young adults with ALL between 21 and 39 years of age. It exhibits adverse clinical features, confers a poor prognosis, and harbors a diverse range of genetic alterations that activate cytokine receptor genes and kinase signaling pathways, making it amenable to treatment with tyrosine kinase inhibitor (TKI) therapy. Multiple groups are currently conducting clinical trials to prospectively screen patients with Ph-like ALL and incorporate the relevant TKI for those harboring ABL-class gene rearrangements or those with JAK-STAT pathway alterations. The success of combinatorial treatment of TKI with chemotherapy in the setting of Ph-positive ALL suggests that this approach may similarly improve outcomes for patients with Ph-like ALL. Hence, Ph-like ALL illustrates the modern treatment paradigm of precision medicine and presents unique opportunities for harnessing international collaborations to further improve outcomes for patients with ALL.
Learning Objectives
To recognize the clinical features and outcomes of Ph-like ALL
To understand the molecular hallmarks of Ph-like ALL
To describe the different Ph-like ALL subgroups and their therapeutic implications
Introduction
Acute lymphoblastic leukemia (ALL) is the commonest childhood cancer and undoubtedly portrays a true success story in the field of pediatric oncology. Since the 1960s, overall survival rates for children with ALL have risen from <10% to nearly 90%.1 This drastic outcome improvement stems from the efficacy of a multiagent chemotherapy regimen, better risk stratification as a result of the improved understanding of the biology of the leukemia, and careful monitoring of minimal residual disease as a measure of treatment response.2 Nevertheless, relapse still occurs in 15% to 20% of patients, and outcomes are dismal after relapse, making ALL the leading cause of cancer-related death among children and young adults.2,3 Over the last decade, advances in cancer genomics have shed important insights into our current understanding of the genomic landscape of ALL and have provided the rationale for novel therapeutic targets to expand the paradigm of precision medicine to this disease. Although ALL genomes are relatively silent compared with other tumor types,4 ALL is characterized by recurrent alterations that carry prognostic significance, allowing for subdivision of ALL into distinct genetic subtypes. The spectrum of these genetic alterations includes aneuploidy, copy number changes, sequence mutations, and chromosomal rearrangements that deregulate gene expression or result in the formation of chimeric fusion proteins.5 Recent large-scale genomic profiling and sequencing studies have led to important discoveries that deepen our understanding of the relationship between gene mutations, outcomes, and potential therapeutic interventions. One example is the identification of a particular subtype of B-ALL that is associated with a high risk of relapse, displays a gene expression profile (GEP) similar to that of Philadelphia (Ph) chromosome–positive ALL, and frequently harbors IKZF1 alterations, but lacks the hallmark BCR-ABL1 oncoprotein, thus known as Ph-like ALL.6-8 This review will highlight the most up-to-date knowledge on Ph-like ALL from the clinical, biologic, and therapeutic perspectives.
Ph-like ALL: the clinical picture
Ph-like ALL comprises 10% of children with standard-risk (SR) B-lineage ALL (age <10 years or white blood cell count ≤50 000/uL at diagnosis) and 15% of children who present with National Cancer Institute (NCI)–defined HR B-ALL (age >10 years or white blood cell count ≥50 000/uL at diagnosis).6 Its frequency increases with age, rising to 21% in adolescents and up to 27% in young adults with B-ALL.6 It is almost 3 times more common than Ph-positive ALL. In contrast to the rising incidence of Ph-positive ALL with age, the incidence of Ph-like ALL seems to peak in adolescents and young adults, and decreases to <10% in older adults between the ages of 40 and 85, as reported in a single German cohort.9 However, recent genomic profiling data from a larger multicentered cohort of >600 adults with B-ALL confirms the high frequency of Ph-like ALL within this population, estimated to be 20% in adults aged 40 to 79 years.10 Males are more commonly affected than females across all age groups, with a male-to-female ratio of 1.5:1 among children, and rising to 4:1 among young adults.6 Ph-like ALL seems to have a predilection among patients of Hispanic descent, which could be partly explained by the high association of CRLF2 rearrangements within this ethnic group.11 Moreover, a recent genome-wide association study identified an inherited susceptibility locus for Ph-like ALL within the GATA3 gene (rs3824662), which is also markedly more common in patients of Hispanic ethnicity and Native American genetic ancestry.12
Patients with Ph-like ALL often exhibit adverse clinical features. These patients generally present with an initial leukocyte count >100 000/uL at diagnosis and elevated minimal residual disease at the end of induction therapy compared with patients with non–Ph-like ALL.6 Notably, high levels of end-of-induction MRD and induction failures appear to be particularly common in Ph-like cases harboring the EBF1-PDGFRB fusion.13 In a multivariable analysis, the presence of a Ph-like GEP constitutes an independent negative prognostic factor.14 This is further supported by the inferior outcomes of Ph-like ALL patients compared with non–Ph-like ALL across all age groups, with a dismal prognosis for young adults with Ph-like ALL (Figure 1).6 Among adolescents with Ph-like ALL, outcomes remain inferior compared with those with other high-risk ALL subtypes such as KMT2A-rearranged ALL and Ph-positive ALL. In the Children’s Oncology Group’s (COG) most recently completed phase 3 randomized controlled trial for HR-ALL, AALL0232, the 5-year event-free survival rate for Ph-like cases was 63% compared with 86% for non–Ph-like cases.14 Importantly, the outcome differences were maintained regardless of the randomized treatment arm for patients with Ph-like ALL, whereas for patients with non–Ph-like ALL, this study demonstrated the superiority of high-dose methotrexate over Capizzi-style methotrexate in the first interim maintenance phase. Outcome differences also exist between different Ph-like ALL subgroups, with JAK2 and EPOR rearrangements having the worst outcome, and Ph-like cases with IKZF1 alteration faring worse than those without IKZF1 alteration.6 Despite the overall poor outcomes in Ph-like ALL, one single-institution study reported comparable outcomes with that of patients with non–Ph-like ALL using an MRD-based risk-directed treatment intensification approach.15 However, the study population was small, with different clinical and biologic characteristics, compared with previous published pediatric Ph-like ALL cohorts, and a significantly higher proportion of patients were treated with hematopoietic stem cell transplantation.
Outcomes of children, adolescents, and young adults with Ph-like ALL. (A) Patients with Ph-like ALL have an inferior outcome compared with patients with non–Ph-like patients treated on AALL0232. (B) Among patients with Ph-like ALL, young adults have the worst prognosis compared with children and adolescents. Adapted from Roberts et al6 and Loh et al,14 with permission.
Outcomes of children, adolescents, and young adults with Ph-like ALL. (A) Patients with Ph-like ALL have an inferior outcome compared with patients with non–Ph-like patients treated on AALL0232. (B) Among patients with Ph-like ALL, young adults have the worst prognosis compared with children and adolescents. Adapted from Roberts et al6 and Loh et al,14 with permission.
Ph-like ALL: the genomic landscape
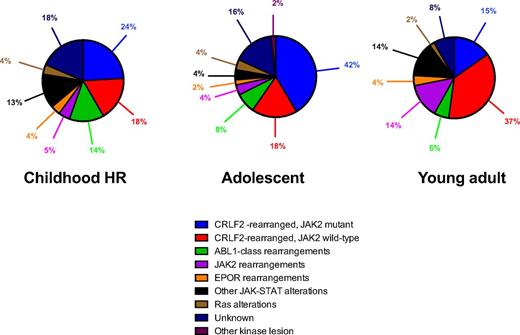
The identification of the Ph-like ALL subset has led to multicollaborative genomic profiling efforts intended to gain insights into the biology of this disease. A recent comprehensive genomic analysis of 154 Ph-like ALL samples generated via different next-generation sequencing platforms, including transcriptome, whole-exome, and whole-genome sequencing, has deciphered the heterogeneous nature of the genomic landscape.9 Despite its complexity, the common molecular hallmark of Ph-like ALL constitutes a myriad of genetic alterations that activate cytokine receptor genes and kinase-signaling pathways, providing the rationale for the use of targeted therapies in this subtype. These alterations can be subdivided into 5 distinct subgroups based on the type of cytokine receptor or kinase fusion present: (1) rearrangements of CRLF2, (2) ABL-class gene rearrangements, (3) JAK2 and EPOR rearrangements, (4) sequence mutations or deletions activating JAK-STAT- or MAPK signaling pathways, and (5) other rare kinase alterations (Figure 2; Table 1).6,16,17
Distribution of Ph-like ALL subgroups among children, adolescents, and young adults. CRLF2 rearrangements are the most prevalent genetic alteration across all age groups. ABL-class gene rearrangements are more frequent in childhood, whereas there is a striking increase in the frequency of JAK2 rearrangements among young adults with Ph-like ALL. HR, high-risk. Adapted from Roberts et al,6 with permission.
Distribution of Ph-like ALL subgroups among children, adolescents, and young adults. CRLF2 rearrangements are the most prevalent genetic alteration across all age groups. ABL-class gene rearrangements are more frequent in childhood, whereas there is a striking increase in the frequency of JAK2 rearrangements among young adults with Ph-like ALL. HR, high-risk. Adapted from Roberts et al,6 with permission.
Repertoire of kinase rearrangements in Ph-like ALL along with their partner genes and potential therapeutic targets
| Kinases . | 5′ partner genes (number of patients) . | Potential TKI . | Clinical trials . |
|---|---|---|---|
| ABL1 | ETV6 (3), NUP214 (6), RCSD1 (1), RANBP2 (1), SNX2 (1), ZMIZ1 (2) | Dasatinib | AALL1131 |
| ABL2 | PAG1 (1), RCSD1 (4), ZC3HAV1 (2) | Dasatinib | AALL1131 |
| PDGFRB | EBF1 (6), SSBP2 (1), TNIP1 (1), ZEB2 (1) | Dasatinib | AALL1131 |
| CSF1R | SSBP2 (4) | Dasatinib | AALL1131 |
| CRLF2 | IGH (19), P2RY8 (11) | Ruxolitinib | AALL1521 |
| JAK2 | ATF7IP (1), BCR (2), EBF1 (1), ETV6 (2), PAX5 (7), PPFIBP1 (1), SSBP2 (2), STRN3 (1), TERF2 (1), TPR (1) | Ruxolitinib | AALL1521 |
| EPOR | IGH (7), IGK (2) | Ruxolitinib | AALL1521 |
| TSLP | IQGAP2 (1) | Ruxolitinib | AALL1521 |
| IL2RB | MYH9 (1) | JAK1/JAK3 inhibitor | N/A |
| TYK2 | MYB (1) | TYK2 inhibitor | N/A |
| NTRK3 | ETV6 (1) | Crizotinib | N/A |
| PTK2B | KDM6A (1), STAG2 (1) | FAK inhibitor | N/A |
| DGKH | ZFAND3 (1) | Unknown | N/A |
| Kinases . | 5′ partner genes (number of patients) . | Potential TKI . | Clinical trials . |
|---|---|---|---|
| ABL1 | ETV6 (3), NUP214 (6), RCSD1 (1), RANBP2 (1), SNX2 (1), ZMIZ1 (2) | Dasatinib | AALL1131 |
| ABL2 | PAG1 (1), RCSD1 (4), ZC3HAV1 (2) | Dasatinib | AALL1131 |
| PDGFRB | EBF1 (6), SSBP2 (1), TNIP1 (1), ZEB2 (1) | Dasatinib | AALL1131 |
| CSF1R | SSBP2 (4) | Dasatinib | AALL1131 |
| CRLF2 | IGH (19), P2RY8 (11) | Ruxolitinib | AALL1521 |
| JAK2 | ATF7IP (1), BCR (2), EBF1 (1), ETV6 (2), PAX5 (7), PPFIBP1 (1), SSBP2 (2), STRN3 (1), TERF2 (1), TPR (1) | Ruxolitinib | AALL1521 |
| EPOR | IGH (7), IGK (2) | Ruxolitinib | AALL1521 |
| TSLP | IQGAP2 (1) | Ruxolitinib | AALL1521 |
| IL2RB | MYH9 (1) | JAK1/JAK3 inhibitor | N/A |
| TYK2 | MYB (1) | TYK2 inhibitor | N/A |
| NTRK3 | ETV6 (1) | Crizotinib | N/A |
| PTK2B | KDM6A (1), STAG2 (1) | FAK inhibitor | N/A |
| DGKH | ZFAND3 (1) | Unknown | N/A |
Adapted from Roberts et al,6 with permission.
CRLF2 rearrangements that result in its overexpression represent the most prevalent alteration in Ph-like ALL, accounting for 47% cases.6 The CRLF2 gene at locus Xp22.3/Yp11.3 encodes the thymic stromal lymphopoetin receptor and heterodimerizes with the IL-7R α-chain to bind its ligand, thymic stromal lymphopoietin (TSLP), leading to downstream signaling that mediates processes such as lymphopoiesis, allergic reaction, and inflammation.18 Three mechanisms of CRLF2 deregulation have been described: (1) translocation of CRLF2 to the immunoglobulin heavy-chain transcriptional enhancer (14q32.3; IGH@-CRLF2); (2) a focal interstitial deletion of the pseudo-autosomal region of the sex chromosomes (Xp22.23 or Yp11.32) centromeric to CRLF2, resulting in the fusion of CRLF2 to the G-protein purinergic receptor P2RY8 gene (P2RY8-CRLF2); and (3) the less common activating CRLF2 point mutation F232C.16,19,20 In addition to Ph-like ALL, CRLF2 rearrangements are frequently observed in Down syndrome–associated ALL and are age dependent, with P2RY8-CRLF2 rearrangements occurring more commonly in younger patients, whereas IGH@-CRLF2 predominate among adolescents and young adults, particularly those of Hispanic ancestry.19,20 Moreover, half of CRLF2-rearranged cases harbor concomitant JAK mutations, the most common of which is the R683G point mutation located in the pseudokinase domain of JAK2.19,21,22
The second major subgroup, comprising 13% of Ph-like ALL, involves rearrangements of ABL-class genes such as ABL1, ABL2, PDGFRB, and CSF1R.6,17 Multiple fusion partners have been identified for each one of these; but all fusions conserve the intact kinase domain of the carboxyl terminal of the ABL-class gene juxtaposed in frame to the amino terminal of the partner gene.
JAK2 and EPOR rearrangements together constitute 11% of Ph-like ALL, but there is a remarkable increase in the prevalence of JAK2 fusions among young adults compared with that in children and adolescents (∼15% vs 5%, respectively).6,16 In Ph-like ALL, JAK2 represents a promiscuous gene with 10 different reported fusion partners. EPOR rearrangements have been identified in ∼4% of Ph-like ALL. Four types of EPOR rearrangements have been described, with each type involving the juxtaposition of the EPOR gene to the enhancer regions of immunoglobulin heavy or κ loci, leading to the deregulated expression of a truncated form of EPOR that has been shown to drive leukemogenesis.23 Both JAK2 and EPOR rearrangements lead to constitutive activation of JAK-STAT signaling pathways, which can be abrogated in vitro and in some preclinical xenograft models by the use of JAK inhibitors such as ruxolitinib.6,17,23,24
Other than kinase fusions, sequence mutations and focal deletions of genes leading to activation of the JAK-STAT pathway are present in 13% of Ph-like ALL.6 The spectrum of alterations within this subgroup includes mutations of IL7R, FLT3, and IL2RB that constitutively activate these cytokine receptors, activating mutations within the JAK kinases themselves (JAK1 and JAK3), and deletion of genes that encode negative regulators of the JAK-STAT pathway (SH2B3). Point mutations in the JAK kinases are usually accompanied by rearrangements of CRLF2, whereas IL7R mutations arise mostly within the transmembrane domain to promote constitutive JAK-STAT signaling. Ras pathway mutations occur in 4% of patients with Ph-like ALL; these include activating mutations of NRAS, KRAS, PTPN11, and NF1, which have also been reported in other ALL subtypes such as hypodiploid ALL,25 KMT2A-rearranged ALL,26 and relapsed ALL.26 Rare kinase fusions involving NTRK3 and DGKH occur in 0.9% of Ph-like ALL. Kinase-activating lesions were not identified in the remaining 5% of Ph-like ALL. Although still considered to be Ph-like, the GEP of patients with kinase fusions (ABL-class, JAK2, and EPOR rearrangements) is distinct from those with other JAK-STAT or Ras pathway alterations. Finally, similarly to Ph-positive ALL, a unifying hallmark of Ph-like ALL is the high frequency of IKZF1 alterations relative to BCR-ABL1–negative non–Ph-like ALL (68% vs 16%, P < .001). IKZF1 alterations occur more frequently in patients with Ph-like ALL, harboring a kinase fusion than in those with a sequence mutation (78% vs 33%, P < .001).6
Ph-like ALL: therapeutic opportunities for precision medicine trials
The recently characterized genomic landscape of Ph-like ALL has revealed a diverse array of targetable kinase-activating lesions that are amenable to tailored kinase inhibitor therapy. Despite its genetic complexity, the majority of Ph-like ALL cases have a targetable lesion involving the ABL or JAK-STAT signaling pathways. Extensive in vitro and ex vivo data have provided compelling evidence to incorporate relevant TKIs in combination with chemotherapy in this ALL subtype. A priori, all ABL-class fusions such as ABL1, ABL2, PDGFRB, and CSF1R rearrangements exhibit exquisite sensitivity to ABL inhibitors (imatinib or dasatinib), whereas JAK2/EPOR rearrangements as well as other activating mutations of the JAK-STAT pathway can be inhibited by JAK inhibitors (eg, ruxolitinib) in both murine pre–B cell lines and patient-derived xenograft models.6,17,27 CRLF2-rearranged Ph-like ALL demonstrates hyperactive JAK-STAT, PI3K, mTOR, and BCL2 signaling, and therapies targeting these pathways are currently being investigated in preclinical and clinical studies.18,27,28 Other rare kinase alterations in Ph-like ALL can also be targeted by crizotinib, FAK inhibitors, and TYK2 inhibitors for NTRK3, PTK2B, and TYK2 fusions, respectively.6 Coupled with these data suggesting the efficacy of TKI therapy are anecdotal reports of patients with Ph-like ALL harboring the EBF1-PDGFRB fusion who have sustained durable responses after treatment with ABL inhibitors in combination with conventional chemotherapy.6,29 In light of the dramatic survival improvement of Ph-positive ALL upon the incorporation of imatinib with chemotherapy,30 the aforementioned lines of evidence provide a strong rationale for the prospective investigation of the efficacy of combinatorial treatment of TKI and chemotherapy in Ph-like ALL with targetable lesions.
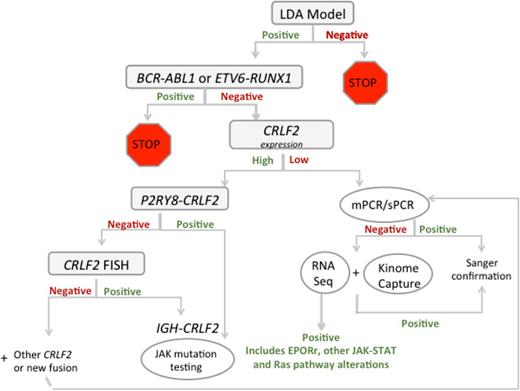
Given the heterogeneity of Ph-like ALL, translation of genomic discoveries into pragmatic real-time clinical algorithms can be challenging. Several pediatric consortiums have adopted different strategies to screen for Ph-like ALL patients to modify their therapy with the relevant TKI. St. Jude’s Children’s Research Hospital uses a next-generation sequencing platform to screen for all patients newly diagnosed with ALL to identify those with Ph-like ALL, whereas the UK group focuses their screen on ABL-class fusions in patients who respond poorly to therapy defined as induction failure, positive MRD at day 28, or persistent MRD at week 14.13 COG has incorporated an algorithm to screen for the Ph-like GEP among all newly diagnosed NCI HR B-ALL patients using a well-validated low-density array card (Figure 3).6,31 Those positively screened for Ph-like GEP during induction will undergo additional genetic testing to identify the associated kinase lesion. Subsequently, dasatinib will be initiated at the beginning of consolidation therapy on a modified BFM-based chemotherapy backbone for patients harboring ABL-class fusions on the current phase 3 trial for HR-ALL, AALL1131. Furthermore, the phase 2 AALL1521 trial aims to determine the optimal dosage of ruxolitinib in combination with chemotherapy, and in addition will assess the efficacy of this combinatorial regimen in Ph-like ALL patients with CRLF2 rearrangements and/or other JAK-STAT pathway alterations. The lack of a uniform molecular definition of Ph-like ALL may account for the variability in the reported frequencies of various genetic lesions. However, it should be emphasized that the ultimate goal is to therapeutically target the underlying kinase lesion rather than identify the Ph-like signature that clusters them. This can be achieved through different approaches, from standard reverse-transcription polymerase chain reaction, fluorescence in situ hybridization, phosphoflow signaling analysis, NanoString assay, or other next-generation genome sequencing techniques, each of which is being pursued actively by different groups of investigators. Other Ph-like subgroups such as those harboring NTRK3 fusions or Ras pathway mutations are infrequent, precluding the possibility of designing a dedicated trial for these lesions. However, because Ras mutations represent a hallmark of near-haploid ALL25 and are frequently found in relapsed ALL,32 one could envision the inclusion of this subgroup into a larger trial designed to study other Ras-mutated ALL subtypes. Direct oncogenic Ras inhibition remains challenging, but targeting downstream Ras pathway effectors such as MEK has demonstrated significant promise and is currently being evaluated prospectively in early-phase trials.32 The ALK inhibitor, crizotinib, is currently being used as up-front therapy or in the setting of disease relapse for several solid tumor types.33 However, one ongoing challenge lies in the integration of these novel therapies into the existing ALL chemotherapy backbone.
Testing algorithm for Ph-like ALL in COG trials. COG, Children’s Oncology Group; LDA, low-density array; mPCR, multiplex polymerase chain reaction; sPCR, singleplex polymerase chain reaction. Courtesy of Shalini Reshmi, Cytogenetics/Molecular Genetics Laboratory, National Children’s Hospital, Columbus, OH.
Testing algorithm for Ph-like ALL in COG trials. COG, Children’s Oncology Group; LDA, low-density array; mPCR, multiplex polymerase chain reaction; sPCR, singleplex polymerase chain reaction. Courtesy of Shalini Reshmi, Cytogenetics/Molecular Genetics Laboratory, National Children’s Hospital, Columbus, OH.
In addition to the aforementioned TKIs, other novel molecular targets have shown promise in preclinical models, suggesting their potential therapeutic application to Ph-like ALL. First, inhibition of heat-shock protein 90 (HSP90), a molecular chaperone that promotes the degradation of wild-type or mutant JAK2 by AUY922 or PU-H71, has demonstrated efficacy in inhibiting proliferation and suppressing downstream signaling in CRLF2-rearranged or JAK-mutant ALL cell lines in addition to murine models that became insensitive to JAK inhibitors or acquired therapy-resistant kinase domain mutations after JAK inhibition.34,35 This serves as a proof-of-concept for further evaluation of HSP90 inhibition combined with JAK inhibitors or other ALL therapies in JAK-dependent ALL. Moreover, preclinical efficacy of retinoids in IKZF1-mutated BCR-ABL1 cell lines and patient-derived xenograft models have been recently described.36 IKZF1 alterations drive stem cell renewal, cause abnormal bone marrow adhesion, and result in decreased TKI sensitivity. Retinoids have been shown to reverse this phenotype and potentiate TKI activity in IKZF1-altered mouse and human BCR-ABL1 ALL. Altogether, HSP90 inhibitors and retinoids represent potential therapeutic candidates given the prevalence of JAK-STAT pathway and/or IKZF1 alterations in Ph-like ALL.
Conclusions and future directions
Ph-like ALL is a newly identified ALL subtype defined by its unique GEP, which is associated with a poor prognosis. Given the high prevalence of targetable kinase-activating lesions identified in patients with Ph-like ALL, there is compelling evidence from both experimental models and clinical observations for the use of relevant TKIs in combination with conventional chemotherapy. Although Ph-like ALL is 3 times more common than Ph-positive ALL, its genetic heterogeneity represents a limiting factor to develop well-designed, statistically powered, randomized controlled trials to establish TKI efficacy for each distinct subset (ABL-class vs JAK-dependent subgroups). This highlights the need to harness international collaborations to systematically study rare, high-risk disease entities to improve outcomes. Successful examples of such international collaborative efforts include studies of KMT2A-rearranged infant ALL and Ph-positive ALL.37 In view of these potential global collaborations, different screening strategies for Ph-like ALL should be standardized among different groups to ensure comparable outcomes. In parallel, future challenges in Ph-like ALL should also focus on investigating mechanisms of resistance, as we learned from the adult chronic myeloid leukemia and Ph-positive ALL experiences regarding the emergence of drug-resistant kinase domain mutations after long-term TKI exposure.38 In vitro saturation mutagenesis screens of Ba/F3 murine cells harboring EBF1-PDGFRB have identified the analogous ABL1 T315I gatekeeper kinase domain (KD) mutation T681I in the majority of dasatinib-resistant clones.39 Furthermore, the recent case report of disease relapse mediated by the T315I KD mutation in a Ph-like ALL patient harboring the ETV6-ABL1 fusion after treatment with dasatinib demonstrates the need for future studies investigating mechanisms of acquired resistance and identifying novel therapies in the relapsed setting.40 Finally, Ph-like ALL illustrates the new paradigm of how genomic discoveries translate into novel therapeutic avenues in the era of precision medicine and presents new opportunities to improve outcomes in high-risk subsets of B-ALL.
Correspondence
Mignon Loh, Loh Laboratory, Helen Diller Family Comprehensive Cancer Center, Box 3112, 1450 3rd St, Room 284, San Francisco, CA 94158; e-mail: mignon.loh@ucsf.edu.
References
Competing Interests
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Author notes
Off-label drug use: Use of TKIs in Ph-like ALL patients with kinase-activating lesions.