Abstract
Substantial progress has been made in our understanding of the pathogenetic basis of myeloproliferative neoplasms. The discovery of mutations in JAK2 over a decade ago heralded a new age for patient care as a consequence of improved diagnosis and the development of therapeutic JAK inhibitors. The more recent identification of mutations in calreticulin brought with it a sense of completeness, with most patients with myeloproliferative neoplasm now having a biological basis for their excessive myeloproliferation. We are also beginning to understand the processes that lead to acquisition of somatic mutations and the factors that influence subsequent clonal expansion and emergence of disease. Extended genomic profiling has established a multitude of additional acquired mutations, particularly prevalent in myelofibrosis, where their presence carries prognostic implications. A major goal is to integrate genetic, clinical, and laboratory features to identify patients who share disease biology and clinical outcome, such that therapies, both existing and novel, can be better targeted.
Learning Objectives
Understand the central role of aberrant JAK-STAT signaling in driving the myeloproliferative phenotype
Appreciate the factors that may influence the earliest stages of MPN emergence
Become familiar with the range of additional mutations in MPN and their role in disease biology
Introduction
The myeloproliferative neoplasms (MPNs) are clonal hematopoietic disorders characterized, in chronic phase, by an overproduction of differentiated hematopoietic cells. The Philadelphia-negative MPNs include 3 main diseases: polycythemia vera (PV), essential thrombocythemia (ET), and myelofibrosis (MF).1 The last decade has witnessed dramatic advances in our understanding of the molecular and cellular basis of excessive myeloproliferation, and this has led to the development and therapeutic use of novel targeted treatments, such as JAK inhibitors. More recently, we have begun to accumulate tantalizing insights into the origins of these cancers, as well as the factors that drive later disease evolution.
Here, we review our current understanding of the evolution of MPNs, from their origins to the emergence of disease and clinical progression. Our evolutionary narrative begins with a review of the molecular basis of myeloproliferation in an established neoplastic clone. We then address the factors that influence disease phenotypes in chronic phase and the basis for disease progression, after which we discuss what is known about how clonal populations of MPN cells might initially emerge. We conclude by considering the potential implications for clinical practice.
Phenotypic driver mutations converge on JAK-STAT signaling
The cardinal and mutually exclusive mutations in MPNs occur in JAK2, CALR, or MPL, referred to herein as the “phenotypic drivers” because of their role in driving the myeloproliferative phenotype. In 2005, a single point mutation resulting in JAK2V617F was identified in most patients with PV and half of those with ET or MF.2-5 JAK2 is intimately associated with the cytoplasmic portions of receptors for key hematopoietic cytokines, such as erythropoietin, thrombopoietin (TPO), and granulocyte colony-stimulating-factor. Normal JAK2 functions to activate intracellular signaling pathways following ligand binding; however, JAK2V617F is rendered constitutively active. The mutation is understood to result in loss of the normal inhibitory function provided by the pseudokinase (JH2) domain upon the active (JH1) kinase domain—in this model, it is unclear whether the disrupted JH1/JH2 interface occurs within an individual JAK2 molecule or between JAK2 dimers.6 The mutation may also result in direct activation of the JH1 domain via an SH2-JH2 linker.7 Subsequent downstream activation of intracellular signaling occurs via signal transducer and activator of transcription (STAT) protein signaling and, to a lesser extent, via mitogen-activated protein kinase and phosphoinositide 3-kinase signaling pathways, which together result in excessive myeloid cell proliferation and differentiation.
Additional genetic aberrations that perturb JAK-STAT signaling are found in JAK2-unmutated MPNs. Mutations in the TPO receptor c-MPL at W515, and less commonly at S505, are found in 5% to 8% of patients with ET and MF.8 These mutations are understood to result in conformation changes to the receptor that mimic the consequences of TPO binding, such that cytoplasmic JAK2 molecules are brought into close proximity conducive for activation, transphosphorylation, and ligand-independent intracellular signaling.9 As might be expected from the normal function of MPL, these mutations are associated with megakaryocyte proliferation, and disease phenotypes of ET and MF are recapitulated in murine models.8
In 2013, mutations in calreticulin (CALR) were identified in most JAK2- or MPL-unmutated patients with ET or MF.10,11 CALR mutations are insertions or deletions affecting the terminal exon of the gene, which all result in a +1 base pair shift to the amino acid reading frame of the DNA sequence, such that the mutant protein acquires a novel C terminus. The identification of such mutations was surprising because CALR was neither a cytokine receptor nor a known participant in JAK-STAT signaling. CALR is best known for its housekeeping function as a chaperone protein in the endoplasmic reticulum (ER), where it aids in the appropriate folding of client protein molecules before their trafficking, either to the cell surface or for extracellular secretion.12 However, knowledge that c-MPL is an ER client protein and that CALR mutations were associated exclusively with only ET or MF (hence resembling MPL-mutated MPNs) suggested the possibility that mutant CALR somehow activates MPL. Consistent with this, mutant CALR can induce an ET/MF phenotype in a variety of mouse models, but only in the presence of MPL.13-17 The mechanism by which mutant CALR complexes with MPL to result in its constitutive activation remains unclear. The extracellular domain of MPL, along with both the N-terminal and mutant C-terminal of CALR, appear necessary for this interaction, which may be facilitated by the newly acquired positive electrostatic charge within the C terminus.13,15 It is unclear why mutant CALR complexes preferentially with MPL and whether mutant CALR–mediated MPL activation occurs after transport from the ER to the cell surface, or whether aberrant activation occurs intracellularly.
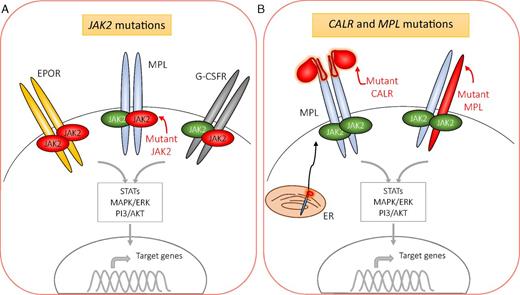
Uncommon variants in phenotypic drivers also lead to constitutive activation of JAK-STAT signaling, such as mutations in exon 12 of JAK2 and the recently identified noncanonical variant MPLS204P.18-20 Occasionally, other proteins that participate in JAK-STAT signaling (eg, SH2B3 and CBL) are also mutated in MPNs, although they are less frequently found as an isolated somatic mutation in patients.21,22 Overall, excepting the ∼10% of patients with MF or ET who have as-yet undiscovered drivers of their disease,10 aberrant activation of intracellular signaling via erythropoietin receptor (EPOR) and/or MPL remains central to the development of the MPN phenotype, regardless of whether the phenotypic driver mutation is in JAK2, CALR, or MPL (Figure 1).
Mutations in JAK2, CALR, and MPL drive excessive myeloproliferation via constitutively active signaling downstream of JAK2. JAK2 associates with the cytoplasmic portion of a variety of receptors, such as those for erythropoietin (EPOR), thrombopoietin (MPL), and granulocyte/macrophage colony-stimulating factor (G-CSFR). JAK2 is also activated in response to additional cytokines (eg, growth hormone and IL-5) (not shown). (A) Mutant JAK2, shown in red, is constitutively active and leads to variable levels of erythroid, megakaryocytic, and, to a lesser degree, granulocytic proliferation and differentiation. It is unclear whether mutant JAK2 dimerizes with mutant or wild-type JAK2 with respect to the individual receptors. (B) Mutations in CALR and MPL result in aberrant activation of signaling downstream of the MPL receptor. Mutant CALR complexes with MPL in the ER. Both mutations in CALR and MPL result in receptor dimerization and activation of JAK2. MAPK/ERK, mitogen-activated protein kinases/extracellular signal-regulated kinases; PI3/AKT, phosphoinositide 3-kinase/serine/threonine kinase Akt; STAT, signal transducer and activator of transcription.
Mutations in JAK2, CALR, and MPL drive excessive myeloproliferation via constitutively active signaling downstream of JAK2. JAK2 associates with the cytoplasmic portion of a variety of receptors, such as those for erythropoietin (EPOR), thrombopoietin (MPL), and granulocyte/macrophage colony-stimulating factor (G-CSFR). JAK2 is also activated in response to additional cytokines (eg, growth hormone and IL-5) (not shown). (A) Mutant JAK2, shown in red, is constitutively active and leads to variable levels of erythroid, megakaryocytic, and, to a lesser degree, granulocytic proliferation and differentiation. It is unclear whether mutant JAK2 dimerizes with mutant or wild-type JAK2 with respect to the individual receptors. (B) Mutations in CALR and MPL result in aberrant activation of signaling downstream of the MPL receptor. Mutant CALR complexes with MPL in the ER. Both mutations in CALR and MPL result in receptor dimerization and activation of JAK2. MAPK/ERK, mitogen-activated protein kinases/extracellular signal-regulated kinases; PI3/AKT, phosphoinositide 3-kinase/serine/threonine kinase Akt; STAT, signal transducer and activator of transcription.
Relationship between phenotypic drivers and clinical presentation
CALR-mutated ET patients display a phenotype that is remarkably similar to those with MPL mutations. Both disease subgroups are characterized by isolated thrombocytosis and no gender bias at presentation.23,24 However, CALR-mutated patients are significantly younger at presentation than both MPL-mutated and JAK2-mutated counterparts. This is reminiscent of the disease characteristics of PV patients with JAK2V617F and JAK2exon12 mutations – those with the latter exhibit more marked erythrocytosis and present at a younger age,18 differences which may reflect differential strengths of aberrant signaling via the EPOR. By analogy, one can speculate that CALR mutations may result in higher levels of MPL signaling compared with those induced by heterozygous mutations in MPL or JAK2. Such a hypothesis accords with the observation that patients with CALR-mutated ET have an increased risk of myelofibrotic transformations,25 particularly because excessive signaling via the MPL receptor is associated with bone marrow fibrosis in transgenic mice26 and increased bone marrow reticulin is seen in patients with ET who have homozygous MPL mutations.27 This would also be consistent with the observation that CALR mutations that result in a more extensive mutant C terminus, for example, the type 1 52-bp deletion (L367fs*46) are more prevalent among patients with MF10,11 and result in a more severe phenotype with frequent MF transformation in a retroviral CALR-mutant mouse model.14
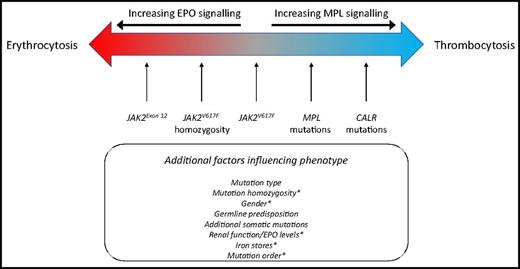
In contrast to CALR or MPL, mutated JAK2 is pleiotropic and results in numerous MPN phenotypes also recapitulated in JAK2V617F mouse models.28 Many factors determine the phenotype that results after a JAK2 mutation. First, specific types of JAK2 mutation, for example, V617I, V617F, or mutations in exon 12, are associated with differing degrees of erythrocytosis vs thrombocytosis.18,29 Recent data demonstrate that this may be due to the differential coupling of certain mutant JAK2 proteins with cytokine receptors.30 Exon 12 mutation K539I has been shown predominantly to bind EPOR, whereas JAK2V617F preferentially complexes with MPL.30 Second, homozygosity for JAK2V617F is a key factor in determining the degree of erythrocytosis that subsequently develops, both in patient samples and animal models.31-33 One can hypothesize that biallelic expression of JAK2V617F may be required for the mutant protein to preferentially complex with EPOR, although this is yet to be demonstrated. While clones bearing homozygous JAK2V617F can be found as minor subclones in patients with ET, they are clonally dominant in patients with PV.34 The factors that influence the degree of expansion of homozygous JAK2V617F clones are not fully understood, but the order in which JAK2V617F is acquired relative to other somatic mutations has been shown to be important.35 Third, patient-specific factors, such as iron status, age, sex, coexistent β-thalassemia traits, and timing of presentation (relative to the disease course in a given patient), may all influence whether a patient harboring JAK2V617F presents with significantly elevated red cell parameters and/or thrombocytosis at diagnosis36 (Figure 2).
Clinical presentation in chronic phase and relationship to phenotypic driver mutation. PV and ET are modeled as a disease spectrum along a biological continuum where different genetic lesions skew the clinical phenotype from that of thrombocytosis to that of additional erythrocytosis (±leukocytosis). CALR mutations result in excessive MPL signaling, in a manner similar to that resulting from MPL mutations. JAK2 mutations signal downstream of multiple cell surface receptors, including MPL, and are thus associated with thrombocytosis but also erythrocytosis and leukocytosis. The exact nature of the phenotypic driver mutation, germline genetic background, and additional somatic mutations influence disease phenotype. *In the context of JAK2V617F, several factors modulate the balance between erythrocytosis and thrombocytosis, including sex, mutation homozygosity, and patient-specific factors such as erythropoietin (EPO) levels, renal function, and iron status.
Clinical presentation in chronic phase and relationship to phenotypic driver mutation. PV and ET are modeled as a disease spectrum along a biological continuum where different genetic lesions skew the clinical phenotype from that of thrombocytosis to that of additional erythrocytosis (±leukocytosis). CALR mutations result in excessive MPL signaling, in a manner similar to that resulting from MPL mutations. JAK2 mutations signal downstream of multiple cell surface receptors, including MPL, and are thus associated with thrombocytosis but also erythrocytosis and leukocytosis. The exact nature of the phenotypic driver mutation, germline genetic background, and additional somatic mutations influence disease phenotype. *In the context of JAK2V617F, several factors modulate the balance between erythrocytosis and thrombocytosis, including sex, mutation homozygosity, and patient-specific factors such as erythropoietin (EPO) levels, renal function, and iron status.
Genetic interactions influence disease presentation and progression
Several additional genetic interactions shape the molecular, cellular, and clinical consequences of phenotypic driver mutations in MPN.
Germline predisposition
Constitutional variation that predisposes to MPNs can be classified into 2 groups: (1) common variants, prevalent in the population, that result in a small predisposition to MPN, and (2) rare variants, often found in familial MPNs, that have a higher penetrance for disease. Well-recognized examples of the former include the JAK2 46/1 haplotype, as well as single nucleotide polymorphisms in the gene TERT, that confer an ∼1.5- to 3.5-fold increased odds of MPN.37 While up to 10% of patients are estimated to have additional affected family members, the germline loci responsible for predisposition in familial MPNs have been ascertained in only a handful of pedigrees.38-41 Two broad mechanisms exist through which germline variation may affect MPN incidence. The observation that JAK2V617F is often found in cis with the 46/1 haplotype in heterozygous individuals initially raised the possibility that this allele was predisposed to acquiring JAK2V617F. However, direct evidence to support such DNA hypermutability has, thus far, not been demonstrated. An alternative model contends that certain germline genetic backgrounds are more permissive for an MPN to emerge. Two recent studies provide evidence for the existence of such an interaction. (1) The association between ET and HBS1L-MYB SNP rs9376092 is understood to be through reduction in MYB expression, which is a recognized driver of thrombocytosis.42 (2) Germline duplication of ATG2B and GSKIP has been shown to enhance sensitivity of megakaryocyte progenitors to TPO, particularly in the presence of JAK2V617F, resulting in highly penetrant MPNs within certain families.38 Several additional predisposition loci for MPNs have been recently identified across genes involved in cell senescence (TERT), JAK-STAT signaling (SH2B3), myeloid differentiation (GFI1B), DNA damage and repair (ATM, CHEK2), and epigenetic regulation (TET2); this suggests that the germline genetic background of an individual may influence diverse biological cellular functions to accentuate the molecular and cellular consequences of nascent MPN clones after their acquisition of phenotypic driver mutations.43
Additional somatic mutations in MPNs
Approximately one third of patients with MPN have additional mutations in known drivers of myeloid malignancies. These mutations alter DNA methylation (DNMT3A, TET2, IDH1/2), chromatin modifications (ASXL1, EZH2, IDH1/2), messenger RNA splicing (U2AF1, SF3B1, SRSF2, ZRSR2), and DNA repair (TP53).10 The presence of certain mutations, such as in ASXL1, SRSF2, IDH1/2, and EZH2, in patients with MF is associated with an increased risk of leukemic transformation and/or reduced survival.44 Investigation of the biological effects of these additional mutations reveal common molecular perturbations, as illustrated in the examples below.
Methylation of cytosines at CpG sites is a mechanism through which information establishing stem cell and differentiation states is retained by cells following cell division. DNMT3A is a de novo methyltransferase, and TET2, a member of the TET family of proteins, hydroxymethylates methylcytosines, which may be viewed simplistically as a process of demethylation. Mutations in both genes are prevalent across MPNs, understood to be loss of function (or dominant-negative effects for DNMT3AR882H),45 and result in a hematopoietic stem cell (HSC) advantage in murine models.46-49 The similarities in loss-of-function consequences despite seemingly opposing functions have recently been reconciled by work demonstrating that DNMT3A cooperates with TET2 to (1) promote the activity of enhancers important in ensuring stem cell fitness through the maintenance of high levels of DNA hydroxymethylation at enhancer centers50 and (2) repress key lineage-specific transcription factors that promote differentiation in hematopoiesis.51 Mutations in IDH1/2 also affect DNA methylation via the generation of 2-hydroxyglutarate that competitively inhibits α-KG–dependent DNA hydroxylases, such as TET2 and Jumonji family histone demethylases.52,53 Mutant IDH1 has also been linked to increased DNA damage via downregulation of the DNA damage sensor ATM.54 Knock-in mice for IDH1R132H suffer expansion of hematopoietic stem/progenitor cells (HSPCs), extramedullary hematopoiesis, and anemia,55 in keeping with this mutation being found in advanced phases of MPN.
Perturbation of polycomb repressor complex 2 (PRC2) is another pathogenic mechanism prevalent in MF. EZH2, the enzymatically active subunit of PRC2, compacts chromatin and represses gene transcription via histone H3 trimethylation at lysine 27 (H3K27). In myeloid malignancies, EZH2 loss-of-function mutations lead to derepression of several target genes, such as the Hox gene family, which enhances HSC self-renewal, and Lin28b/ Hmga2, which promotes fibrosis and reduces erythropoiesis in a JAK2V617F context.56-58 Loss of EZH2 can also be a consequence of other genetic perturbations, such as loss of heterozygosity at chromosome 7q, as well as mutations in spliceosome components U2AF1 and SRSF2.59,60 Mutations in ASXL1 are present in up to a quarter of patients with MF and also lead to reduced PRC2 recruitment, although other additional pathogenic effects have been reported.61-63
Order of mutation acquisition
Akin to other cancers, MPNs are clonally heterogeneous, reflecting the ongoing interplay between somatic mutation, cellular adaptation to the changing microenvironment, and selection of tumor subclones. The sequence of acquisition of somatic mutations can be inferred from the genotypes of detectable subclones. For instance, if some tumor cells have JAK2V617F, and others from the same patient bear JAK2V617F with an additional somatic mutation, then this indicates that JAK2V617F came first. Genotyping individual hematopoietic colonies has shown that the order of acquisition of JAK2V617F, relative to mutations in TET2 or DNMT3A, influences subclonal composition within HSPCs and mature cell compartments, disease presentation, and clinical outcome.35,64 In JAK2-first patients, the HSPC compartment is dominated by double-mutant cells, and such patients present at a younger age, often with PV.35 Conversely, in TET2-first patients, the HSPC compartment is dominated by single-mutant cells, and such patients present at an older age, usually with ET. Studies of mutation order have also revealed that DNMT3A mutations, often thought to be early events, frequently occur after an initial JAK2 mutation. This situation is difficult to detect in the absence of clonal assays because only small numbers of JAK2 single-mutant colonies may be present, probably reflecting their outcompetition by double-mutant clones. At least 3 mechanisms, not mutually exclusive, may underlie these observations: (1) The first mutation may alter a cell’s response to the second mutation. Thus, the transcriptional response to TET2 is altered by the presence of a prior JAK2V617F mutation.35 (2) The first mutation may alter HSC differentiation and give rise to altered target cell populations in which the second mutation can arise. (3) The first mutation may alter the number and function of mature progeny and thus affect the bone marrow microenvironment. Further studies are required to address which mechanisms are most important in driving the phenotypic differences in patients with different temporal patterns of mutation acquisition, and whether phenotype in chronic phase, or disease progression, is influenced by the order of acquisition of other pairs of common mutations.
Disease initiation and clonal outgrowth in MPNs
The conventional view of hematopoiesis envisions a self-renewing pool of long-term repopulating HSCs producing terminally differentiated cells via a series of intermediates. However, recent data from mice suggest that steady-state hematopoiesis does not routinely rely on HSCs but predominantly reflects the successive recruitment of thousands of transiently active progenitors.65,66 Furthermore, there is now compelling evidence for heterogeneity in the differentiation potential of individual cells within the HSC compartment in mice.67-69
Direct evidence for a clonal stem-cell origin of MPNs came 40 years ago when Adamson and colleagues demonstrated that females with PV who were heterozygous for 2 glucose-6-phosphate dehydrogenase alleles expressed only 1 allele in their blood cells.70 Both JAK2V617F and CALR-mutated cells are readily detectable in immunophenotypically defined HSPCs and across all myeloid lineages, confirming that these mutations arise in cells close to the apex of the hematopoietic hierarchy.10,71 It is tempting to speculate that after the acquisition of a driver mutation, platelet-biased HSCs might be particularly prone to giving rise to an MPN, but direct evidence for this is lacking. In some studies but not others, JAK has been found in the lymphoid compartments of patients with MPN, raising the possibility that HSCs with differing degrees of lymphoid potential are targets of somatic mutation in different patients.72-76 However, alternative explanations include the presence of other somatic mutations or the long-lived nature of mature lymphocytes, which means it may be many years before an HSC mutation becomes detectable in lymphocyte populations. Regardless of the uncertainty over the precise nature of the cell or cells of origin, for MPNs to emerge requires that (1) these cells acquire phenotypic driver mutations and (2) this results in persistent and significant clonal outgrowth.
Acquisition of mutations in hematopoiesis
Human cells accumulate somatic mutations throughout their lifetime as a result of cell-intrinsic mutational processes and exposure to external mutagens. The accumulated DNA changes in a single cell can be viewed as a “barcode” that can then be used to estimate tissue-specific mutation rates, trace the cell’s developmental origins, and understand the nature of DNA-damaging processes. Under normal circumstances, these genetic changes are unique to individual cells, making their study possible only through single-cell interrogation techniques. However, the clonal expansion of tumors renders these aberrations detectable in bulk tissue. Study of these mutation spectra in cancer reveal many mutational patterns, so-called mutation signatures, that in turn inform us of the biological processes that drive mutation acquisition in normal tissues as well as in cancer.77,78
Compared with solid tumors, hematopoietic malignancies are relatively mutation sparse. A key study by Welch and colleagues demonstrated that the mutation rate in acute myeloid leukemia (AML) was not elevated relative to age-matched hematopoietic progenitors from healthy individuals, suggesting that most mutations present in AML reflect those acquired before oncogenic transformation.79 Two mutational patterns have been recognized to contribute to background mutation acquisition in myeloid cells. The first (signature 1) is understood to be the consequence of spontaneous deamination of methylcytosines, which results in an age-associated genome-wide accumulation of C>T transition mutations at CpG dinucleotides.77,79 The second pattern of mutations (signature 5) exhibits various nucleotide changes at low frequency, but with a bias toward T>C mutations occurring on the transcribed DNA strand.77 The cause of this mutation signature is not known. Both mutational signatures are observed ubiquitously across somatic tissues as well as in the germline.80
Whether these background mutational processes are sufficient to explain the acquisition of specific oncogenic mutations remains the subject of ongoing research. Two observations suggest that the frequency of acquisition of oncogenic drivers may occur at higher rates than conventionally thought. First, some patients with MPN have been shown to harbor multiple oncogenic driver mutations in ancestrally unrelated clones, suggesting that these mutations can arise multiple independent times in the same individual.81,82 Second, in a recent study of familial MPN, two thirds of carriers of a germline duplication (involving ATG2B and GSKIP) developed an MPN harboring somatic mutations in JAK2, CALR, or MPL. The germline duplication enhanced the outgrowth of mutant cells, and there was no coexistent evidence of hypermutability that might otherwise have increased mutation acquisition in these patients.38 Together, these data suggest that acquisition of phenotypic driver mutations in cells capable of initiating MPNs might be relatively frequent but that clones either remain small (and thus are undetected in analyses of cell populations) or do not normally survive, unless additional conditions facilitate their clonal expansion.
Factors influencing clonal outgrowth of cells with phenotypic driver mutations
Approximately half of patients with a JAK2V617F-mutated MPN in chronic phase lack additional somatic mutations in oncogenic drivers. However, studies in mice suggest that although JAK2V617F is capable of driving an MPN phenotype, it may be less able to initiate clonal expansion in an individual HSC. Three different knockin mouse models have shown that JAK2V617F HSCs are no better or are less adept than wild-type HSCs in initiating clonal expansion, as assayed by competitive repopulation transplantation assays.28 Furthermore, transplantation of single JAK2V617F HSCs into recipient mice only rarely results in an MPN phenotype.83 Therefore, additional factors may be required for an initiating JAK2 mutation to result in an expanded mutant clone. These may include interaction with the constitutional genetic background of a patient or a physiological stochastic expansion of individual HSCs that is independent of the driver mutation. Moreover, there is increasing evidence that aging and the bone marrow microenvironment may play important roles.
Aging is a strong risk factor for MPN. Recent studies demonstrate that with increasing age, clonal hematopoietic expansions occur in the absence of overt hematological disease.84-87 Mutations leading to clonal hematopoiesis (CH) commonly involve DNMT3A, TET2, ASXL1, and PPM1D, but also JAK2V617F. Of note, mutations in CALR have not yet been reported in CH. This may reflect the inherent challenges in detecting such mutations from standard exome sequencing due to poor sequencing coverage and alignment efficiency over the mutated region. However, it also raises the possibility that CALR mutations have a higher penetrance for the development of overt disease. Several observations suggest that CH does not simply reflect the increased risk of mutation acquisition with age, but that aging itself favors clonal expansion. First, CH is rare under the age of 40 years but increases exponentially in the elderly, a pattern inconsistent with a linear rate of somatic mutation accumulation over time.79 Second, some mutations (eg, SF3B1) are associated with CH only in more advanced decades of life87 ; it would be unlikely that specific mutations are acquired at different rates in different age groups. Third, no known oncogenic drivers can be identified from analysis of whole-exome or whole-genome sequencing in up to half of individuals with CH, suggesting that oncogene-independent mechanisms, such as clonal drift or reduced bone marrow genetic diversity, can lead to clonal expansion.86,88 It is recognized that HSCs undergo a wide range of age-related biochemical and functional changes, and it seems plausible that some of these may favor the outgrowth of cells that acquire a driver mutation.89 Moreover, both cell-intrinsic and environmental influences are likely to play a role.
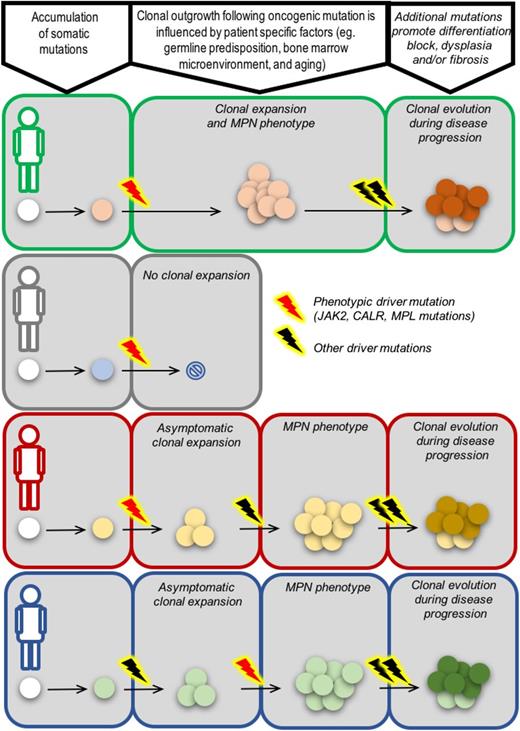
Several lines of evidence point to the importance of environmental effects. Medical interventions, immune dysregulation, and inflammation have all been shown to alter selective pressures for mutant clones within the bone marrow. In TP53-mutated therapy-related MDS, TP53-mutated cells have been demonstrated to preexist at low levels before any exposure to therapy but with outgrowth occurring only in the context of postchemotherapy bone marrow.90 Furthermore, CH is prevalent in the bone marrow of patients with aplastic anemia, where one can envisage that immune-mediated destruction of normal bone marrow cells favors selection for clones more adept at evading cell death, such as those bearing PIG-A mutations, deletion of the major histocompatibility locus on chromosome 6p or mutations in TP53-inhibitor PPM1D.91 There is also increasing evidence that inflammation has a key role in promoting MPN initiation and influencing disease evolution. The secretion of proinflammatory cytokines by bone marrow stromal cells, such as interleukin-6 (IL-6), IL-33, fibroblast growth factor, C-X-C motif ligand 10, IL-33, and tumor necrosis factor-α, have been shown to preferentially promote the expansion of JAK2V617F-mutated clones.92-94 Furthermore, elevated levels of NFE-2 (itself induced by inflammatory cytokine IL-1β) drive MPN proliferation and progression through modulation of inflammatory cascades, including elevated IL-8 expression.95,96 Overall, it remains unclear exactly how a single phenotypic driver mutation initiates a clonal expansion that will evolve into an overt MPN, but current evidence suggests that the constitutional genetic background, aging process, and tumor microenvironment are key factors, with each exerting cell-intrinsic and environmental effects (Figure 3).
From origins to outcomes. Different evolutionary paths to MPN and disease progression in 4 patients, each with a unique genetic background (green, gray, red, blue). In the first patient (green), a phenotypic driver mutation acquired in an HSC results in clonal expansion and the emergence of an MPN phenotype as a consequence of favorable cell-intrinsic and/or environmental factors. The MPN in this context has no additional oncogenic driver mutations, as is common for patients in chronic phase. Additional driver mutations, such as those that perturb polycomb repressor 2 function (EZH2, ASXL1 mutations), spliceosome components (SRSF2, SF3B1, U2AF1), or DNA damage repair (TP53), can lead to cells gaining a further clonal advantage and disease progression. In the second patient (gray), the cell-intrinsic and/or environmental context is not favorable, and a cell acquiring a phenotypic driver mutation does not have a clonal advantage relative to competing normal cells. In some circumstances, a phenotypic driver mutation may be insufficient to result in abnormal blood counts and an overt MPN but can instead result in a clonal expansion. Additional mutations or cell-extrinsic changes may be required to result in emergence of disease (patient in red). Finally, in some patients, phenotypic driver mutations may not be the first event. Clonal hematopoiesis as a result of mutations in, for example, TET2, DNMT3A, ASXL1 may be the required backdrop for a phenotypic driver mutation to result in an overt MPN (patient in blue).
From origins to outcomes. Different evolutionary paths to MPN and disease progression in 4 patients, each with a unique genetic background (green, gray, red, blue). In the first patient (green), a phenotypic driver mutation acquired in an HSC results in clonal expansion and the emergence of an MPN phenotype as a consequence of favorable cell-intrinsic and/or environmental factors. The MPN in this context has no additional oncogenic driver mutations, as is common for patients in chronic phase. Additional driver mutations, such as those that perturb polycomb repressor 2 function (EZH2, ASXL1 mutations), spliceosome components (SRSF2, SF3B1, U2AF1), or DNA damage repair (TP53), can lead to cells gaining a further clonal advantage and disease progression. In the second patient (gray), the cell-intrinsic and/or environmental context is not favorable, and a cell acquiring a phenotypic driver mutation does not have a clonal advantage relative to competing normal cells. In some circumstances, a phenotypic driver mutation may be insufficient to result in abnormal blood counts and an overt MPN but can instead result in a clonal expansion. Additional mutations or cell-extrinsic changes may be required to result in emergence of disease (patient in red). Finally, in some patients, phenotypic driver mutations may not be the first event. Clonal hematopoiesis as a result of mutations in, for example, TET2, DNMT3A, ASXL1 may be the required backdrop for a phenotypic driver mutation to result in an overt MPN (patient in blue).
Clinical implications and future directions
The JAK inhibitor ruxolitinib has been a valuable addition to the therapeutic armamentarium for MPNs, in particular for patients with MF who have splenomegaly and/or disease-related symptoms.97,98 However, allogeneic stem cell transplantation still remains the only potentially curative treatment of MPNs, an approach limited by age-related comorbidities and high treatment-related mortality. Demonstrable disease-modifying activity for conventional therapeutic agents is lacking. Ruxolitinib and interferon-α have been associated with reductions in allele burdens of phenotypic drivers mutations in some cases, but molecular response is variable and unpredictable. Among the multitude of novel agents tested in clinical trials, many have shown clinical responses but only in a minority of patients, or there are dose-limiting toxicities.99 Patient heterogeneity may be a key factor contributing to the lack of demonstrable clinical efficacy for many agents. Therefore, a major challenge is how we use our emerging understanding of the pathogenetic basis of MPNs to identify groups of patients with shared disease biology and clinical outcome, such that both existing therapies, as well as novel agents, can be better targeted to specific patient groups. Novel paradigms for both disease classification and predictions of clinical outcome may be needed to meet this goal.
The 2016 revision to the World Health Organization classification of MPNs retains the traditional distinction among PV, ET, and MF and distinguishes between prefibrotic and fibrotic MF.1 However, such a conceptual schema has significant weaknesses. First there is a fundamental challenge associated with defining diseases as discrete entities on the basis of continuous variables, such as hemoglobin level or amount of bone marrow fibrosis.100 Second there is an emphasis on the identification of cytological nuances (especially megakaryocyte morphology) that are subjective in their interpretation and that have poor interobserver consensus.101-103 Third, despite the importance of somatic mutations, there remains substantial molecular overlap between, and heterogeneity within, MPN categories.10 Furthermore, there is significant overlap between the spectrum of mutations found in MPN and myelodysplasia (MDS), and it is unclear whether patients with MF who lack mutations in JAK2, CALR, or MPL have disease biology more akin to MDS than MPN. Accurate classification is critical because it predetermines the therapeutic strategy used by clinicians. Therefore, there is a rationale for large-scale sequencing studies that incorporate patients with MPN, MDS, and related myeloid conditions, combined with clinical and laboratory parameters, in order to provide a more biological basis for the way we classify these disorders in the future.
Another major goal for the future is to identify, at an early stage, patients at risk of a poor outcome. The International Prognostic Scoring System (IPSS)104 has been valuable for considering the timing of ASCT in patients with MF. However, there are several ongoing challenges. First, current prognostic scoring systems apply to patients with MF, and there are no predictive models for use in patient with chronic-phase MPN. Second, multiple scoring systems are available for patients with MF, depending on whether risk stratification is performed at diagnosis or later,105 whether MF is primary or post-ET/post-PV,106-108 and the age of the patient.105 Third, both the IPSS and Dynamic IPSS (DIPSS) rely on potentially subjective scores, such as the presence or absence of constitutional symptoms. Recently, patient age and JAK2, CALR, and MPL status have been used to build a simple yet objective predictive model for patients with MF.109 However, such a model does not take into account the presence of additional somatic mutations that strongly associate with outcome.110 Comprehensive sequencing of patients, combined with clinical parameters and longitudinal follow-up, are now required to elucidate how the various genetic and nongenetic factors contribute to the different possible outcomes for patients with MPN. Such a “knowledge bank” approach has been developed in AML111 and has the potential to deliver a universal and clinically relevant model for predicting outcome for all patients with MPN.
Acknowledgments
The authors thank Jacob Grinfeld for valuable comments on the manuscript.
J.N. is supported by a Cancer Research UK Clinician Scientist Fellowship and European Haematology Association research award. Work in the Green laboratory is supported by the Wellcome Trust, the Medical Research Council, Bloodwise, Cancer Research UK, and the National Institute for Health Research Cambridge Biomedical Research Centre.
Correspondence
Anthony R. Green, University of Cambridge Department of Haematology, Cambridge Institute of Medical Research, Hills Rd, Cambridge CB2 0XY, United Kingdom; e-mail: arg1000@cam.ac.uk.
References
Competing Interests
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Author notes
Off-label drug use: None disclosed.
This article was selected by the Blood and Hematology 2017 American Society of Hematology Education Program editors for concurrent submission to Blood and Hematology 2017. It is reprinted with permission from Blood 2017, Volume 130.