Abstract
Allogeneic hematopoietic cell transplantation (alloHCT) often represents the only curative treatment for various malignant and nonmalignant disorders. Initially, the only suitable donors were considered human leukocyte antigen (HLA)–matched or partially matched relatives. The founding of international unrelated donor and umbilical cord blood registries expanded unrelated donor options and access for patients. In the absence of a matched sibling donor (MSD) with 13% to 51% availability, the current consensus recommends use of a matched unrelated donor (MUD) at HLA-A, B, C, and DRB1 with consideration of matching at HLA-DPB1 and -DQB1. MUD donor availability (donor willing and available to donate) ranges from 29% to 78% with African American patients on the lower end and white non-Hispanic patients with the highest likelihood of a match. Recent studies comparing donor to no-donor treatment options in malignant disease consistently point to substantially better outcomes following alloHCT. In the absence of an MSD or MUD, alternative donor choices turn to haploidentical related (Haplo), mismatched unrelated donor (MMUD), and umbilical cord blood (UCB). Novel strategies for alloHCT, including the use of posttransplant cyclophosphamide-based graft vs host disease prophylaxis, have expanded the safety and effectiveness of transplant procedures across HLA barriers using Haplo and MMUD. The less restrictive matching requirements for UCB transplant are well documented and allow for transplant across multiply mismatched HLA alleles. When all donor options are considered, nearly all patients have an available donor. Here we discuss the likelihood of donor availability, complete HLA match by available donor type, and current controversies warranting future research.
Learning Objectives
Describe the degree of HLA matching and association of matching/mismatching on outcomes for available allogeneic donor options
Understand the likelihood of availability and hierarchy of donor type selection for allogeneic hematopoietic cell transplantation
CLINICAL CASE
A 60-year-old Hispanic man presents with de novo acute myeloid leukemia (M2;inv 3/t(3;3)). Induction and consolidation therapy achieved complete remission with no measurable residual disease based on next-generation sequencing. Family studies identified 1 sibling as a full human leukocyte antigen (HLA) match. However, they have a history of malignant melanoma and were deemed ineligible to donate. One additional (65-year-old) sibling is haploidentical to the patient and is willing and eligible to donate. An unrelated donor search identifies multiple young (<30 years old) well-matched (8/8-considering HLA-A, B, C, and DRB1) and single mismatched (7/8) unrelated donors and multiple cord blood units available.
Introduction
Allogeneic hematopoietic cell transplantation (alloHCT) often represents the only curative treatment for various malignant and nonmalignant disorders. Over the approximately 50 years since the first alloHCTs were attempted and demonstrated engraftment and curative potential, the definition of optimal matching has continued to evolve. Initially, the only suitable donors were considered HLA matched or partially matched relatives. The founding of the National Marrow Donor Program (NMDP), Anthony Nolan Registry, and other international donor registries expanded the potential to identify suitable donors outside of the patient's immediate family. Unrelated donor registries have grown to include over 40 million volunteer donors and cord blood registries to over 800 000 banked umbilical cord blood units worldwide (World Marrow Donor Association stats). HLA testing technologies and matching strategies have evolved significantly as well, making the characterization of match between unrelated individuals more routine.1 The advent of DNA-based typing technologies and enhanced databases of well-characterized HLA allele sequences have increased the precision and accuracy of typing, leading to improved matching (Table 1). Yet, the question remains, what is the optimal match? The answer is not straightforward, as the optimal HLA match for a given patient will vary based on the donor options available at the time of need and clinical factors (eg, timing of transplant, patient size/weight, and prior HLA sensitization).
HLA typing techniques and resulting resolution
| Methodology . | Approach . | Interpretation . | Resolution . | Application . | Results . |
|---|---|---|---|---|---|
| Serology | Cellular assay based on complement fixation by HLA-specific antibodies | Cell death—yes/no | Low | Family screening Null allele confirmation | A2, A24 |
| Sequence-specific primers (PCR-SSP) | HLA sequence-specific PCR primers | Amplification—yes/no | Low to high, dependent on DNA sequence coverage | Family screening Verification typing | Low—A*02:XX, A*24:XX or A*02AB, A*24:BC High—A*0201g, A*24:02g |
| Sequence-specific oligonucleotide probes (PCR-SSOP) | HLA sequence-specific oligonucleotide probes that bind to polymorphic sequences of amplified DNA | Probe binding—yes/no | Low to high depending on DNA sequence coverage | Family screening Verification typing | Low—A*02:XX, A*24:XX or A*02AB, A*24:BC High—A*02:01G, A*24:02G |
| Sanger sequence-based typing (SBT) | HLA amplicon sequencing using base termination | Base pair reads and consensus alignment | High to allele level depending on coverage | All | High—A*02:01G, A*24:02G Allele—A*02:01:01:03, A*24:02:01:01 |
| Next-generation sequencing (NGS) | Multiple platforms, based on massive parallel sequencing reactions | Base pair calling and consensus alignment | High to allele-level depending on coverage | All | High—A*02:01G, A*24:02G Allele—A*02:01:01:03, A*24:02:01:01 |
| Methodology . | Approach . | Interpretation . | Resolution . | Application . | Results . |
|---|---|---|---|---|---|
| Serology | Cellular assay based on complement fixation by HLA-specific antibodies | Cell death—yes/no | Low | Family screening Null allele confirmation | A2, A24 |
| Sequence-specific primers (PCR-SSP) | HLA sequence-specific PCR primers | Amplification—yes/no | Low to high, dependent on DNA sequence coverage | Family screening Verification typing | Low—A*02:XX, A*24:XX or A*02AB, A*24:BC High—A*0201g, A*24:02g |
| Sequence-specific oligonucleotide probes (PCR-SSOP) | HLA sequence-specific oligonucleotide probes that bind to polymorphic sequences of amplified DNA | Probe binding—yes/no | Low to high depending on DNA sequence coverage | Family screening Verification typing | Low—A*02:XX, A*24:XX or A*02AB, A*24:BC High—A*02:01G, A*24:02G |
| Sanger sequence-based typing (SBT) | HLA amplicon sequencing using base termination | Base pair reads and consensus alignment | High to allele level depending on coverage | All | High—A*02:01G, A*24:02G Allele—A*02:01:01:03, A*24:02:01:01 |
| Next-generation sequencing (NGS) | Multiple platforms, based on massive parallel sequencing reactions | Base pair calling and consensus alignment | High to allele-level depending on coverage | All | High—A*02:01G, A*24:02G Allele—A*02:01:01:03, A*24:02:01:01 |
HLA typing for search and match determination
To facilitate HLA match assessment, patients and potential donors (related and unrelated) should be high-resolution HLA typed using DNA-based methods for HLA-A, B, C, DRB1, and DPB1.2 Extended typing can include HLA-DRB3/4/5, DQB1, DQA1, and DPA1, especially in the case of highly sensitized patients to avoid mismatches that may be a target of anti-HLA antibodies (eg, donor-specific antibodies) to minimize the risk of graft failure.3 Traditional DNA-based techniques for HLA typing focused on the antigen recognition domain (ARD).2 Technological advances now make the routine sequencing of the entire HLA gene feasible. A recent study found that mismatching outside of the ARD was associated with increased risk of acute graft-vs-host disease (GVHD) grades II to IV but with no increased risk of transplant-related mortality or decreased overall survival (OS),4 suggesting that matching for allelic variation outside the ARD has limited impact in the matched unrelated donor (MUD) setting.
HLA typing should be verified via confirmatory testing prior to finalizing donor selection for alloHCT through an American Society of Histocompatibility and Immunogenetics or European Federation for Immunogenetics accredited laboratory, preferably with guidance from an accredited histocompatibility and immunogenetics laboratory director to assist the clinical team with interpretation of the typing results and match assessment between the prospective donor(s) and patient.
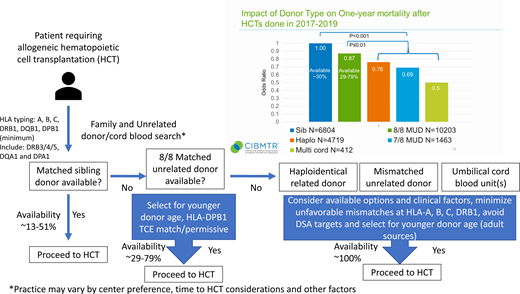
What are the available donor options and likelihood of availability?
The conventional optimal donor choice is an HLA-matched family member, most often a matched sibling. However, the rate of matched sibling donor (MSD) availability can vary substantially based on ethnicity and age of the patient with rates ranging from 13% to 51%.5 For those patients without a matched sibling, the first-choice alternative donor option is an MUD. This recommendation is supported by the findings of the annual Center for International Blood and Marrow Transplant Research (CIBMTR) Center Specific Analysis. The 2021 report included >25 000 unrelated and related alloHCTs performed between 2017 and 2019 in the United States with follow-up through 1 year after alloHCT. A comparison of alternative donor types can be assessed using the odds ratio of OS at 1 year compared with MSD as the baseline with an odds ratio (OR) for MUD of 0.87 (95% confidence interval [CI], 0.78-0.97, P = .010), haploidentical related (Haplo) OR of 0.76 (0.68-0.84, P < .001), mismatched unrelated donor (MMUD) (7/8) OR of 0.69 (0.59-0.81, P < .001), and umbilical cord blood (UCB) (multiple UCB ≥4/6) OR of 0.50 (0.39-0.64, P < .001) (see the visual abstract). This very large, contemporary, multicenter analysis of real-world data clearly demonstrates the hierarchy for donor selection prioritizing matched siblings, MUDs, and then mismatched graft sources.
Despite the large international pool of >40 million volunteer unrelated donors, MUD availability varies and is most substantially affected by the ethnic background of the patient. MUD donor availability (donor willing and available to donate) ranges from 29% to 78%, with African American patients on the lower end and white non-Hispanic patients with the highest likelihood of a match (NMDP internal analysis). In the absence of an MUD, alternative donor choices turn to Haplo, MMUD, and UCB. Novel strategies for alloHCT, including the use of posttransplant cyclophosphamide-based graft-vs-host disease prophylaxis, have expanded the safety and effectiveness of transplant procedures across HLA barriers using Haplo6 and MMUD.7,8 The less restrictive matching requirements for UCB transplant are well documented and allow for transplant across multiply mismatched HLA alleles.9 When all donor options are considered, there is a high likelihood that all patients will have an available donor.10
How should alternative donor options be prioritized? Varying reports suggest situations where 1 donor type may be preferred over others (eg, Haplo vs double UCB in adult patients receiving alloHCT for lymphoma or acute leukemia based on BMT CTN 1101,11 younger alternative donors in the context of older MSD, MMUD vs Haplo), but much controversy remains. The BMT CTN 1702 protocol, “Clinical Transplant-Related Long-Term Outcomes of Alternative Donor Allogeneic Transplantation (CTRL-ALT-D)” (NCT03904134), is investigating the outcomes of alloHCT following biological assignment to an MUD or alternative donor (Haplo, UCB, or MMUD) based on likelihood of MUD availability. This trial reflects current consensus that an MUD, if available in a timely manner, is the preferred alternative to an MSD and will shed further light on alternative donor choice and outcomes. This includes an evaluation of the urgency of need and potential to obtain various donor types.
Matched sibling donors
As noted above, an MSD is the benchmark against which all alternative donor options are evaluated. Donor choice is generally limited in the MSD setting and with donor age tightly correlated to patient age. Given that multiple studies suggest that lower donor age is associated with better OS in both the matched unrelated and haploidentical alloHCT setting,12-14 questions have been raised about whether an older sibling donor is still the best option when multiple donor choices are available. A CIBMTR study of older MSD (≥50 years old) vs younger MUD (<50 years old) alloHCT for lymphoma and leukemia reported superior outcomes in MSD.15 A recent analysis comparing older MSD (≥50 years old) to younger MUD (≤35 years old) alloHCT for myelodysplastic syndromes found that MUD was associated with lower relapse and improved disease-free survival (DFS), suggesting that in certain circumstances (eg, high risk of relapse), MUD may be advantageous.16 In addition, availability of younger Haplo or MMUD may warrant investigation as an alternative to an older matched sibling as outcomes for all donor types continue to improve.
One potential concern in older donors is the higher rates of clonal hematopoiesis of indeterminant potential (CHIP) associated with increasing age.17 Prior studies differ on the findings regarding risk/benefit of CHIP in the alloHCT setting. Frick et al18 reported increased risk of chronic GVHD following alloHCT with a related donor harboring CHIP, while Gibson et al19 reported potential benefits of DNMT3A variants in faster engraftment and better disease control. While these data raise potential concerns about the suitability of older donors for optimal transplant outcomes, there are scarce data to support prioritizing alternative donors over an available MSD at present.
HLA matching in MUD
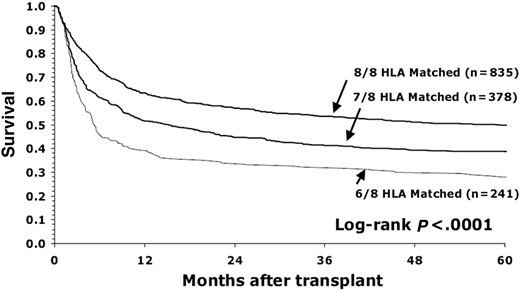
These data were reviewed and summarized in the NMDP/CIBMTR donor selection guidelines published in Blood in 2019 and provided recommendations for optimal MUD matching in the context of conventional calcineurin inhibitor-based GVHD prophylaxis.20 The optimal level of match in the MUD setting was established in the seminal publication by Lee et al.21 This pivotal study demonstrated that matching for HLA-A, B, C, and DRB1 was associated with superior OS and lower rates of acute GVHD21 (Figure 1). This level of match is often referred to as 8/8 matching. HLA-DQB1 match was not associated with any outcomes as an isolated mismatch and did not confer any additional risk when paired with HLA-A, B, C, or DRB1. Inclusion of DQB1 in matching is often referred to as 10/10 matching. Most (>95%) cases matched at 8/8 will also be matched at 10/10, so generally bring along a match at HLA-DQB1. These findings were replicated in subsequent analyses in a more contemporary population,22 as well as by graft type,23 nonmalignant disease,24 and international cohorts.25
Survival of patients with early-stage disease depending on degree of HLA matching (8/8, 7/8, and 6/8) for HLA-A, B, C, and DRB1. (Figure courtesy of Lee et al.21 )
Survival of patients with early-stage disease depending on degree of HLA matching (8/8, 7/8, and 6/8) for HLA-A, B, C, and DRB1. (Figure courtesy of Lee et al.21 )
HLA-DPB1 mismatching in the context of an 8/8 or a 10/10 match is associated with increased risk of nonrelapse mortality driven by higher GVHD and lower risk of relapse with no association with OS.21,22 A model pioneered by Fleischhauer et al26 based on the immunogenicity of HLA-DPB1 in the context of 3 T-cell epitope (TCE) groups provides a methodology for mitigating the risks associated with HLA-DPB1 mismatching. In multiple analyses, the selection of HLA-DPB1 TCE permissive mismatches was associated with improved OS and reduced risk of GVHD, further adding to the strategy for selecting an optimal 8/8 MUD.22,26,27 In addition, a recent analysis found that the TCE3 group could be further categorized into core and noncore alleles to reduce the risk of GVHD and transplant- related mortality compared with nonpermissive mismatches.28 The potential to find an MUD with an HLA-DPB1 allele match may be limited, but the potential to identify a permissive mismatch is highly likely and imputable to support identification of donors with missing DPB1 typing29,30 (Table 2).
HLA matching in matched unrelated donor allogeneic hematopoietic cell transplantation
| Reference . | Consensus guidelines . | |||
|---|---|---|---|---|
| Dehn et al20 (2019) | 1. 8/8 match at ARD level for HLA-A, B, C, DRB1 2. Younger donor age 3. Match or permissive HLA-DPB1 TCE 4. Minimize mismatches at HLA-DRB3/4/5 and DQB1 5. Avoid DSA targets including DQA1 and DPA1 | |||
| New research published, since release of guidelines | ||||
| Reference | Population | Study type | Comparison | Key findings |
| Mayor et al4 (2021), ultra-high resolution | N = 5140 10/10 MUD, first alloHCT for ALL, AML, or MDS. Myeloablative or RIC | Observational—multicenter | 12/12 Ultra-high resolution vs ≤11/12 | UHR matching associated with lower risk of acute GVHD II-IV, no associations with survival outcomes |
| Arrieta-Bolaños et al28 (2022), TCE core | N = 5140 10/10 MUD, first alloHCT for ALL, AML, or MDS. Myeloablative or RIC | Observational—multicenter | 10/10 MUD HLA-DPB1 permissive core alleles vs noncore alleles vs nonpermissive | 10/10 MUD HLA-DPB1 TCE3 permissive core associated with reduced risk of acute GVHD II-IV and TRM compared to nonpermissive mismatches |
| Reference . | Consensus guidelines . | |||
|---|---|---|---|---|
| Dehn et al20 (2019) | 1. 8/8 match at ARD level for HLA-A, B, C, DRB1 2. Younger donor age 3. Match or permissive HLA-DPB1 TCE 4. Minimize mismatches at HLA-DRB3/4/5 and DQB1 5. Avoid DSA targets including DQA1 and DPA1 | |||
| New research published, since release of guidelines | ||||
| Reference | Population | Study type | Comparison | Key findings |
| Mayor et al4 (2021), ultra-high resolution | N = 5140 10/10 MUD, first alloHCT for ALL, AML, or MDS. Myeloablative or RIC | Observational—multicenter | 12/12 Ultra-high resolution vs ≤11/12 | UHR matching associated with lower risk of acute GVHD II-IV, no associations with survival outcomes |
| Arrieta-Bolaños et al28 (2022), TCE core | N = 5140 10/10 MUD, first alloHCT for ALL, AML, or MDS. Myeloablative or RIC | Observational—multicenter | 10/10 MUD HLA-DPB1 permissive core alleles vs noncore alleles vs nonpermissive | 10/10 MUD HLA-DPB1 TCE3 permissive core associated with reduced risk of acute GVHD II-IV and TRM compared to nonpermissive mismatches |
ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; DSA, donor-specific anti-HLA antibodies; MDS, myelodysplastic syndromes; RIC, reduced intensity conditioning; TRM, transplant-related mortality; UHR, ultrahigh resolution; 8/8, high-resolution match at HLA-A, B, C, and DRB1; 10/10, high-resolution match at HLA-A, B, C, DRB1, and DQB1; 12/12, HLA match at HLA-A, B, C, DRB1, DQB1, and DPB1.
The 1 non-HLA donor characteristic that consistently associates with improved survival after alloHCT is younger donor age and is prioritized above extended matching at HLA-DPB1 and other loci in the current donor selection guidelines.12,13,20 International donor registries now prioritize recruitment of younger donors (≤40 years old), and NMDP has established donor age as a metric of product quality emphasizing the use of donors ≤35 years old.
Further studies are required to validate the effects of extended HLA matching and younger donor age in MUD in the context of novel GVHD prophylaxis strategies, such as posttransplant cyclophosphamide (ptCy), abatacept, and advanced graft engineering.
HLA matching in mismatched graft sources
Related haploidentical donors
The selection and prioritization of Haplo donors is limited by family size. The European Blood and Marrow Transplantation group recently published consensus recommendations for donor selection in Haplo alloHCT focused mainly on non-HLA donor characteristics.31 Novel HLA match and mismatch associations with Haplo alloHCT outcomes were published after these recommendations. Solomon et al. reported an association between mismatching at HLA class II loci and decreased relapse and improved OS in a single-center study of T-replete Haplo alloHCT using ptCy.32 A recent large multicenter study reported HLA locus-specific associations with various outcomes and proposed a model for Haplo donor selection to optimize DFS. The study evaluated overall degree of high-resolution matching, impact of individual loci, HLA-DPB1 TCE matching, and matching for the conserved exon 1 leader sequence of HLA-B (B leader) previously found to associate with acute GVHD in the MMUD setting.33 The model recommends prioritizing HLA-B leader match, HLA-DPB1 TCE nonpermissive mismatch (opposite of MUD), DRB1 mismatch, and DQB1 match for optimal DFS and was codified in an online tool (Table 3).34
HLA matching in haploidentical related donor allogeneic hematopoietic cell transplantation
| Reference . | Consensus guidelines . | |||
|---|---|---|---|---|
| Ciurea et al31 (2020) | T-replete Haplo using ptCy-based GVHD prophylaxis: 1. Avoid DSA (MFI <1000) 2. Younger donor over older 3. Male donor for male recipient 4. Sibling or offspring over parent 5. Father preferred over mother 6. ABO match over minor over major mismatch | |||
| New research published, since release of guidelines | ||||
| Study | Population | Trial/study design | Comparison(s) | Key findings |
| Solomon et al32 (2020) | N = 208 consecutive first alloHCT for hematologic malignancy receiving T-cell replete Haplo with ptCy GVHD prophylaxis | Observational—single center | 1. Total number of mismatches 2. Individual locus match/mismatch effects on outcomes | 1. HLA-DRB1 and HLA-DPB1 nonpermissive mismatch independently associated with improved OS 2. HLA-A mismatch increased chronic GVHD |
| Fuchs et al34 (2022) | N = 1484, first alloHCT for ALL, AML, or MDS, adult patients receiving T-cell replete Haplo with ptCy-based GVHD prophylaxis | Observational—multicenter | 1. Total number of mismatches 2. Individual locus match/mismatch effects on outcomes | 1. No association with number of mismatches 2. HLA-B leader match improves OS and DFS; HLA-DRB1 GVH vector mismatch decreases relapse and improves DFS; HLA-DPB1 nonpermissive mismatch improves OS and DFS; HLA-C match decreases chronic GVHD |
| Reference . | Consensus guidelines . | |||
|---|---|---|---|---|
| Ciurea et al31 (2020) | T-replete Haplo using ptCy-based GVHD prophylaxis: 1. Avoid DSA (MFI <1000) 2. Younger donor over older 3. Male donor for male recipient 4. Sibling or offspring over parent 5. Father preferred over mother 6. ABO match over minor over major mismatch | |||
| New research published, since release of guidelines | ||||
| Study | Population | Trial/study design | Comparison(s) | Key findings |
| Solomon et al32 (2020) | N = 208 consecutive first alloHCT for hematologic malignancy receiving T-cell replete Haplo with ptCy GVHD prophylaxis | Observational—single center | 1. Total number of mismatches 2. Individual locus match/mismatch effects on outcomes | 1. HLA-DRB1 and HLA-DPB1 nonpermissive mismatch independently associated with improved OS 2. HLA-A mismatch increased chronic GVHD |
| Fuchs et al34 (2022) | N = 1484, first alloHCT for ALL, AML, or MDS, adult patients receiving T-cell replete Haplo with ptCy-based GVHD prophylaxis | Observational—multicenter | 1. Total number of mismatches 2. Individual locus match/mismatch effects on outcomes | 1. No association with number of mismatches 2. HLA-B leader match improves OS and DFS; HLA-DRB1 GVH vector mismatch decreases relapse and improves DFS; HLA-DPB1 nonpermissive mismatch improves OS and DFS; HLA-C match decreases chronic GVHD |
MFI, mean fluorescence intensity.
Mismatched unrelated donors
Recommendations for the prioritization of HLA match and mismatch in MMUD are predominately based on past experience in the setting of calcineurin inhibitor–based GVHD prophylaxis strategies.20 While multiple studies investigated the potential to apply algorithms that prioritize mismatches based on structural similarity or peptide binding affinity, most have failed validation in large multicenter studies.35 Fernandez- Viña et al36 described a permissive mismatch at HLA-C where the alleles (C*03:03 and C*03:04) only differ outside the antigen recognition domain, but the extension of these findings to other mismatches is limited. Hurley et al37 reported that mismatches at HLA-A, B, C, and DRB1 limited to the host-vs-graft direction were well tolerated and similar to a full match but only applicable in the setting of mismatching at a homozygous locus in the patient. HLA-B leader matches associated with lower risk of acute GVHD.33 Prioritizing HLA-DPB1 TCE permissive mismatches and minimizing overall level of mismatch at extended HLA class II loci can also contribute to better outcomes, including OS in the MMUD setting26,38 (Table 4).
HLA matching in mismatched unrelated donor allogeneic hematopoietic cell transplantation
| Reference . | Consensus guidelines . | |||
|---|---|---|---|---|
| Dehn et al20 (2019) | 1. 7/8 match at ARD level for HLA-A, B, C, DRB1 2. Younger donor age 3. Match or permissive HLA-DPB1 TCE 4. Minimize mismatches at HLA-DRB3/4/5 and DQB1 5. Avoid DSA targets including DQA1 and DPA1 | |||
| New research published, since release of guidelines | ||||
| Study | Population | Trial/study design | Comparison | Results |
| Petersdorf et al33 (2020) | N = 33 982 total cohort with N = 1457 HLA-B mismatched, first alloHCT for multiple diseases, CNI-based GVHD prophylaxis | Observational study | Exon 1 B leader match vs mismatch in 9/10 MMUD | B leader match associated with decreased acute GVHD, no association between match status and survival |
| Shaw et al7 (2021) | N = 80 7/8-4/8 MMUD alloHCT for hematologic malignancies, ptCy-based GVHD prophylaxis with bone marrow grafts | Prospective trial | 7/8 matched vs <7/8 matched | No differences in any outcomes |
| Reference . | Consensus guidelines . | |||
|---|---|---|---|---|
| Dehn et al20 (2019) | 1. 7/8 match at ARD level for HLA-A, B, C, DRB1 2. Younger donor age 3. Match or permissive HLA-DPB1 TCE 4. Minimize mismatches at HLA-DRB3/4/5 and DQB1 5. Avoid DSA targets including DQA1 and DPA1 | |||
| New research published, since release of guidelines | ||||
| Study | Population | Trial/study design | Comparison | Results |
| Petersdorf et al33 (2020) | N = 33 982 total cohort with N = 1457 HLA-B mismatched, first alloHCT for multiple diseases, CNI-based GVHD prophylaxis | Observational study | Exon 1 B leader match vs mismatch in 9/10 MMUD | B leader match associated with decreased acute GVHD, no association between match status and survival |
| Shaw et al7 (2021) | N = 80 7/8-4/8 MMUD alloHCT for hematologic malignancies, ptCy-based GVHD prophylaxis with bone marrow grafts | Prospective trial | 7/8 matched vs <7/8 matched | No differences in any outcomes |
CNI, calcineurin inhibitor; 7/8, single high-resolution mismatch at HLA-A, B, C, and DRB1; 9/10, single high-resolution mismatch at HLA-A, B, C, DRB1, and DQB1.
The recently described associations between HLA match and mismatch in the multiply mismatched Haplo ptCy setting warrant investigation in MMUD.34 The expanded donor pool in the MMUD setting can support additional selection criteria (eg, donor-specific anti-HLA antibodies in highly sensitized patients and potential to prioritize favorable match/mismatches).
Umbilical cord blood
Traditionally, UCB match is considered at HLA-A, B at the antigen level and HLA-DRB1 at high resolution to achieve a minimum of a 4/6 match to the patient. In the single UCB alloHCT setting, matching at HLA-C and at high resolution for HLA-A, B, C, and DRB1 is associated with improved outcomes.39 This is not always practical in the setting of UCB HCT where there is a stronger emphasis placed on achieving a minimum total nucleated and/or CD34 cell dose, limiting UCB choice. Expert recommendations from the NMDP/CIBMTR20 and American Society of Transplant and Cellular Therapy9 suggest selecting the best match considering HLA-A, B, C, and DRB1 and avoiding UCB <4/8 matched at high resolution or <4/6 using the traditional match standards (Table 5).
HLA matching in umbilical cord blood allogeneic hematopoietic cell transplantation
| Reference . | Consensus guidelines . |
|---|---|
| Dehn et al20 (2019) | ≥4/6 HLA-A and -B antigen match, HLA-DRB1 high-resolution match, ≥4/8 high-resolution match meeting minimum cryopreserved cell dose for single unit alloHCT of TNC ≥2.5 × 107/kg and CD34+ cells ≥1.5 × 105/kg and double-unit alloHCT TNC ≥1.5 × 107/kg and CD34+ cells ≥1.0 × 105/kg per unit |
| Politikos et al9 (2020) | ≥4/6 HLA-A and -B antigen match, HLA-DRB1 high-resolution match, ≥4/8 high-resolution match meeting minimum cryopreserved cell dose for single-unit alloHCT of TNC ≥3.0 × 107/kg and CD34+ cells ≥2.0 × 105/kg and double-unit alloHCT CD34+ cells ≥1.5 × 105/kg per unit |
| Reference . | Consensus guidelines . |
|---|---|
| Dehn et al20 (2019) | ≥4/6 HLA-A and -B antigen match, HLA-DRB1 high-resolution match, ≥4/8 high-resolution match meeting minimum cryopreserved cell dose for single unit alloHCT of TNC ≥2.5 × 107/kg and CD34+ cells ≥1.5 × 105/kg and double-unit alloHCT TNC ≥1.5 × 107/kg and CD34+ cells ≥1.0 × 105/kg per unit |
| Politikos et al9 (2020) | ≥4/6 HLA-A and -B antigen match, HLA-DRB1 high-resolution match, ≥4/8 high-resolution match meeting minimum cryopreserved cell dose for single-unit alloHCT of TNC ≥3.0 × 107/kg and CD34+ cells ≥2.0 × 105/kg and double-unit alloHCT CD34+ cells ≥1.5 × 105/kg per unit |
TNC, total nucleated cell count.
Changing landscape in HCT and implications for optimal alternative donor selection
As transplant continues to evolve and growth inevitably occurs in the HLA mismatched setting thanks to safer, more effective approaches to minimize acute and chronic GVHD without risking increased relapse and infections, we will gain more insights into the matching/mismatching to optimize outcomes. Prioritization of alternative donor types (Haplo, MMUD, and UCB) in the absence of an MSD or MUD remains controversial, with varying programs preferring one approach over others. Until these data mature, the optimal match will vary based on the donor type available to a given patient and the choice of donors within that selection pool.
CLINICAL CASE (Continued)
The patient was enrolled on BMT CTN 1702 and biologically assigned to the MUD donor arm due to their good search prognosis score.40 The MUD donor options included multiple 8/8 donors either matched or permissively mismatched for HLA-DPB1 TCE and matched for HLA-DQB1. The transplant team selected the youngest 10/10 HLA-DPB1 TCE permissive mismatched donor available and successfully proceeded to transplant.
Acknowledgments
The author thanks Bronwen Shaw, Jeffery Auletta, and Steven Devine for helpful discussions and critical review of the manuscript.
Conflict-of-interest disclosure
Stephen R. Spellman: no competing financial interests to declare.
Off-label drug use
Stephen R. Spellman: nothing to disclose.