Skip Nav Destination
Close Modal
Update search
- Title
- Author
- Full Text
- Abstract
- Keyword
- DOI
- Title
- Author
- Full Text
- Abstract
- Keyword
- DOI
- Title
- Author
- Full Text
- Abstract
- Keyword
- DOI
- Title
- Author
- Full Text
- Abstract
- Keyword
- DOI
- Title
- Author
- Full Text
- Abstract
- Keyword
- DOI
- Title
- Author
- Full Text
- Abstract
- Keyword
- DOI
NARROW
Publications
Format
Subjects
Article Type
Topics
Date
Availability
1-20 of 5685
Follow your search
Access your saved searches in your account
Would you like to receive an alert when new items match your search?
1
Sort by
Journal Articles
Gene transfer and genome editing of T cells for cancer immunotherapy: from allogeneic HSCT to TCR gene editing
Available to Purchase
Journal:
Hematology
Hematology Am Soc Hematol Educ Program (2025) 2025 (1): 1–14.
Published: 2025
Images
Challenges for adoptive T-cell therapy and proposed gene transfer and genom...
Available to Purchase
in Gene transfer and genome editing of T cells for cancer immunotherapy: from allogeneic HSCT to TCR gene editing
> Hematology
Published: 2025
Figure 1. Challenges for adoptive T-cell therapy and proposed gene transfer and genome-editing solutions. Several challenges need to be faced to fully exploit the ability of T lymphocytes to eradicate cancer cells. Several of these challenges can be addressed by gene transfer (gene addition) and... More about this image found in Challenges for adoptive T-cell therapy and proposed gene transfer and genom...
Images
Timeline of adoptive T-cell therapy with engineered T cells. This figure r...
Available to Purchase
in Gene transfer and genome editing of T cells for cancer immunotherapy: from allogeneic HSCT to TCR gene editing
> Hematology
Published: 2025
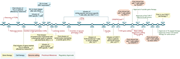
Figure 2. Timeline of adoptive T-cell therapy with engineered T cells. This figure represents a timeline of some of the key milestones in adoptive T-cell therapy with engineered cells for cancer treatment. ADA SCID, adenosine deaminase severe combined immunodeficiency; allo-HSCT, allogeneic hema... More about this image found in Timeline of adoptive T-cell therapy with engineered T cells. This figure r...
Images
TCRs and CARs. This picture depicts the advantages (green arrow) and disad...
Available to Purchase
in Gene transfer and genome editing of T cells for cancer immunotherapy: from allogeneic HSCT to TCR gene editing
> Hematology
Published: 2025
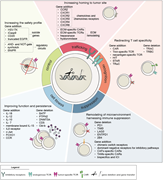
Figure 3. TCRs and CARs. This picture depicts the advantages (green arrow) and disadvantages (red arrow) of chimeric antigen receptors (CARs) and T-cell receptors (TCRs) for adoptive T-cell therapy for cancer. HLA, human leukocyte antigen. More about this image found in TCRs and CARs. This picture depicts the advantages (green arrow) and disad...
Images
Genome-editing tools: mode of action. Several genome-editing tools, with d...
Available to Purchase
in Gene transfer and genome editing of T cells for cancer immunotherapy: from allogeneic HSCT to TCR gene editing
> Hematology
Published: 2025
Figure 4. Genome-editing tools: mode of action. Several genome-editing tools, with different characteristics and modes of action, have been developed and tested for their efficiency in engineering the genome of mammalian cells, including T lymphocytes. While zinc finger nucleases, TALEN, and CRI... More about this image found in Genome-editing tools: mode of action. Several genome-editing tools, with d...
Images
Manufacturing disease-specific engineered T-cell–based cellular products. ...
Available to Purchase
in Gene transfer and genome editing of T cells for cancer immunotherapy: from allogeneic HSCT to TCR gene editing
> Hematology
Published: 2025
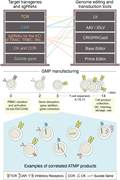
Figure 5. Manufacturing disease-specific engineered T-cell–based cellular products. Based on disease diagnosis, antigen repertoire, and knowledge of the characteristics of the tumor microenvironment, different biotechnological reagents can be combined to generate T-cell–based cellular products w... More about this image found in Manufacturing disease-specific engineered T-cell–based cellular products. ...
Journal Articles
From treatment to biology and back: managing iron overload in transfused hemoglobinopathies
Available to Purchase
Journal:
Hematology
Hematology Am Soc Hematol Educ Program (2025) 2025 (1): 103–110.
Published: 2025
Journal Articles
Buckle up! Managing surgery in patients with bleeding disorder of unknown cause
Available to Purchase
Journal:
Hematology
Hematology Am Soc Hematol Educ Program (2025) 2025 (1): 111–115.
Published: 2025
Journal Articles
Diagnosis of bleeding disorder of unknown cause: how many tests are enough to diagnose BDUC?
Available to Purchase
Journal:
Hematology
Hematology Am Soc Hematol Educ Program (2025) 2025 (1): 116–119.
Published: 2025
Journal Articles
Predictors for future bleeding in bleeding disorder of unknown cause
Available to Purchase
Journal:
Hematology
Hematology Am Soc Hematol Educ Program (2025) 2025 (1): 120–126.
Published: 2025
Journal Articles
(Un) Diagnosing von Willebrand disease
Available to Purchase
Journal:
Hematology
Hematology Am Soc Hematol Educ Program (2025) 2025 (1): 127–130.
Published: 2025
Journal Articles
What's new in hereditary hemorrhagic telangiectasia?
Available to Purchase
Journal:
Hematology
Hematology Am Soc Hematol Educ Program (2025) 2025 (1): 131–136.
Published: 2025
Journal Articles
When it's not Glanzmann thrombasthenia or Bernard-Soulier syndrome: diagnosing other qualitative platelet disorders
Available to Purchase
Journal:
Hematology
Hematology Am Soc Hematol Educ Program (2025) 2025 (1): 137–146.
Published: 2025
Journal Articles
Long-term outcome and management of complement-mediated thrombotic microangiopathy/aHUS
Available to Purchase
Journal:
Hematology
Hematology Am Soc Hematol Educ Program (2025) 2025 (1): 147–153.
Published: 2025
Journal Articles
The varieties of therapeutic experience: navigating treatment options for patients with PNH
Available to Purchase
Journal:
Hematology
Hematology Am Soc Hematol Educ Program (2025) 2025 (1): 154–163.
Published: 2025
Journal Articles
Cryptogenic stroke: definitions and management
Available to Purchase
Journal:
Hematology
Hematology Am Soc Hematol Educ Program (2025) 2025 (1): 15–21.
Published: 2025
Journal Articles
Update in the diagnosis of complement-mediated thrombotic microangiopathy/atypical hemolytic uremic syndrome
Available to Purchase
Journal:
Hematology
Hematology Am Soc Hematol Educ Program (2025) 2025 (1): 164–175.
Published: 2025
Journal Articles
Anticoagulation in malignancy: the patient who bleeds and clots simultaneously
Available to Purchase
Journal:
Hematology
Hematology Am Soc Hematol Educ Program (2025) 2025 (1): 176–182.
Published: 2025
Journal Articles
JAK2 wild-type erythrocytosis: concept, differential diagnosis, diagnostic steps, and treatment approaches
Available to Purchase
Journal:
Hematology
Hematology Am Soc Hematol Educ Program (2025) 2025 (1): 183–190.
Published: 2025
Journal Articles
The untransfusable patient
Available to Purchase
Journal:
Hematology
Hematology Am Soc Hematol Educ Program (2025) 2025 (1): 191–198.
Published: 2025
1


![Genome-editing tools: mode of action. Several genome-editing tools, with different characteristics and modes of action, have been developed and tested for their efficiency in engineering the genome of mammalian cells, including T lymphocytes. While zinc finger nucleases, TALEN, and CRISPR/Cas9 promote gene disruption and/or gene correction or gene substitution through the generation of DNA double-strand break (DSB), base editors and prime editors modify the genome in the absence of DNA DSB. Epigenome editing tools induce transcription inhibition or activation. Depending on the mechanisms of action, the different tools are characterized by a variable risk of genotoxicity (translocation between the different DNA DSB sites, off-target base insertion and/or deletions [INDELs], large deletions, or delayed proliferation through p53 activation).155 Cas9, caspase 9; pegRNA, prime editing guide RNA; sgRNA, single guide RNA; UGI, uracil DNA glycosylase inhibitor.](https://ash.silverchair-cdn.com/ash/content_public/journal/hematology/2025/1/10.1182_hematology.2025000681/4/s_hem2025000681figure4.png?Expires=2147483647&Signature=UYAzy4B0XeychVJcAeq6EK0Y013qb-Q3aDA96KQS7NWxtlCP9s-UgKAJRq1soEvSVABVHONRd5-zt42vglK56W3qNfKTJ1KRLeofQ6KjzK6IsFzJbku47qAjLCqBZkTBDkSznf3IYpiwSGMaSYgEkY6ufkrdraJppZ1vr4ol9V5JlIZ7cgHfi3yCJWYOiKJqBG7H2NE~Dgr606vF1NlGf3xv118LU3xwpQN1sQOuXmcOy298hoByOTHAaOCQOBXJ0iqKcgjQAUsvIbE2Te2obJIlE57VI7Qbpn9o4WH5~I2vXTQwQictjoPkivCgTPwCjp6nJvufukY~dW7~VKuyoA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)