Key Points
A roadmap of F9 missense mutations highlights hot spots of amino acid substitutions associated with qualitative defects in FIX.
Treatment responses were delineated in cross-reactive material–positive hemophilia B patients with suboptimal outcomes to replacement therapy.
Visual Abstract
Hemophilia B, a rare bleeding disorder, is caused by genetic variations in F9. Although quantitative factor IX (FIX) deficiencies are successfully treated by protein replacement therapy, qualitative defects may result in dysfunctional proteoforms in the circulation, potentially interfering with prophylactically or therapeutically administered recombinant FIX (rFIX) concentrates. To delineate the F9 missense variants associated with qualitative defects, we integrated genotype and phenotype from the European Association for Haemophilia and Allied Disorders F9 Coagulation Factor Variant Database. Of the 663 patients for whom activity (FIX:Act) and antigen (FIX:Ag) data were available, 40% of patients with severe hemophilia (n = 248), 50% of patients with moderate hemophilia (n = 244), and 47% of patients with mild hemophilia (n = 171) had cross-reactive material defined as FIX:Ag ≥40%. Variants associated with qualitative defects were predominantly localized (1) at proteolytic sites for FIX processing and activation, (2) within exosite II of the serine protease domain, and (3) at calcium ion coordinating residues within the Gla/EGF-1 domain of the FIX light chain. To study the effect of dysfunctional FIX proteoforms on thrombin generation in the presence of rFIX, we investigated 2 individuals with hemophilia B harboring a variant of unknown significance with an unexplained bleeding phenotype despite prophylaxis. Ex vivo therapeutic monitoring using patient plasma supplemented with rFIX concentrates, bypassing agent or emicizumab, enabled head-to-head comparison and revealed limited normalization of the prolonged initiation phase in coagulation. Alignment of information on genotype with functional proteotype may clarify the heterogeneity in bleeding phenotype and treatment response and provide a stepping stone for personalized care.
Introduction
Hemophilia B is a rare X-linked congenital bleeding disorder resulting from a functional defect in coagulation factor IX (FIX). Genetic variations that impair messenger RNA transcription, protein translation, and protein secretion into the blood result in quantitative FIX deficiency, whereas genetic variations that induce the expression and secretion of endogenous FIX proteoforms with altered functionality result in qualitative FIX deficiency.1 Patients with hemophilia B are classified as severe (<1%), moderate (1%-5%), and mild (6%-40%) based on the residual plasma FIX activity (FIX:Act), and patients are treated accordingly by prophylactic administration of plasma-derived or recombinant FIX (rFIX) concentrates to prevent joint bleeds.2-4
The clinical bleeding phenotype in hemophilia B does not always correspond with residual FIX:Act levels.5 In addition, hemostatic responses to administered FIX concentrates vary considerably, and breakthrough bleeds frequently occur.6 Recently, this unexplained treatment failure has been suggested to result from competition of the patients’ nonfunctional FIX proteoform, also referred to as cross-reactive material (CRM) positive (CRM+), with the therapeutically administered FIX concentrate in the extravascular space.7 Importantly, the full catalytic potential of FIXa requires a variety of macromolecular interactions, including activation of the zymogen through the intrinsic (FXIa) or extrinsic pathway (tissue factor [TF]-FVIIa) and phospholipid membrane, cofactor (FVIIIa), and substrate binding (FX).8-11 Recent studies on F9 missense mutations resulting in residual FIX proteoform expression have highlighted the potential of variant pathogenicity prediction based on altered protein-protein interactions.11-14 We hypothesize that treatment failure in patients with CRM+ hemophilia B can result from competition mechanisms beyond those in the extravascular space and that individuals with hemophilia B may benefit from alternative therapeutic strategies that take the underlying functional defect into account. This hypothesis was substantiated by Lee et al who revealed that the bispecific antibody emicizumab improves the procoagulant activity of FIXa proteoforms with impaired FVIIIa interaction.15 Taken together, there is a medical need to better understand the prevalence and functional effect of missense variants associated with qualitative defects in hemophilia B, to ultimately improve individual CRM+ patient treatment strategies.
The aim of this study was to address the genotype-proteotype knowledge gap in CRM+ hemophilia B by linking variant hot spots to qualitative defects in FIX. To this end, we analyzed the extent of dysfunctional circulating proteoforms using the open-access F9 Coagulation Factor Variant Database established by the European Association for Haemophilia and Allied Disorders (EAHAD) containing information on F9 genotypes and associated clinical phenotype of >4000 patients with hemophilia B.16-19 Based on this analysis, we set out to reveal mechanistic insights of a not-previously described variant p.(Gln96Arg) of unknown significance. We deep phenotyped 2 siblings with hemophilia B, with an apparent discrepancy between diagnostic activity levels, mild disease classification, unexplained severity of bleeding phenotype, and poor response to prophylactic treatment by in silico prediction, in vitro hemostatic profiling, and ex vivo thrombin generation combined with therapeutic spiking.
Methods
F9 Coagulation Factor Variant Database analysis
Genotype and laboratory data from the EAHAD F9 Coagulation Factor Variant Database were extracted for genotype-phenotype analysis.19 The initial database contained 4715 entries (accessed on 6 May 2024) and was filtered for cases with missense mutations in F9 resulting in amino acid substitutions (n = 3127). Data were subsequently cleaned for completeness in FIX:Act (n = 2527) and FIX:Ag (n = 663) data, resulting in 663 hemophilia B cases available for analysis. Unique F9 missense mutations (n = 241) provided by the Genome Aggregation Database (version 4.1.0) were summarized for allele frequency in the general population. The ClinVar germ line pathogenicity classification was simplified into 4 groups: benign, pathogenic, conflicting pathogenicity, and not available (NA).
Hemophilia B cases
A family of a mother carrier (II.I) and her 2 sons with hemophilia B (III.I) and (III.II) participated in this study. At the time of study participation, the siblings were 13 and 14 years old and received 27 and 38 IU/kg body weight Alprolix (eftrenonacog α; Bioverativ Therapeutics Inc, Sanofi) twice weekly as prophylaxis.
Plasma collection
During a regular visit to the Amsterdam University Medical Center, blood samples were drawn in 3.2% (v/v) sodium citrate tubes for coagulation testing. Platelet-poor plasma was obtained by centrifugation at 3000g for 5 minutes, followed by a second centrifugation at 14 000g for 5 minutes. Informed consent was provided for additional blood draws for extended functional coagulation testing and mass spectrometry–based proteomic screening for research purposes.
Routine laboratory testing
Coagulation testing included prothrombin time, activated partial thromboplastin time, thrombin time, fibrinogen, von Willebrand factor activity, von Willebrand factor antigen, and coagulation factor activity testing (FII:Act, FV:Act, FVII:Act, FVIII:Act, FIX:Act, FX:Act, FXI:Act, FXIII:Ac). Platelet function analysis agonist adenosine diphosphate and platelet function analysis epinephrine were determined in whole blood. All conventional laboratory tests were performed in the Amsterdam UMC clinical chemistry laboratory according to standard operating procedures. FIX:Act was determined by a 1-stage clotting assay (FIX:Act; Actin FS, Siemens Healthineers), and FIX antigen (FIX:Ag) was determined by enzyme-linked immunosorbent assay (Asserachrom; Stago). The presence of inhibitors was evaluated by the Nijmegen Bethesda inhibitor assay method.20
Thrombin-generation experiments with in vitro supplementation
Thrombin generation in platelet-poor plasma was measured as previously described, and coagulation was initiated with 1 pM TF in the presence of 4 μM phospholipids.21 Thrombin-generation tests were performed with plasma collected before and after rFIX replacement therapy (eftrenonacog α, Alprolix, Swedish Orphan Biovitrum). In addition, patient plasma was in vitro supplemented with 3 recombinant FIX concentrates (Alprolix; nonacog γ, Rixubis, Baxalta; nonacog α, Benefix, Pfizer), activated prothrombin complex concentrate (factor eight inhibitor bypassing activity, Baxter), or the bispecific antibody emicizumab (Hemlibra, Roche). Pooled normal human plasma (>250 individuals, Amsterdam UMC) and plasma collected from the mother of the 2 studied siblings with hemophilia B were used as controls.
Coagulation and fibrinolysis proteomic screening by LC-MS/MS
Quantitative plasma proteomics targeting 30 proteins involved in coagulation and fibrinolysis was performed according to an in-house–developed research use only liquid chromatography (LC) coupled to tandem mass spectrometry (MS/MS)–based proteomics assay, as detailed in the supplemental Methods.
Data analysis
All laboratory and genotype data were analyzed in R (version 4.1.2). FIX:Act and FIX:Ag data from the EAHAD F9 Coagulation Factor Variant Database containing strings “<” or “>” were converted to their cutoff value (eg, 0.01, 1, or 5) to obtain numerical data for FIX:Act/FIX:Ag ratio calculation. FIX amino acid residues and protein domains were annotated based on the UniProt database (including the 28 amino acid signal peptide and 18 amino acid propeptide sequence). LC-MS/MS data were processed in Skyline (version 24.1.0.199), and the area under the curve of tryptic peptides relative to the corresponding stable isotope-labled peptide was interpolated in the calibration curve to report results in IU/dL, U/dL, or g/L units. For the in silico structure-function analysis, the variant was visualized (PyMOL Molecular Graphics System, version 2.5.7, Schrödinger, LLC) using an FIX zymogen model incorporating the EGF-1 crystal structure (protein data bank: 1EDM).22,23 The pathogenicity of p.(Gln96Arg) substitution was predicted with the Polymorphism Phenotyping version 2 (PolyPhen-2) tool (scale 0.0 [tolerated] to 1.0 [deleterious]).24
Results
Overrepresentation of arginine residue substitutions in hemophilia B compared with the general population
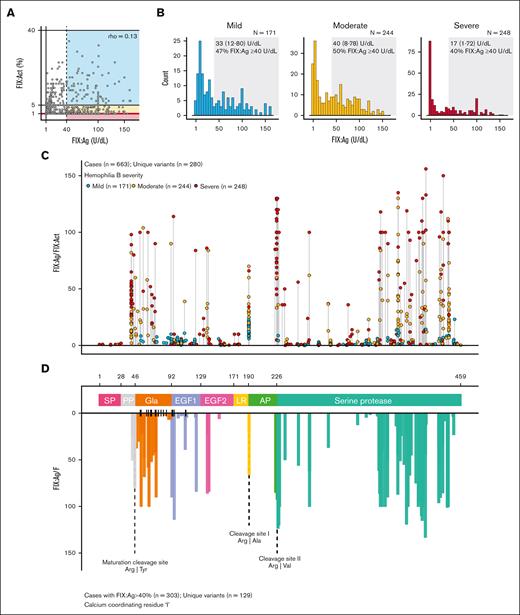
In the EAHAD F9 coagulation database, the most frequently reported genetic variations in hemophilia B are missense mutations (66%), including 628 unique variants across 3127 registered cases (Figure 1A). Missense variants associated with hemophilia B are localized across the FIX light and heavy chain. The most common variants are p.(Gly106Ser) (n = 150) in the EGF-1 domain and p.(Thr342Met) (n = 322) in the protease domain, which are associated with mild and moderate disease severity, respectively. Interestingly, among the 13 unique variants reported in >25 cases, substitutions of arginine residues were overrepresented. The reference population is characterized by the well-known polymorphism p.(Thr194Ala) with a reported allele frequency of 27% (Figure 1B). Remarkably, this database contains additional variants (n = 241 in total), with allele frequencies of ≤1%, some of which are classified as pathogenic or likely pathogenic (Figure 1B) with a PolyPhen score of >0.5 (supplemental Figure 1).
Overview of FIX variants associated with hemophilia B disease phenotype. (A) Most hemophilia B cases listed in the EAHAD F9 Variant Database harbored a missense mutant (n = 3127 cases; 66% of all 4715 cases; accessed on 6 May 2024). The number of cases per unique variant (n = 628 cases) is displayed over the FIX protein sequence to identify genotype hot spots associated with hemophilia B. (B) Allele frequencies of F9 missense mutations in the reference population reported in the Genome Aggregation Database (n = 241 missense mutations, accessed on 8 December 2024). Variants are displayed over the functional domains of FIX and colored by ClinVar germ line classification simplified in benign/likely benign, pathogenic/likely pathogenic, conflicting pathogenicity, and NA. AP, activation peptide; EGF, epidermal growth factor–like domain; Gla, Gla domain; LR, linking region; NA, not available; PP, propeptide; SP, signal peptide.
Overview of FIX variants associated with hemophilia B disease phenotype. (A) Most hemophilia B cases listed in the EAHAD F9 Variant Database harbored a missense mutant (n = 3127 cases; 66% of all 4715 cases; accessed on 6 May 2024). The number of cases per unique variant (n = 628 cases) is displayed over the FIX protein sequence to identify genotype hot spots associated with hemophilia B. (B) Allele frequencies of F9 missense mutations in the reference population reported in the Genome Aggregation Database (n = 241 missense mutations, accessed on 8 December 2024). Variants are displayed over the functional domains of FIX and colored by ClinVar germ line classification simplified in benign/likely benign, pathogenic/likely pathogenic, conflicting pathogenicity, and NA. AP, activation peptide; EGF, epidermal growth factor–like domain; Gla, Gla domain; LR, linking region; NA, not available; PP, propeptide; SP, signal peptide.
FIX missense variant hot spots associated with qualitative defects in CRM+ hemophilia B
To evaluate the genotypes that are associated with functional protein defects, we studied patients with CRM+ hemophilia B in the EAHAD F9 Coagulation Factor Variant Database. We first selected the cases with available FIX:Act and FIX:Ag data (n = 663 cases harboring 280 unique F9 missense mutations). The proportions of severe, moderate, and mild hemophilia B within this population were 37% (n = 248), 37% (n = 244), and 26% (n = 171), respectively. We found a poor relationship between FIX:Act and FIX:Ag (Spearman ρ = 0.13) values and observed a high variability in FIX:Ag distribution (Figure 2A). Interestingly, 46% of this population had FIX:Ag levels of 40 IU/dL or higher, including 40% of severe hemophilia B cases, 50% of moderate hemophilia B cases, and 47% of mild hemophilia B cases (Figure 2B). Taken together, a significant proportion of the patients were considered CRM+, irrespective of disease severity, revealing the extent of dysfunctional FIX proteoforms in hemophilia B.
Contribution of CRM in hemophilia B and FIX variant hot spots associated with qualitative defects. (A) Relation between FIX:Act and FIX:Ag reported for patients with hemophilia B harboring a missense mutation (n = 663 cases with FIX:Act and FIX:Ag data available). Each dot represents 1 hemophilia B case, and the colored rectangles indicate the disease classification based on FIX:Act (blue = mild hemophilia, yellow = moderate hemophilia, and red = severe hemophilia). Cases with CRM+ with FIX:Ag ≥40 U/dL (dashed line) were indicated by the dashed line and colored rectangles of disease severity. (B) Distribution of reported FIX:Ag levels in patients with mild (n = 171), moderate (n = 244), or severe (n = 248) hemophilia B based on their FIX activity levels. Median and interquartile range of FIX:Ag and proportion (%) of patients with FIX:Ag ≥40 U/dL are provided per severity subclass. (C) Skyline of variant location hot spots associated with qualitative defects in FIX. For hemophilia B cases with FIX:Act and FIX:Ag levels available (n = 663), the FIX:Ag/FIX:Act ratio was determined to unveil variants associated with qualitative defects. Each dot represents an individual patient case, disease severity is indicated by color, and cases are arranged by variant location within FIX. (D) Hemophilia B CRM+ cases with residual systemic levels of FIX:Ag ≥40% were selected (n = 303), and the median FIX:Ag/FIX:Act was provided per variant location. Data are colored by the domains signal peptide (SP), propeptide (PP), Gla domain (Gla), EGF-like domains (EGF-1 and -2), linker sequence, activation peptide (AP), and serine protease domain. Residues located in the Gla and EGF-1 domains involved in calcium ion positioning are marked ( | ), and proteolytic cleavage sites are visualized by dashed lines. All data are based on the EAHAD F9 Variant Database (accessed on 6 May 2024).
Contribution of CRM in hemophilia B and FIX variant hot spots associated with qualitative defects. (A) Relation between FIX:Act and FIX:Ag reported for patients with hemophilia B harboring a missense mutation (n = 663 cases with FIX:Act and FIX:Ag data available). Each dot represents 1 hemophilia B case, and the colored rectangles indicate the disease classification based on FIX:Act (blue = mild hemophilia, yellow = moderate hemophilia, and red = severe hemophilia). Cases with CRM+ with FIX:Ag ≥40 U/dL (dashed line) were indicated by the dashed line and colored rectangles of disease severity. (B) Distribution of reported FIX:Ag levels in patients with mild (n = 171), moderate (n = 244), or severe (n = 248) hemophilia B based on their FIX activity levels. Median and interquartile range of FIX:Ag and proportion (%) of patients with FIX:Ag ≥40 U/dL are provided per severity subclass. (C) Skyline of variant location hot spots associated with qualitative defects in FIX. For hemophilia B cases with FIX:Act and FIX:Ag levels available (n = 663), the FIX:Ag/FIX:Act ratio was determined to unveil variants associated with qualitative defects. Each dot represents an individual patient case, disease severity is indicated by color, and cases are arranged by variant location within FIX. (D) Hemophilia B CRM+ cases with residual systemic levels of FIX:Ag ≥40% were selected (n = 303), and the median FIX:Ag/FIX:Act was provided per variant location. Data are colored by the domains signal peptide (SP), propeptide (PP), Gla domain (Gla), EGF-like domains (EGF-1 and -2), linker sequence, activation peptide (AP), and serine protease domain. Residues located in the Gla and EGF-1 domains involved in calcium ion positioning are marked ( | ), and proteolytic cleavage sites are visualized by dashed lines. All data are based on the EAHAD F9 Variant Database (accessed on 6 May 2024).
Next, we displayed the FIX:Ag/FIX:Act ratio per location of amino acid substitution and per reported hemophilia B case in relation to disease severity (Figure 2C). Here, quantitative defects can be recognized by agreement in FIX:Ag/FIX:Act ratio (≈1), whereas qualitative defects can be recognized by increased FIX:Ag/FIX:Act ratio. The frequently reported variants associated with qualitative defects exhibited a heterogeneous severity phenotype, including p.(Arg379Gln) (n = 2 severe, n = 16 moderate, and n = 2 mild). In addition, multiple variants were associated with severe disease phenotype, despite FIX:Ag levels of >100 IU/dL. We subsequently selected the CRM+ cases with FIX:Ag levels of ≥40 U/dL (n = 303) to highlight regions in FIX resulting in hemophilia B due to variants with qualitative defects. Therefore, we determined the median FIX:Ag/FIX:Act ratio for cases stratified per variant amino acid position (Figure 2D) and focused on variants in which the qualitative defect was most evident (FIX:Ag/FIX:Act ratio of ≥20). This analysis revealed a variety of hot spots for dysfunctional proteoforms. A notable category of variants with qualitative defects includes residues between p.Gly355 and p.Ile443, spanning exosite 2 of the protease domain (Figure 2D; supplemental Figure 2). Variants in the protease domain associated with qualitative defects included the frequently reported variants p.(Arg379Gln) (n = 20) and p.(Ile443Thr) (n = 27), but not p.(Thr342Met) (n = 45). A second category of variants was the arginine substitutions at p.Arg43 and p.Arg46 representing the furin cleavage site, rendering the propeptide uncleavable, and p.Arg191 and p.Arg226, representing the zymogen activation sites (Figure 2D; supplemental Figure 2). The third notable category consisted of multiple variants distributed over the Gla, EGF-1, and EGF-2 domains (FIX light chain), which we further investigated.
Calcium ion coordinating residues associated with qualitative defects in hemophilia B
The overview of variants causing qualitative defects highlighted residues within the FIX light chain which were associated with patients with CRM+ hemophilia B with mild to severe phenotype (Figure 3A). Within the Gla domain, amino acid substitutions of glutamic acids p.(Glu54Asp), p.(Glu67Lys), p.(Glu72Gln), p.(Glu73Asp), and p.(Glu73Lys), which are modified into γ-carboxyglutamic acid for calcium ion coordination, were associated with qualitative defects (Figure 3B). Likewise, within the EGF-1 and EGF-2 domains, only selective amino acid residues were associated with qualitative defects, namely those at positions Asp93, Gln96, Asp110, Glu124, Asn138, and Arg140 (Figure 3C). Of these, the negatively charged Asp93 and β-hydroxylated Asp110 are part of a consensus sequence of the EGF-1 domain, known to contribute to calcium ion coordination, and Glu124 and Arg140 form a salt bridge between the EGF-1 and EGF-2 domains (Figure 3C). This analysis highlights that in addition to residues in the Gla domain, which coordinate calcium ions essential for FIX membrane adherence (Figure 3D), residues in the consensus sequence Asp-Gly-Asp-Glu-Cys (DGDQC), which play a pivotal role in calcium ion coordination within the EGF-1 domain, affect FIXa function (Figure 3E-F).
Qualitative defects observed for patients with hemophilia B with variants at residues coordinating calcium ions in the Gla and EGF-1 domains. (A) Hemophilia B cases harboring an amino acid substitution variant in the Gla, EGF-1, or EGF-2 domain classified by disease severity and evaluated for their residual FIX:Ag level in the plasma. (B) FIX:Ag/FIX:Act determined per case and grouped per unique variant in the Gla domain. Dots represent the individual case values, bars represent median values per unique variant, and residues involved in calcium coordination are marked by asterisks. (C) FIX:Ag/FIX:Act determined per case and grouped per unique variant in the EGF-1 and EGF-2 domains. (D) Zymogen FIX model with incorporation of the EGF-1 crystal structure.23 Visualization in PyMOL with colors representing functional domains and calcium ion coordination residues in the EGF-1 is in orange. (E) Graphical overview of calcium ion coordinating residues in the EGF-1 domain. (F) Amino acid sequence homology of EGF-1 domain between FIX, FVII, and FX (National Center for Biotechnology Information basic local alignment search tool EGF-1 FIX; Homo sapiens) with residues in the consensus sequence indicated by an asterisk. All hemophilia B case data are based on the EAHAD F9 Variant Database (accessed on 6 May 2024).
Qualitative defects observed for patients with hemophilia B with variants at residues coordinating calcium ions in the Gla and EGF-1 domains. (A) Hemophilia B cases harboring an amino acid substitution variant in the Gla, EGF-1, or EGF-2 domain classified by disease severity and evaluated for their residual FIX:Ag level in the plasma. (B) FIX:Ag/FIX:Act determined per case and grouped per unique variant in the Gla domain. Dots represent the individual case values, bars represent median values per unique variant, and residues involved in calcium coordination are marked by asterisks. (C) FIX:Ag/FIX:Act determined per case and grouped per unique variant in the EGF-1 and EGF-2 domains. (D) Zymogen FIX model with incorporation of the EGF-1 crystal structure.23 Visualization in PyMOL with colors representing functional domains and calcium ion coordination residues in the EGF-1 is in orange. (E) Graphical overview of calcium ion coordinating residues in the EGF-1 domain. (F) Amino acid sequence homology of EGF-1 domain between FIX, FVII, and FX (National Center for Biotechnology Information basic local alignment search tool EGF-1 FIX; Homo sapiens) with residues in the consensus sequence indicated by an asterisk. All hemophilia B case data are based on the EAHAD F9 Variant Database (accessed on 6 May 2024).
Clinical phenotype of hemophilia B cases harboring a p.(Gln96Arg) variant in the EGF-1 domain
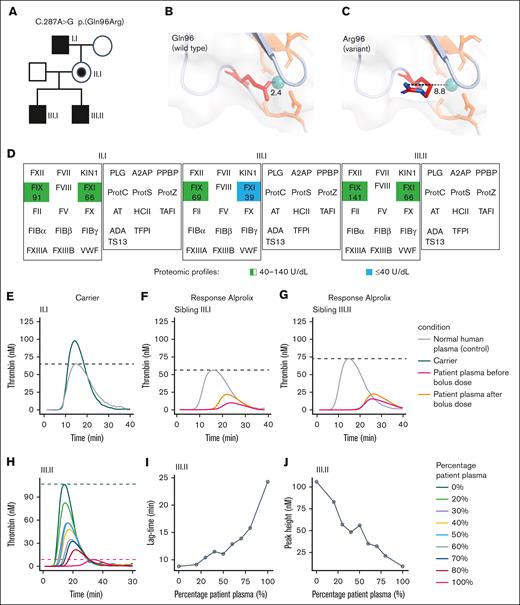
To demonstrate the importance of these calcium-coordinating residues in the EGF-1 domain, we studied 2 siblings (III.I and III.II) with hemophilia B in our medical center who harbored an F9 C.287 A>G missense mutation resulting in p.(Gln96Arg) in the EGF-1 domain (Figure 4A). The variant p.(Gln96Arg) was considered of unknown significance, as this specific amino acid substitution was not reported in the ClinVar database (accessed on 6 May 2024). However, the PolyPhen score of 0.97 predicted this variant as probably damaging, and another variant at this position, p.(Gln96Pro), was listed in the EAHAD database with antigen-activity discordance indicative of a qualitative defect (Figure 3C). In silico mutagenesis of uncharged Gln substitution into a positively charged Arg residue at this position indicated unfavorable calcium ion coordination as a consequence of repulsive forces driving p.Arg96 side chain toward the solvent (Figure 4B-C).
Hemophilia B caused by an F9 mutant, initially classified as variant of unknown significance, which results in a qualitative defect and impaired thrombin generation. (A) Pedigree with inherited F9 C.287A>G mutant resulting in p.(Gln96Arg) variant. (B-C) The p.(Gln96Arg) variant is located in the EGF-1 domain and introduces a positively charged arginine within the calcium ion binding cavity. (D) Other deficiencies or abnormalities in proteins contributing to the severe bleeding phenotype were excluded by quantitative mass spectrometry–based screening of plasma proteins corresponding to the tier 1 diagnostic gene list,25 updated and endorsed at the International Society of Thrombosis and Hemostasis 2023 conference. Normal concentration of coagulation and fibrinolysis proteins in the plasma was confirmed, except for mild FXI deficiency (39 U/dL) identified in 1 sibling (III.1). (E) Thrombin-generation profile with plasma from the carrier (II.I). (F-G) Thrombin generation with plasma from siblings (III.I) and (III.II) before and 1 hour after 25 U/kg Alprolix as prophylaxis. (H) Inhibitor testing by mixing normal plasma pool and patient plasma (III.II) followed by thrombin generation with (I) lag time and (J) peak height. Data were obtained by calibrated automated thrombography (CAT) assay with 1 pM TF for coagulation initiation. Dashed lines indicate the peak height in the control condition (pooled normal human plasma).
Hemophilia B caused by an F9 mutant, initially classified as variant of unknown significance, which results in a qualitative defect and impaired thrombin generation. (A) Pedigree with inherited F9 C.287A>G mutant resulting in p.(Gln96Arg) variant. (B-C) The p.(Gln96Arg) variant is located in the EGF-1 domain and introduces a positively charged arginine within the calcium ion binding cavity. (D) Other deficiencies or abnormalities in proteins contributing to the severe bleeding phenotype were excluded by quantitative mass spectrometry–based screening of plasma proteins corresponding to the tier 1 diagnostic gene list,25 updated and endorsed at the International Society of Thrombosis and Hemostasis 2023 conference. Normal concentration of coagulation and fibrinolysis proteins in the plasma was confirmed, except for mild FXI deficiency (39 U/dL) identified in 1 sibling (III.1). (E) Thrombin-generation profile with plasma from the carrier (II.I). (F-G) Thrombin generation with plasma from siblings (III.I) and (III.II) before and 1 hour after 25 U/kg Alprolix as prophylaxis. (H) Inhibitor testing by mixing normal plasma pool and patient plasma (III.II) followed by thrombin generation with (I) lag time and (J) peak height. Data were obtained by calibrated automated thrombography (CAT) assay with 1 pM TF for coagulation initiation. Dashed lines indicate the peak height in the control condition (pooled normal human plasma).
Although the 2 brothers were classified as having mild hemophilia B based on their residual FIX activity levels of 5% and 6% at the time of diagnosis, they had both traumatic and spontaneous joint bleeds, which were confirmed by physical examination and ultrasound. The boys’ ages at their first joint bleeds were 3.1 and 6 years, and prophylaxis was started at 4.3 and 5.5 years of age, respectively. However, despite administration of pharmacokinetic-dosed prophylactic FIX concentrate twice a week with a trough level of 10 IU/dL (27 and 38 IU/kg body weight Alprolix, respectively), the boys continued to experience nontraumatic joint bleeds and associated acute and chronic synovitis of ankle joints. These joint bleeds occurred both spontaneously and after mild activity. With lifetime annualized joint bleeding rates of 0.6 and 0.9, indicating an average joint bleed every 20 and 13 months, respectively, the phenotype was considered moderate to severe.25 Taken together, this FIX missense variant resulting in an amino acid substitution in the calcium ion binding cavity of EGF-1 was associated with a hemophilia B phenotype characterized by a significant bleed despite prophylaxis.
Laboratory phenotype and ex vivo thrombin generation of hemophilia B cases harboring a p.(Gln96Arg) variant in the EGF-1 domain
In agreement with the roadmap annotation, the p.(Gln96Arg) variant was associated with a qualitative defect in FIX, as indicated by the disagreement in FIX activity (FIX:Act = 12%-18%) and FIX antigen levels (FIX:Ag > 40 IU/dL) for both siblings. Besides the low FIX:Act levels, no clinical relevant abnormalities in routine hemostatic laboratory testing were found, except for mildly reduced FXI:Act of 42% for 1 sibling (III.I) (supplemental Table 1). To further exclude other proteins contributing to the moderate-to-severe bleeding phenotype, we screened the plasma proteome of the siblings (III.I and III.II) and their mother (II.I) for rare deficiencies in coagulation factor or abnormal levels of coagulation enzyme regulators by quantitative mass spectrometry. Again, no quantitative hemostatic protein abnormalities were detected, other than FXI of 39 IU/dL for 1 sibling (III.I) (Figure 4D; supplemental Table 2). Furthermore, thrombin generation in the mother was normal (Figure 4E) but severely impaired and not in agreement with FIX:Act levels exceeding 5% in both siblings (Figure 4F-G). Moreover, there was no indication for the presence of inhibitors (Figure 4H-J), and thrombin-generation profiles of the siblings were characterized by a prolonged lag time, suggesting an effect of the p.(Gln96Arg) FIX variant on the initiation phase of coagulation (Figure 4I).
Ex vivo and in vitro evaluation of therapeutics on coagulation potential by concentrate supplementation in plasma from 2 patients with CRM+ with variant in EGF-1
We studied the therapeutic response to rFIX concentrates by FIX activity and quantity measurements combined with assessment of the overall coagulation potential using thrombin-generation assays. Upon a 25 IU/kg Alprolix bolus dose, the FIX:Act improved to 36% for 1 sibling (III.I) and to 31% for the other sibling (III.II). This corresponded with an increase of FIX:Ag based on enzyme-linked immunosorbent assay of 29% (from 85 to 110 IU/dL) within 1 hour for 1 sibling (III.I) and 12% (106 to 119 IU/dL) for the other sibling (III.II). By complementary LC-MS/MS analysis, the FIX concentration increased from 141 to 159 IU/mL and from 69 to 179 IU/mL FIX for siblings III.I and III.II, respectively (supplemental Table 2). Thrombin generation remained reduced in peak height for 1 sibling (III.I), and the lag time remained prolonged for both siblings (Figure 4F-G).
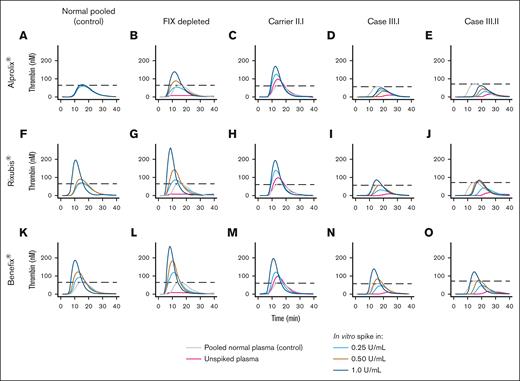
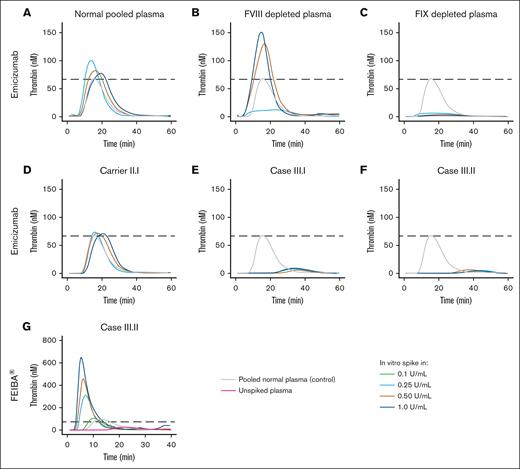
To assess therapeutic efficacy in vitro, we supplemented normal pooled plasma, FIX-depleted plasma, and plasma from the carrier and sons, from whom blood was collected just before Alprolix bolus dose, with increasing concentrations of Alprolix (Figure 5A-E) and 2 alternative rFIX concentrates, Rixubis (Figure 5F-J) and Benefix (Figure 5H-O), and subsequently measured thrombin generation. To evaluate the effect of rFIX concentrates on thrombin generation in the condition of normal FIX levels, Alprolix, Rixubis, and Benefix were supplemented to normal pooled plasma. In vitro supplementation of Rixubis and Benefix in normal pooled plasma resulted in an enhanced thrombin-generation potential, whereas Alprolix did not (Figure 5A,F,K). Furthermore, in FIX-deficient plasma, the in vitro thrombin-generation response was more evident for Rixubis and Benefix compared with Alprolix (Figure 5B,G,L). In the plasma from the 2 boys, which was collected just before the next 25 IU/kg Alprolix prophylactic bolus dose, Benefix and Rixubis also outperformed Alprolix by means of thrombin-generation peak height improvement after in vitro supplementation. Next, we assessed whether the impaired FIX activity due to p.(Gln96Arg) variant in the EGF-1 domain could be rescued by bypassing FVIII binding by supplementation of the bispecific antibody emicizumab. In vitro supplementation of emicizumab indeed restored the thrombin generation in FVIII-deficient plasma, but not in FIX-deficient plasma, nor in the plasma of the siblings (Figure 6A-F). Bypassing FIX activation by supplementation of factor eight inhibitor bypassing activity resulted in an enhanced in vitro thrombin-generation response in patient plasma, overshooting the thrombin generation in normal pooled plasma from lowest concentration for in vitro spike in Figure 6G.
Evaluation of in vitro efficacy of recombinant FIX concentrates on thrombin generation in patient plasma. Thrombin-generation profiles on in vitro supplementation of Alprolix in (A) normal pooled plasma, (B) FIX-depleted plasma, (C) plasma of a hemophilia B carrier harboring p.(Gln96Arg), and (D-E) plasma of her 2 sons with hemophilia B. Thrombin-generation profiles on in vitro supplementation of Rixubis in (F) normal pooled plasma, (G) FIX-depleted plasma, (H) plasma of a hemophilia B carrier harboring p.(Gln96Arg), and (I-J) plasma of her 2 sons with hemophilia B. Thrombin-generation profiles on in vitro supplementation of Benefix in (K) normal pooled plasma, (L) FIX-depleted plasma, (M) plasma of a hemophilia B carrier harboring p.(Gln96Arg), and (N-O) plasma of her 2 sons with hemophilia B. Data were obtained by CAT assay with 1 pM TF for coagulation initiation. Dashed lines indicate the peak height in the control condition (pooled normal human plasma).
Evaluation of in vitro efficacy of recombinant FIX concentrates on thrombin generation in patient plasma. Thrombin-generation profiles on in vitro supplementation of Alprolix in (A) normal pooled plasma, (B) FIX-depleted plasma, (C) plasma of a hemophilia B carrier harboring p.(Gln96Arg), and (D-E) plasma of her 2 sons with hemophilia B. Thrombin-generation profiles on in vitro supplementation of Rixubis in (F) normal pooled plasma, (G) FIX-depleted plasma, (H) plasma of a hemophilia B carrier harboring p.(Gln96Arg), and (I-J) plasma of her 2 sons with hemophilia B. Thrombin-generation profiles on in vitro supplementation of Benefix in (K) normal pooled plasma, (L) FIX-depleted plasma, (M) plasma of a hemophilia B carrier harboring p.(Gln96Arg), and (N-O) plasma of her 2 sons with hemophilia B. Data were obtained by CAT assay with 1 pM TF for coagulation initiation. Dashed lines indicate the peak height in the control condition (pooled normal human plasma).
Evaluation of in vitro efficacy of emicizumab and inhibitor bypassing coagulation complex in patient plasma. Thrombin-generation profiles on in vitro supplementation of emicizumab (Hemlibra) in (A) normal pooled plasma, (B) FVIII-depleted plasma, (C) FIX-depleted plasma, (D) plasma of the carrier II.I, (E) plasma of sibling III.I harboring p.(Gln96Arg), and (F) plasma of sibling III.II harboring p.(Gln96Arg). (G) Thrombin-generation profiles on in vitro supplementation of anti-inhibitor coagulant complex factor eight inhibitor bypassing activity in patient (III.II) plasma. Data were obtained by CAT assay with 1 pM TF for coagulation initiation. Dashed lines indicate the peak height in the control condition (pooled normal human plasma).
Evaluation of in vitro efficacy of emicizumab and inhibitor bypassing coagulation complex in patient plasma. Thrombin-generation profiles on in vitro supplementation of emicizumab (Hemlibra) in (A) normal pooled plasma, (B) FVIII-depleted plasma, (C) FIX-depleted plasma, (D) plasma of the carrier II.I, (E) plasma of sibling III.I harboring p.(Gln96Arg), and (F) plasma of sibling III.II harboring p.(Gln96Arg). (G) Thrombin-generation profiles on in vitro supplementation of anti-inhibitor coagulant complex factor eight inhibitor bypassing activity in patient (III.II) plasma. Data were obtained by CAT assay with 1 pM TF for coagulation initiation. Dashed lines indicate the peak height in the control condition (pooled normal human plasma).
Discussion
The overview of F9 missense mutations that are associated with distinct qualitative defects in FIX highlighted variants at the (1) propeptide and activation site peptide cleavage, (2) calcium ion–coordinating residues in the Gla and EGF-1 domains, and (3) exosite residues of the serine protease domain. This variant overview facilitated the interpretation of a variant of unknown significance in 2 siblings with CRM+ hemophilia B who experienced breakthrough bleeds despite prophylaxis with FIX concentrates. Counterintuitively, almost half of the patients with available data in the EAHAD F9 Coagulation Factor Variant Database were reported to be CRM+ with FIX:Ag > 40 IU/dL, irrespective of disease severity. Because these so-called FIX proteoforms affect molecular structure and function, we anticipate that they may exert dominant-negative effects on specific properties of FIX, suggesting clinical relevance of CRM+ hemophilia B beyond the risk of inhibitor development.16,19,26,27 In addition, we anticipate that these dysfunctional FIX proteoforms in circulation may be of significance for the bleeding phenotype for girls and women who are carriers of an F9-mutant allele. To improve our understanding of qualitative defects in hemophilia B and to appropriately address bleeding episodes in female patients, it will be essential to include female F9 variant carriers in clinical trial design for fit-for-purpose prophylactic or treatment options.28
Importantly, the presence of FIX proteoforms may explain the discrepancy between activity assays initiated by either the intrinsic or extrinsic pathway29 and the lack of correlation between laboratory results and bleeding phenotype, both of which affect disease severity classification.1,30 In addition, circulating FIX proteoforms may compete with exogenous rFIX concentrates in terms of phospholipid membrane binding and macromolecular complex formation, thus rendering FIX replacement therapy less efficient. We envision that it would therefore be valuable to evaluate new treatment strategies, including emicizumab, anti-tissue factor pathway inhibitor, and gene therapy-based approaches in subclasses of CRM+ hemophilia B which express FIX proteoforms with distinct molecular defects.9,29-33 The potential of tailoring treatment strategies is exemplified by the observation that the procoagulant activity of (recombinant) FIX proteoforms that affect FVIII binding was improved by emicizumab.15 Similarly, variants affecting zymogen-to-enzyme conversion using the intrinsic (FXIa mediated) or extrinsic (TF-FVIIa mediated) pathways are hypothesized to particularly benefit from FIX activation bypassing strategies.
The variant roadmap highlighted an interesting group of single amino acid alterations in the Gla and EGF-1 domains resulting in moderate-to-severe CRM+ hemophilia B. This is in agreement with previous reports on the role of the EGF-1 domain in mediating multiple protein-protein interactions, including those with TF-FVIIa, FXIa, and cofactor VIIIa, and in correctly aligning the catalytic triad within the protease domain.8,9,14,23 The overview of variants associated with qualitative defects is also in line with previous reports on FIX variant pathogenicity in this region,13,31,32 including a recent study that used molecular modeling of zymogen FIX with FXIa to explain a mild bleeding phenotype associated with the hydrophobic residue substitution p.(Val92Ala) at the intersection of the Gla and EGF-1 domains.12 Here, we suggest that, in particular, variants in the EGF-1 domain that affect calcium coordination are associated with a moderate-to-severe bleeding phenotype. Although calcium coordination in the Gla domain mainly involves Glu residues,33 the calcium ion binding cavity in the EGF-1 domain involves the Asp-Gly-Asp-Gln-Cys motif, which is conserved between serine proteases FVII and FX, together with the β-hydroxylated Asp110 residue and Asp111 and Gly124.9,31,34,35 Together with the salt bridge between EGF-1 and EGF-2, these residues maintain the conformation that allows for TF-FVIIa and cofactor FVIII binding.9,11,32,34-36 We hypothesize that variants in the calcium ion binding cavity will predominantly affect FIX activation by TF-FVIIa and will have limited effect on FIX activation by FXIa, as demonstrated for variants at position Gly94.32 Combined with previous studies, the CRM+ cases with p.(Gln96Arg) described here emphasize the intricate relationship between defective calcium ion binding in EGF-1 domain and the moderate-to-severe hemophilia B clinical phenotype.37,38 This is supported by a CRM+ (FIX:Ag 114 U/dL) patient in the EAHAD database with a p.(Gln96Pro)(“New London”) variant who displays extremely slow FIX activation potential but normal FIXa protease activity.38 In an equivalent FIX variant database (https://www.factorix.org/; accessed on June 2024), 4 additional amino acid substitutions have been reported at position 96, namely p.(Gln96Lys), p.(Gln96His), p.(Gln96Glu), and Gln96∗.16 Importantly, it should be emphasized that the nature of the amino acid substitutions may result in different molecular consequences and thus potentially different clinical phenotypes.
This study highlights the value of the open-access EAHAD F9 Coagulation Factor Variant Database for understanding qualitative defects in patients with hemophilia B worldwide.19 Some observations in our study, however, need careful interpretation, because they are based on a reported database rather than an observational cohort or clinical study. First, for only 663 of 3127 cases (21%) with F9 missense variant entries in the EAHAD F9 Coagulation Factor Variant Database, sufficient data were available for inclusion in our analysis, mainly due to the limited availability of FIX:Ag data. We therefore expect that the described prevalence of qualitative defects in this study may be biased toward those cases for which FIX:Ag levels were reported, which underlines the importance of data completeness in prospective or retrospective studies to prevent selection bias in future studies. Second, the reported FIX:Ag and FIX:Act values have been acquired by different analytical methodologies, which may confound comparison of absolute values between laboratories globally. As information on both the preanalytical and analytical phases was not systematically reported, we were unable to adjust for laboratory methodology. Third, for some cases, only 1% FIX:Ag was reported, despite the annotation of mild hemophilia B, which we considered unlikely. Last, in this study, we assume that sufficient washout period was taken into account before FIX:Ag and FIX:Act testing.
Taken together, this study argues for careful evaluation of qualitative defects in FIX in patients with hemophilia B with a discordant activity-to-antigen ratio in combination with a poor response to replacement therapy. The presence of endogenous “anticoagulant” FIX proteoforms in circulation may not only contribute to the disease phenotype but may also compete with or induce dominant-negative effects on replacement therapy. We hypothesize that particularly variants affecting the TF-FVIIa-complex interaction may be most sensitive to proteoform competition with FIX concentrates, by impairing both the initiation and amplification phases of coagulation.14,39-41 Ex vivo supplementation of therapeutics into patient plasma holds great potential for the assessment of novel strategies in CRM+ patients with different molecular subtype.21 Our study highlights the potential of hemophilia B disease classification beyond FIX:Act testing, to better align with clinical bleeding phenotype.5 In conclusion, this study provided overview of missense variants associated with qualitative defects in FIX, emphasizing the potential of future research on hemophilia B patient subclassification in relation to replacement therapy efficacy or alternative novel tailored treatment strategies.
Acknowledgments
This study makes use of data collected by the European Association for Haemophilia and Allied Disorders (EAHAD) F9 Coagulation Factor Variant Database. The authors gratefully acknowledge the participation of all patients with hemophilia B and the medical team taking care of these patients. They express their gratitude to the EAHAD staff and volunteers for their contribution to the Coagulation F9 Variant database. They thank the care providers and parents of the 2 siblings with hemophilia B for participation in the case study. They thank Kyra N. Smit for contribution in patient case discussions and for supervision of diagnostic laboratory analysis.
Authorship
Contribution: T.T.v.D. initiated the study, conducted mass spectrometry, analyzed and interpreted the data, and wrote the manuscript; S.G. initiated the study and contributed to patient care; I.K. and N.F. analyzed the data; A.K. conducted mass spectrometry; W.F.K. performed TG assays; M.B. contributed to patient care; H.M.H. supervised diagnostic laboratory analysis; K.F. contributed to patient care and interpreted the data; J.C.M.M. designed the study, supervised TG analysis, and interpreted the data; M.v.d.B. initiated the study and design and interpreted the data; and all authors reviewed and approved the manuscript.
Conflict-of-interest disclosure: T.T.v.D. received research grants from The Society for the Advancement of Science, Medicine and Surgery (Het Genootschap ter bevordering van Natuur- Genees- en Heelkunde); and the Professor Heimburger award from Commonwealth Serum Laboratories Behring. The institution of K.F. has received unrestricted research grants from Commonwealth Serum Laboratories Behring, Sobi, and Novo Nordisk; and consultancy fees from F. Hoffmann-La Roche, Sanofi, Sobi, and Novo Nordisk. J.C.M.M. received consultancy fees from Alveron Pharma and Synapse Research Institute. The remaining authors declare no competing financial interests.
Correspondence: Maartje van den Biggelaar, Sanquin Research, Plesmanlaan 125, 1066 CX, Amsterdam, The Netherlands; email: m.vandenbiggelaar@sanquin.nl.
References
Author notes
Population-based data were obtained by open access databases from the Genome Aggregation Database and Coagulation Factor Databases of the European Association for Haemophilia and Allied Disorders. Original data and scripts are available from the corresponding author, Maartje van den Biggelaar (m.vandenbiggelaar@sanquin.nl), on request.
The full-text version of this article contains a data supplement.