Abstract
Systemic iron balance must be tightly regulated to prevent the deleterious effects of iron deficiency and iron overload. Hepcidin, a circulating hormone that is synthesized by the liver, has emerged as a key regulator of systemic iron homeostasis. Hepcidin inhibits the absorption of dietary iron from the intestine and the release of iron derived from red blood cells from macrophages. Therefore, variation in hepcidin levels modifies the total amount of iron stored in the body and the availability of iron for erythropoiesis. The production of hepcidin by the liver is modulated by multiple physiological stimuli, including iron loading, inflammation, and erythropoietic activity. Investigation of the functions of the gene products mutated in inherited iron disorders using tissue-culture systems and animal models has provided valuable insights into the mechanisms by which these hepcidin responses are mediated. This review focuses on recent advances in our understanding of the molecular mechanisms underlying the regulation of systemic iron homeostasis.
The role of hepcidin in systemic iron balance
Each day, the adult human body requires approximately 25 mg of iron for hemoglobin synthesis in erythropoiesis. The majority of this iron is supplied by the recycling of senescent erythrocytes by macrophages within the reticuloendothelial system. Only 1-2 mg of this iron is obtained through the absorption of dietary iron, which serves to replace insensible iron losses such as the sloughing of epithelial cells and minor bleeding. Once released into the plasma, iron circulates bound to the glycoprotein transferrin. Excess iron that is not consumed in erythropoiesis or other cellular processes is stored primarily in the liver and in reticuloendothelial macrophages. In settings of altered iron demands, such as the increased needs of pregnancy, the body can modulate the absorption of dietary iron; however, no regulated mechanism for iron excretion from the body has been identified. Therefore, to ensure sufficient availability of iron for hemoglobin synthesis and other metabolic processes while avoiding the oxidative damage to cells that can result from excess free iron, iron balance must be tightly regulated (reviewed in Hentze1 ).
The collective work of several investigators has established that hepcidin, a circulating hormone produced by the liver, plays a central role in the regulation of systemic iron homeostasis (reviewed in Ganz2 ). Hepcidin is a 25–amino acid peptide with homology to members of the defensin family of antimicrobial peptides. Hepcidin binds to ferroportin, a widely expressed transmembrane protein that functions to export iron from cells. Ferroportin is highly expressed at sites where iron is released from cells into the plasma, including the basolateral membrane of enterocytes and the plasma membrane of macrophages. Because hepcidin binding leads to the internalization and degradation of ferroportin in lysosomes, hepcidin acts to decrease the absorption of dietary iron and the release of recycled iron from macrophage stores. The essential role of hepcidin in the maintenance of systemic iron balance has been demonstrated in mouse models. Mice lacking hepcidin expression develop systemic iron overload, whereas transgenic mice overexpressing hepcidin exhibit severe iron deficiency anemia. In humans, loss-of-function mutations in the hepcidin gene HAMP (hepcidin antimicrobial peptide) cause juvenile hemochromatosis, an autosomal recessive disorder characterized by severe iron deposition in multiple organs, including the liver, heart, and endocrine tissues, that typically presents in the first through the third decades of life.
Hepcidin production by the liver is modulated in response to several physiological stimuli. Hepcidin expression is induced by iron loading, thereby providing a means to limit further iron uptake from the diet. In contrast, hepcidin expression is repressed by anemia and hypoxia, thereby providing a means to increase iron availability for erythropoiesis. Hepcidin levels are also increased by inflammatory stimuli. This review emphasizes recent advances in our understanding of the molecular mechanisms through which hepcidin expression is modulated to influence systemic iron balance.
Hepcidin regulation by iron
Studies conducted in mice initially demonstrated that hepcidin expression is induced in response to dietary and parenteral iron loading. The physiological importance of the regulation of hepcidin by iron is evident from the phenotypes of several inherited disorders of iron balance that are characterized by hepcidin dysregulation (reviewed in Pietrangelo3 ). Hepcidin deficiency is a feature of the known recessive forms of hereditary hemochromatosis, which are caused by mutations in the genes HFE, transferrin receptor 2 (TFR2), hemojuvelin (HFE2, also known as HJV), and, as described above, hepcidin. Due to the insufficient hepcidin production in these disorders, the body is unable to limit the absorption of dietary iron appropriately, leading to a progressive increase in body iron stores. In contrast, a failure to appropriately down-regulate hepcidin production is seen in iron-refractory iron deficiency anemia (IRIDA), an inherited disorder caused by loss-of-function mutations in the gene TMPRSS6. In IRIDA, hepcidin excess results in impaired iron absorption and impaired ability to utilize iron that has been reclaimed by macrophages. The fact that the genetic defects in these disorders cannot be compensated for to maintain systemic iron balance indicates that the encoded gene products play key roles in the hepcidin response to systemic iron status. Recent evidence suggests that the actions of these gene products, which are all expressed in the liver, may converge at a key signaling pathway promoting hepcidin expression by hepatocytes, as described in detail below.
Positive regulation of hepatic BMP signaling
Signaling by bone morphogenetic proteins (BMPs), members of the TGF-β superfamily of ligands, has emerged as a key pathway promoting hepcidin expression by the liver (Figure 1). Binding of BMPs to paired serine/threonine kinase receptors results in phosphorylation of receptor-associated SMAD (sons of mothers against decapentaplegic homologue) proteins, which after complexing with the common mediator protein SMAD4, ultimately modulate gene transcription by binding to specific sequences in the promoters of BMP target genes. HJV, a glycosylphosphatidylinositol-anchored cell-associated protein of the RGM (repulsive guidance molecule) family, functions as a coreceptor for BMPs, which augments hepatic BMP signaling to induce hepcidin expression (reviewed in Corradini4 ). HJV appears to play a central role in the hepatic BMP signaling pathway that promotes hepcidin expression, because patients with hereditary hemochromatosis due to HJV mutations have severe hepcidin deficiency and exhibit a juvenile hemochromatosis phenotype with severity similar to disease due to mutation in the hepcidin gene itself. Mice with liver-specific disruption of the gene encoding the common mediator SMAD4 also exhibit hepcidin deficiency and severe iron overload, further supporting the key role of hepatic BMP signaling in hepcidin production.5
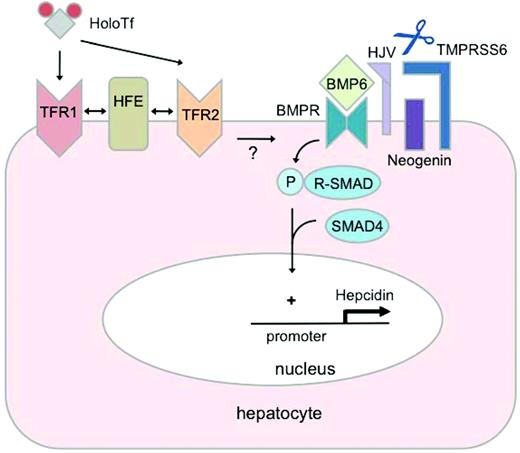
Regulation of hepcidin by iron. Binding of BMP6 to receptors (BMPRs) at the hepatocyte plasma membrane results in phosphorylation (P) of receptor-associated SMAD proteins (R-SMADs), which form a complex with the common mediator SMAD4. This SMAD4 complex translocates to the nucleus to bind specific regulatory elements in the promoter of the hepcidin gene, increasing hepcidin transcription. The glycosylphosphatidylinositol-linked protein HJV acts as a BMP coreceptor that promotes hepcidin signaling through this pathway. Neogenin may interact with HJV and enhance BMP signaling. In contrast, the transmembrane serine protease TMPRSS6 inhibits hepcidin signaling through the BMP pathway by cleaving HJV from the plasma membrane. Hepatic BMP6 production increases in response to chronic iron loading, suggesting an autocrine or paracrine mechanism for regulating hepcidin synthesis to limit the progression of systemic iron loading. Binding of holo-transferrin (HoloTf) to TFR1 displaces HFE, which may interact with TFR2 to promote hepcidin expression. Whereas evidence suggests that HFE modulates hepcidin signaling through the BMP pathway, how HFE and its interactions with TFR1 and TFR2 may relate functionally to the BMP pathway is not yet known.
Regulation of hepcidin by iron. Binding of BMP6 to receptors (BMPRs) at the hepatocyte plasma membrane results in phosphorylation (P) of receptor-associated SMAD proteins (R-SMADs), which form a complex with the common mediator SMAD4. This SMAD4 complex translocates to the nucleus to bind specific regulatory elements in the promoter of the hepcidin gene, increasing hepcidin transcription. The glycosylphosphatidylinositol-linked protein HJV acts as a BMP coreceptor that promotes hepcidin signaling through this pathway. Neogenin may interact with HJV and enhance BMP signaling. In contrast, the transmembrane serine protease TMPRSS6 inhibits hepcidin signaling through the BMP pathway by cleaving HJV from the plasma membrane. Hepatic BMP6 production increases in response to chronic iron loading, suggesting an autocrine or paracrine mechanism for regulating hepcidin synthesis to limit the progression of systemic iron loading. Binding of holo-transferrin (HoloTf) to TFR1 displaces HFE, which may interact with TFR2 to promote hepcidin expression. Whereas evidence suggests that HFE modulates hepcidin signaling through the BMP pathway, how HFE and its interactions with TFR1 and TFR2 may relate functionally to the BMP pathway is not yet known.
Whereas several BMP ligands are able to promote hepcidin transcription in vitro, the finding that Bmp6-knockout mice exhibit severe systemic iron overload suggests that BMP6 is a key endogenous BMP ligand for hepcidin signaling.6,7 BMP6 is expressed in the liver, and hepatic levels of Bmp6 mRNA increase in response to chronic dietary iron loading in a manner similar to hepcidin mRNA.8 These findings have led to a model in which hepatocytes modulate BMP6 secretion in response to changing iron stores, thereby regulating hepcidin synthesis in an autocrine or paracrine manner. The mechanism by which iron modulates BMP6 transcription has yet to be elucidated. This process, however, does not appear to require the hereditary hemochromatosis proteins HFE, TFR2, or HJV,9,10 and hepatic Bmp6 mRNA expression does not appear to be induced by acute changes in serum transferrin saturation.11
Compared with HJV, the mechanisms by which the hemochromatosis proteins HFE and TFR2 contribute to hepcidin synthesis are less well understood. HFE, a major histocompatibility complex class I-like molecule, competes with transferrin for binding to TFR1 (transferrin receptor 1), a transmembrane protein that mediates uptake of transferrin-bound iron, particularly during erythroid cell development. Transgenic mouse models suggest that HFE induces hepatic hepcidin expression when dissociated from TFR1.12 TFR2 is a homolog of TFR1 that has lower affinity for iron-bound transferrin, and in vitro studies suggest that interaction of TFR2 with HFE is necessary for the induction of hepcidin expression by iron-loaded transferrin.13 These findings have suggested an “iron-sensing” model in which HFE, TFR1, and TFR2 modulate hepcidin signaling in response to changes in the circulating level of iron-bound transferrin. Mice with combined mutations in Hfe and Tfr2 exhibit hepcidin deficiency and iron overload that are more severe than in mice with mutations in either gene alone.14 Therefore, HFE and TFR2 each appear to retain some capacity to promote hepcidin production in the absence of the other protein. These results are compatible with clinical observations that patients with hemochromatosis associated with HFE mutations typically present as adults and exhibit a relative hepcidin deficiency that is less severe than the marked hepcidin suppression observed in juvenile hemochromatosis due to mutations in HJV or HAMP. The expression of HFE and TFR2 is not restricted exclusively to hepatocytes, and the potential relationship of their extrahepatic functions to iron balance requires further study.
Recent work suggests that HFE may play a role in the modulation of BMP signaling. In Hfe-knockout mice, a model of HFE hemochromatosis, hepatic-signaling responses induced by the BMP6 ligand may be blunted. The livers of Hfe-knockout mice have elevated Bmp6 mRNA, as would be expected in the setting of increased iron stores, but also phosphorylation of receptor-associated SMAD proteins and expression of BMP target genes that appear impaired.10,15 Conversely, forced overexpression of Hfe in mouse liver increases the expression of BMP target genes16 in a manner that requires HJV.17 These findings support a model in which HFE intersects with the BMP pathway downstream of HJV to promote hepcidin transcription. A recent study found that after iron depletion, the induction of hepcidin expression in response to an acute iron challenge was impaired in Hfe-knockout mice and was nearly absent in mice harboring mutations in Tfr2, Hjv, or Bmp6, suggesting that all 4 proteins contribute to the hepcidin response induced by acute iron changes.9 How HFE and its interactions with TFR1 and TFR2 relate functionally to the BMP pathway, however, remains to be determined.
Another protein that may interact with the BMP pathway to promote hepcidin expression is neogenin, a member of the DCC (deleted in colorectal cancer) family of tumor-suppressor proteins. Mice homozygous for a targeted disruption of neogenin develop severe hepatic iron overload that is associated with decreased levels of hepcidin and reduced markers of hepatic BMP signaling, demonstrating an essential role for neogenin in iron homeostasis.18 In addition, primary cultured hepatocytes derived from neogenin mutant mice show an impaired ability to induce SMAD phosphorylation and hepcidin transcription in response to treatment with the hepcidin-inducing ligand BMP2. Interestingly, neogenin has been shown to interact with HJV. However, the functional relationship between neogenin and HJV and the mechanism by which neogenin mutation leads to hepcidin repression are currently unclear.
Negative regulation of hepatic BMP signaling
Whereas reducing the availability of the BMP ligand represents one means of modulating hepcidin expression by the liver, additional mechanisms have been identified that have the capacity to inhibit hepcidin signaling through the BMP pathway in vitro or in vivo. In addition to its membrane-bound form, HJV appears to exist as circulating form in plasma (soluble HJV) that results from proteolytic processing by a proprotein convertase such as furin. Several studies have suggested a model in which soluble HJV acts as a decoy receptor that sequesters BMPs, thereby suppressing hepcidin production by reducing hepatic signaling through the BMP pathway (reviewed in Corradini4 ). In vitro, the proprotein convertase–mediated cleavage of HJV is increased by iron deficiency and hypoxia.19 Because HJV is expressed not only in the liver but also at high levels in skeletal muscle, the release of soluble HJV from skeletal muscle into the circulation has been proposed as a mechanism to suppress hepatic hepcidin expression and thus increase iron availability for muscle needs during differentiation and periods of hypoxia resulting from physical activity. However, mice in which the Hjv gene was selectively disrupted in skeletal muscle showed no overt disruption of iron homeostasis, whereas liver-specific disruption of Hjv recapitulated the iron overload phenotype exhibited by mice with global Hjv deletion.20 The extrahepatic functions of HJV and the potential role of the soluble form remain unknown.
TMPRSS6 (also known as matriptase-2), a transmembrane serine protease produced by the liver, appears to function as a key negative regulator of hepatic BMP signaling. The key role of TMPRSS6 in hepcidin suppression was revealed by the phenotype of a chemically induced Tmprss6 mouse mutant exhibiting iron deficiency anemia and increased hepatic hepcidin mRNA.21 This finding facilitated the identification of TMPRSS6 mutations as a genetic cause of IRIDA in humans. In vitro, TMPRSS6 has been shown to cleave HJV from the plasma membrane22 and, compatible with this result, mice with genetic loss of Tmprss6 show up-regulated BMP signaling that is dependent upon the presence of HJV.23,24 The cleaved form of HJV generated by TMPRSS6 is distinct from that generated by proprotein convertase activity.25 Interestingly, TMPRSS6 protein levels in rat liver increase in response to acute iron deficiency,26 and in hepatoma cells, hypoxia increases TMPRSS6 expression and results in a concurrent decrease in membrane-bound HJV.27 The mechanisms controlling TMPRSS6 activity in vivo require further study.
Another molecule that may negatively regulate hepatic BMP signaling for hepcidin production is SMAD7, an inhibitory SMAD protein that is induced by TGF-β signaling. SMAD7 appears to antagonize TGF-β signaling by acting in a negative-feedback loop. Similar to the expression of hepcidin and Bmp6, hepatic mRNA levels of Smad7 increase with chronic dietary iron loading.8 In primary murine hepatocytes, SMAD7 expression is induced by BMP6, whereas SMAD7 overexpression greatly reduces hepcidin mRNA levels and abolishes the hepcidin response to BMP6.28 These findings suggest that SMAD7 may function to provide negative feedback to dampen BMP signaling for hepcidin production. The relative contribution of SMAD7 activity to hepcidin regulation in vivo has yet to be established.
Hepcidin regulation by inflammation
Excessive hepcidin production may occur in several acquired disease states, including infections, malignancies, and inflammatory conditions. Hepcidin elevation leads to impaired utilization of iron from macrophage stores, reduced absorption of dietary iron, and a resulting hypoferremic state that impairs the delivery of iron to maturing erythroid cells in the BM. This process contributes to the anemia of chronic disease (also known as the anemia of inflammation), an anemia that is usually normochromic and normocytic and that is associated with erythropoietin hyporesponsiveness, reduced proliferation of erythroid precursors, and decreased red blood cell survival. Whereas several inflammatory stimuli have been identified that induce hepcidin expression, the mechanism by which such stimuli promote hepcidin expression has been most clearly defined for IL-6. Several studies have demonstrated that IL-6 activates hepcidin expression through the JAK/STAT signaling cascade, which induces binding of the transcriptional activator STAT3 to specific target elements in the hepcidin promoter (reviewed in Hentze1 ). The ability of other inflammatory agents, such as lipopolysaccharide (LPS), IL-1, and TNF-α, to promote hepcidin expression may reflect their IL-6–inducing properties. It has been hypothesized that hepcidin induction by these stimuli may reflect an evolutionary response that limits iron availability to pathogens.
Several studies have suggested that hepcidin signaling in inflammation may require cooperative activity of the BMP pathway. For example, in mice with liver-specific deletion of Smad4, the ability of IL-6 injection to induce hepatic hepcidin expression is lost.5 In mice that were pretreated with a small-molecule inhibitor of BMP signaling, the ability of IL-6 to induce hepcidin production was reduced.29 However, Bmp6-knockout mice retain an ability to increase hepatic hepcidin expression after IL-6 injection.7 The exact mechanism by which hepcidin signaling pathways in inflammation interact with the BMP pathway remains to be clarified.
Endoplasmic reticulum (ER) stress, a cellular state arising from the accumulation of improperly folded proteins, also induces hepcidin expression using a signaling program known as the unfolded protein response (UPR). The UPR is also activated by LPS and pro-inflammatory cytokines such as IL-6 and IL-1β, suggesting that hepcidin induction by ER stress may cooperate with JAK/STAT signaling responses during inflammation. Treatment of a hepatoma cell line with chemical agents that promote ER stress by interfering with disulfide bond formation was found to modulate hepcidin expression. Later phases of the UPR were associated with a decline in levels of the transcription factor CHOP (CCAAT/enhancer-binding protein [C/EBP] homologous protein), which acts to negatively regulate the activity of C/EBPα, another nuclear factor known to bind to specific elements in the hepcidin promoter to increase transcription.30 CREBH (cyclic AMP response element–binding protein H), a transcription factor that is activated by ER stress, inflammatory cytokines, and LPS to promote the expression of acute-phase response genes, has also been shown to activate the hepcidin promoter in vitro. The physiological relevance of this induction was revealed in Crebh-knockout mice, which fail to elevate their hepatic hepcidin expression to wild-type levels after treatment with an ER-stressing agent or LPS.31
Hepcidin regulation by erythropoiesis
Hepcidin production is suppressed under conditions of increased erythropoietic drive. Proposed mediators of this hepcidin suppression include hypoxia and circulating factors secreted by erythroid precursors in the BM. Hepcidin suppression is a prominent feature of congenital iron-loading anemias that are characterized by ineffective erythropoiesis, such as β-thalassemia. In these disorders, hepcidin levels remain inappropriately low relative to increased body iron stores, perpetuating intestinal iron absorption in the face of systemic iron overload.
Two products secreted by erythroid precursors that may contribute to the hepcidin suppression observed in iron-loading anemias are the TGF-β superfamily members GDF15 (growth differentiation factor-15) and TWSG1 (twisted gastrulation protein-1). In an in vitro model of human erythropoiesis, GDF15 and TWSG1 mRNA levels were up-regulated during erythroblast maturation.32 GDF15 protein levels have been found to be elevated in serum of individuals with β-thalassemia, refractory anemia with ringed sideroblasts, and congenital dyserythropoietic anemia types I and II. GDF15 treatment was shown to have some capacity to mediate hepcidin suppression in hepatoma cells and primary human hepatocytes, although the mechanism mediating the hepcidin suppression was not demonstrated. TWSG1 expression was increased in a mouse model of β-thalassemia. TWSG1 may function as an inhibitor of hepatic BMP signaling, because TWSG1 was shown to inhibit the ability of BMP2 and BMP4 to induce hepcidin expression in hepatoma cells and primary human hepatocytes.33 The relative contribution of GDF15 and TWSG1 to hepcidin suppression in vivo remains to be clarified. In addition, it remains unclear whether either molecule contributes to hepcidin regulation outside of the pathological setting of ineffective erythropoiesis.
Hepcidin expression is also modulated by hypoxia, an effect that was originally demonstrated in hepatoma cells cultured under hypoxic conditions and in mice housed in hypobaric chambers. The ability of hypoxia to influence hepcidin expression may reflect its ability to induce erythropoietin production by the kidney and thus promote erythropoietic activity. Indeed, healthy humans studied under normoxic conditions showed a marked reduction in their urinary hepcidin levels within 48 hours after administration of recombinant erythropoietin.34 In animal models, the ability of erythropoietin administration to decrease hepcidin expression is blocked by administration of cytotoxic agents that suppress BM activity, suggesting that hepcidin suppression in response to erythropoietin is dependent upon erythropoietic activity. In a cohort of healthy humans studied at sea level and after acute and chronic exposure to high altitude, acute hypoxia exposure caused a rapid and marked increase in serum erythropoietin; serum hepcidin was reduced by acute hypoxia exposure and showed a further decrease after several days of hypoxic exposure.35 Interestingly, serum levels of hepcidin and ferritin were correlated at all time points in this study, suggesting that the suppression of hepcidin observed may have been mediated by either iron itself or by the kinetics of iron utilization in response to hypoxia.
Hypoxia may also exert a local effect on hepcidin production in the liver through the actions of hypoxia inducible factors (HIFs), heterodimeric transcription factors that promote the expression of genes that mediate responses to hypoxia. Under normoxic conditions, the regulatory α-subunit of HIF is hydroxylated by prolyl-hydroxylases, which triggers its degradation through the ubiquitin-proteosome pathway. However, under conditions of hypoxia or iron depletion, hydroxylation of the α-subunit is inhibited, leading to its stabilization, which facilitates HIF-mediated transcriptional responses. Accordingly, one study found that hepatic levels of the HIF-1α isoform increased in mice fed an iron-deficient diet. The investigators demonstrated a requirement for HIF-1α to achieve the full degree of hepcidin suppression induced by a low-iron diet, and they showed that HIF-1α could bind to the hepcidin promoter in mouse liver.36 However, others have found no evidence for dysregulated systemic iron homeostasis in mice lacking HIF-1α in the liver,37 and the ability of HIF-1 to directly bind the hepcidin promoter in vivo has been debated.38 As described above, it is possible that hypoxia may affect hepcidin expression locally in the liver through other mechanisms, such as by increasing levels of soluble HJV19 and/or by increasing the expression of TMPRSS6.27
Influence of cellular regulators on systemic iron balance
Whereas the central role of the hepcidin-ferroportin axis in the regulation of systemic iron balance is well established, distinct mechanisms that regulate iron balance at the cellular level have also been identified. Recent studies have suggested that these cellular and systemic pathways interact to influence iron balance.1 For example, mice harboring an intestinal-specific deletion of the HIF-2α subunit were found to exhibit decreased serum and liver iron levels, even though their hepatic hepcidin expression was markedly decreased.39 HIF-2α was shown to directly bind to the promoter of DMT1 (divalent metal transporter 1), which encodes the principal apical transporter than imports ferrous iron into enterocytes.37,39 Conversely, mice harboring an intestine-specific deletion of the H-subunit of the iron-storage protein ferritin showed increased body iron stores even though their hepcidin expression by the liver was induced.40 These findings demonstrate that under certain circumstances, hepatic regulation of hepcidin expression cannot fully compensate for defects in cellular iron balance in enterocytes to maintain normal systemic iron balance.
The recent finding that hepcidin expression can be modulated at the cellular level by an miRNA adds another layer of complexity to the regulation of systemic iron balance. miRNAs are a class of short, noncoding RNAs that bind to complementary sequences on target messenger RNA transcripts to negatively regulate gene expression at the posttranscriptional level. In mice, injection of an antagonist of the liver-specific miR-122 was found to induce systemic iron deficiency and increase levels of the mRNAs encoding hepcidin, HFE, and HJV. In vitro studies suggested that miR-122 may bind to specific sites in the 3′-untranslated region of the Hfe and Hjv mRNAs.41 Because antagonism of miR-122 did not alter downstream markers of BMP-signaling responses, the mechanism by which modulation of miR-122 alters hepcidin mRNA levels remains uncertain.
Conclusion
Production of the iron-regulatory hormone hepcidin is regulated in response to several physiological stimuli, including iron loading, inflammation, and erythropoiesis. By altering hepcidin levels, these stimuli modulate iron availability for erythropoiesis and other physiological processes and regulate the level of body iron stores. The collective work of many investigators has begun to unravel the complexity of the molecular mechanisms mediating these hepcidin responses. However, many key questions regarding these mechanisms of hepcidin regulation remain to be addressed, and additional gene products that participate in hepcidin regulation are likely to be identified in the future. By expanding our knowledge of iron physiology, continued investigation of the molecular mechanisms regulating systemic iron balance may direct the development of new diagnostic tests and targeted molecular therapies that may improve therapeutic outcomes for patients with iron disorders.
Acknowledgments
This work was supported in part by National Institutes of Health grant K08 DK084204 from the National Institute of Diabetes and Digestive and Kidney Diseases. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Institute of Diabetes and Digestive and Kidney Diseases or the National Institutes of Health.
Disclosures
Conflict-of-interest disclosure: The author declares no competing financial interests. Off-label drug use: None disclosed.
Correspondence
Karin E. Finberg, Medical Instructor in Pathology, Duke University Medical Center, Box 3712, Durham, NC 27710; Phone: (919) 668-0992; Fax: (919) 681-1394; e-mail: karin.finberg@duke.edu.