Abstract
The genetics of acute lymphoblastic leukemia are becoming well understood and the incidence of individual chromosomal abnormalities varies considerably with age. Cytogenetics provide reliable risk stratification for treatment: high hyperdiploidy and ETV6-RUNX1 are good risk, whereas BCR-ABL1, MLL rearrangements, and hypodiploidy are poor risk. Nevertheless, some patients within the good- and intermediate-risk groups will unpredictably relapse. With advancing technologies in array-based approaches (single nucleotide polymorphism arrays) and next-generation sequencing to study the genome, increasing numbers of new genetic changes are being discovered. These include deletions of B-cell differentiation and cell cycle control genes, as well as mutations of genes in key signaling pathways. Their associations and interactions with established cytogenetic subgroups and with each other are becoming elucidated. Whether they have a link to outcome is the most important factor for refinement of risk factors in relation to clinical trials. For several newly identified abnormalities, including intrachromosomal amplification of chromosome 21 (iAMP21), that are associated with a poor prognosis with standard therapy, appropriately modified treatment has significantly improved outcome. After the successful use of tyrosine kinase inhibitors in the treatment of BCR-ABL1–positive acute lymphoblastic leukemia, patients with alternative ABL1 translocations and rearrangements involving PDGFRB may benefit from treatment with tyrosine kinase inhibitors. Other aberrations, for example, CRLF2 overexpression and JAK2 mutations, are also providing potential novel therapeutic targets with the prospect of reduced toxicity.
Background
Acute lymphoblastic leukemia (ALL) is the most common cancer in children, but is rare in adolescents and adults. Significant advances have been made in the successful treatment of ALL, with an overall survival rate of 85% in children.1 Although outcome has improved in adults, it remains inferior to that in children, with a long-term survival of only 45%.2 These advances have emerged from developments in treatment regimens and the introduction of risk stratification to modulate the intensity of therapy based on the risk of treatment failure. The important risk factors used in stratification for treatment include age and WBC count, which indicate the National Cancer Institute (NCI) risk status, and cytogenetics, as well as treatment response measured by the level of minimal residual disease. However, these features fail to accurately detect all patients who go on to relapse and no new drugs have been introduced into ALL therapy in recent years, so survival improvements on current therapies are reaching their limits. It is also a major consideration that the drugs used are highly toxic, often inducing severe acute and late side effects, with some patients being refractory to treatment. Therefore, to achieve the goal of curing all patients with ALL and reducing toxicity, there is a need for new therapies to target the underlying molecular pathology of the disease, which forms the crux of leukemia research at this time. The focus of this review is B-cell precursor ALL (BCP-ALL). The article highlights those cytogenetic and genetic changes that have affected treatment decisions to date and explores the potential of novel therapies emerging from the current genomic landscape of BCP-ALL.
Cytogenetic subgroups and outcome
Good-risk cytogenetic abnormalities
The acquired chromosomal abnormalities occurring in BCP-ALL are well understood. and there are strong associations between different cytogenetic subgroups and outcome that are used for risk stratification on most treatment protocols.3,4 The distribution of the commonly occurring established cytogenetic subgroups within childhood BCP-ALL is indicated in Figure 1A. High hyperdiploidy (51-65 chromosomes) and t(12;21)(p13;q22)/ETV6-RUNX1 (seen almost exclusively in young children) are good-risk subgroups.3 Routinely, patients with these abnormalities are treated on standard-risk protocols. Because of their excellent outcome, for ETV6-RUNX1–positive patients with good-risk clinical features (NCI standard risk: < 10 years old with a WBC count < 50 × 109/L), therapy reductions are under consideration.
Distribution of cytogenetic abnormalities from data collected from UK childhood ALL treatment trials. (A) Relative incidence of the established cytogenetic abnormalities among childhood BCP-ALL. (B) Distribution of different cytogenetic subgroups according to age. The abnormalities are color coded and the incidence of each abnormality indicated according to age group. The relative incidence of T-ALL is also shown. IGH@ indicates all translocations except with CRLF2; CRLF2, IGH@-CRLF2 and P2RY8-CRLF2; hap/hypo, hypodiploidy (< 44 chromosomes); t(9;22), BCR-ABL1 positive; and HeH, high hyperdiploidy.
Distribution of cytogenetic abnormalities from data collected from UK childhood ALL treatment trials. (A) Relative incidence of the established cytogenetic abnormalities among childhood BCP-ALL. (B) Distribution of different cytogenetic subgroups according to age. The abnormalities are color coded and the incidence of each abnormality indicated according to age group. The relative incidence of T-ALL is also shown. IGH@ indicates all translocations except with CRLF2; CRLF2, IGH@-CRLF2 and P2RY8-CRLF2; hap/hypo, hypodiploidy (< 44 chromosomes); t(9;22), BCR-ABL1 positive; and HeH, high hyperdiploidy.
Established poor-risk cytogenetic abnormalities
Those abnormalities associated with a high risk of relapse are the Philadelphia chromosome (Ph) translocation, t(9;22)(q34;q11)/BCR-ABL1, rearrangements of the MLL gene at 11q23, and hypodiploidy of less than 44 chromosomes, including both near haploidy (< 30 chromosomes) and low hypodiploidy (30-39 chromosomes). In early studies, the translocation t(1;19)(q23;p13)/TCF3-PBX1 was associated with a poor outcome (for review, see Moorman5 ). This opinion has been moderated by the more aggressive therapy of modern protocols. However, the rare variant t(17;19)(q22;p13)/TCF3-HLF continues to be associated with a dismal outcome, with all reported patients being known to have relapsed while on therapy and subsequently dying.5 In adults, karyotype complexity comprising 5 or more abnormalities is an additional high-risk abnormality.5
More recently identified poor-risk abnormalities
iAMP21.
iAMP21 is an intriguing chromosomal aberration characterized by gross chromosomal instability of chromosome 21. It manifests as a highly abnormal chromosome 21, with considerable structural variation between patients as shown by cytogenetics and single-nucleotide polymorphism arrays.6 Despite this heterogeneity, iAMP21 defines a distinct cytogenetic subgroup of older children (median age, 9 years; Figure 1B) with BCP-ALL. Strikingly, it is associated with a dismal outcome, with a high risk of both early and late relapses, when patients are treated as standard risk. Outcome is dramatically improved when these patients are treated on high-risk regimens (Figure 2A).7
Kaplan-Meier curves showing survival for different cytogenetic abnormalities. (A) Event-free survival of children with iAMP21 treated as standard risk on the UK childhood ALL trial ALL97/99 (n = 28) compared with those treated as high risk on ALL2003 (n = 53). (B) Event-free survival of childhood BCP-ALL patients from ALL97/99 according to cytogenetic risk groups (adapted from Moorman et al3 ). Good risk includes high hyperdiploidy and ETV6-RUNX1; poor risk includes t(9;22)(q34;q11.2), iAMP21, MLL translocations, near haploidy, low hypodiploidy, t(17;19)(q23;p13), abnormal 17p, and loss of 13q; intermediate risk includes normal karyotype and all other abnormalities. (C) Overall survival of adults with ALL treated on the adult trial UKALLXII/ECOG2993 (adapted from Moorman et al5 ). Percentages are overall survival at 7 years. High-risk abnormalities include: t(4;11)(q21;q23), IGH@ translocations, CRLF2 rearrangements, low hypodiploidy/near-triploid, and complex karyotype. Philadelphia positive/t(9;22)(q34;q11) patients were sufficiently prevalent to be shown separately. Standard risk includes all other abnormalities. The improved outcome of high hyperdiploidy is indicated separately.
Kaplan-Meier curves showing survival for different cytogenetic abnormalities. (A) Event-free survival of children with iAMP21 treated as standard risk on the UK childhood ALL trial ALL97/99 (n = 28) compared with those treated as high risk on ALL2003 (n = 53). (B) Event-free survival of childhood BCP-ALL patients from ALL97/99 according to cytogenetic risk groups (adapted from Moorman et al3 ). Good risk includes high hyperdiploidy and ETV6-RUNX1; poor risk includes t(9;22)(q34;q11.2), iAMP21, MLL translocations, near haploidy, low hypodiploidy, t(17;19)(q23;p13), abnormal 17p, and loss of 13q; intermediate risk includes normal karyotype and all other abnormalities. (C) Overall survival of adults with ALL treated on the adult trial UKALLXII/ECOG2993 (adapted from Moorman et al5 ). Percentages are overall survival at 7 years. High-risk abnormalities include: t(4;11)(q21;q23), IGH@ translocations, CRLF2 rearrangements, low hypodiploidy/near-triploid, and complex karyotype. Philadelphia positive/t(9;22)(q34;q11) patients were sufficiently prevalent to be shown separately. Standard risk includes all other abnormalities. The improved outcome of high hyperdiploidy is indicated separately.
IGH@ translocations.
A range of genes have been reported as translocation partners of the immunoglobulin heavy chain locus IGH@, including the gene encoding cytokine receptor-like factor 2 (CRLF2, also known as thymic stromal lymphopoietin receptor, TLSPR), the CCAAT enhancer-binding protein (CEBP) family of transcription factors, the DNA-binding protein inhibitor ID4, the cytokine receptor for erythropoietin (EPOR), and the microRNA miR-125b.8 Although, currently, there appears to be no functional link between these partners, IGH@ translocations predominate in adolescents and young adults (Figure 1B) and are associated with an inferior survival.4
Association of chromosomal abnormalities with age
Although virtually all chromosomal abnormalities occur in both childhood and adult ALL, as indicated for some of the specific abnormalities in the above section, there is a significant difference in incidence of most cytogenetic subgroups according to age, as shown in Figure 1B. For example, MLL rearrangements [including t(4;11)(q21;q23) and other MLL rearrangements] dominate in infants up to 1 year of age; ETV6-RUNX1 and high hyperdiploidy predominate in young children, with very few ETV6-RUNX1 adult cases, whereas the incidence of BCR-ABL1 increases dramatically with age.
Additional genetic aberrations
CRLF2
The majority of chromosomal abnormalities described in “Cytogenetic subgroups and outcome” are considered to be primary genetic changes driving leukemia initiation, with additional genetic events required for the development of overt ALL. Genomic studies and next-generation sequencing (targeted and genome wide) have revealed the presence of many additional genetic abnormalities. In BCP-ALL, rearrangements involving CRLF2 are found in ∼6% of childhood and adult BCP-ALL,4,9 although they are more prevalent in Down syndrome (54%). In addition to the translocation IGH@-CRLF2, deletions within the pseudoautosomal region (PAR1) of the sex chromosomes results in the fusion, P2RY8-CRLF2. Both rearrangements link the full-length coding sequence of CRLF2 to alternative transcriptional control, either through the IGH@ enhancer or the P2RY8 promoter, resulting in overexpression of CRLF2.10,11 Interestingly, CRLF2 overexpression sometimes occurs in the absence of these rearrangements and the rare activating mutations of CRLF2, F232C substitution in the juxtamembrane region.12 The prognosis associated with CRLF2 rearrangements is protocol dependent,4,9,13 likely reflecting the range of cohorts studied, that many have not incorporated comprehensive analysis of CRLF2 expression, and that the presence of associated genetic alterations has not been considered. Approximately 40% of CRLF2-rearranged BCP-ALL harbor activating mutations of JAK1 and JAK2, primarily at residue R683 in the pseudokinase domain of JAK2. Both CRLF2 overexpression and JAK mutations result in constitutive activation of the JAK-STAT pathway.
IKZF1 and other key deletions
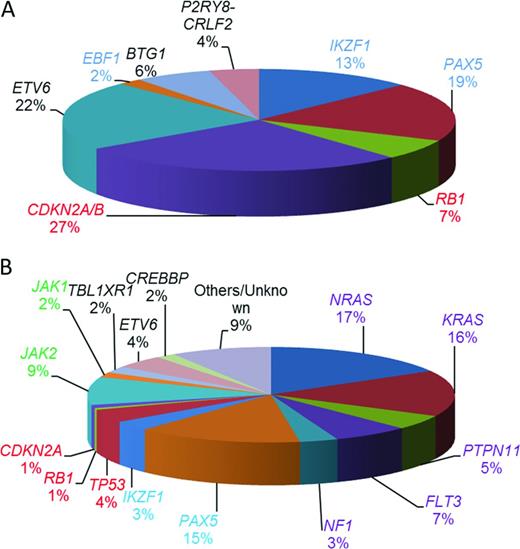
Other recurrent copy number abnormalities include focal gene deletions within transcription factors important in B-cell development and differentiation and cell cycle regulators.14 Those most frequently involved in BCP-ALL include: PAX5, IKZF1, EBF1, RB1, CDKN2A/B, ETV6, and BTG1. Their relative incidence is indicated in Figure 3A. IKZF1 encodes the transcription factor IKAROS. It is of clinical interest because IKZF1 deletions are the only copy number changes that have universally been associated with an inferior outcome. They may either remove the entire gene, often resulting from large deletions of the short arm of chromosome 7, leading to haploinsufficiency and reduced Ikaros protein levels, or they are intragenic. The most common intragenic deletion includes exons 4 to 7, leading to in-frame splicing of exon 3 to exon 8 and the production of the Ik6 isoform with dominant-negative activity. Other types of intragenic deletions also occur, the functional consequences of which are less clear.
Estimated relative incidences of key copy number changes and mutations in childhood BCP-ALL (not mutually exclusive). (A) Relative incidences of 1363 deletions of genes commonly deleted in BCP-ALL calculated from multiplex ligation–dependent probe amplification analysis of 1427 BCP-ALL patients entered into UK childhood ALL treatment trials.22 (B) Estimated distribution of mutations in the common pathways among high-risk childhood BCP-ALL patients.17 Shown are the coding regions and untranslated regions of 125 genes sequenced in 187 high-risk childhood BCP-ALL in the Children's Oncology Group trial COG P9906. Those genes involved in B-cell development and differentiation are color coded in blue and those involved in cell cycle regulation are color coded in red. Mutations affecting the RAS signaling pathway are purple and those affecting the JAKs are green. The transcriptional regulators and others are color coded in black.
Estimated relative incidences of key copy number changes and mutations in childhood BCP-ALL (not mutually exclusive). (A) Relative incidences of 1363 deletions of genes commonly deleted in BCP-ALL calculated from multiplex ligation–dependent probe amplification analysis of 1427 BCP-ALL patients entered into UK childhood ALL treatment trials.22 (B) Estimated distribution of mutations in the common pathways among high-risk childhood BCP-ALL patients.17 Shown are the coding regions and untranslated regions of 125 genes sequenced in 187 high-risk childhood BCP-ALL in the Children's Oncology Group trial COG P9906. Those genes involved in B-cell development and differentiation are color coded in blue and those involved in cell cycle regulation are color coded in red. Mutations affecting the RAS signaling pathway are purple and those affecting the JAKs are green. The transcriptional regulators and others are color coded in black.
Key sequence mutations
Recurrent sequence mutations have also been identified within genes involved in different signaling pathways, including RAS signaling, B-cell development and differentiation, and cell cycle control17 (Figure 3B). Sequence mutations or deletions in the TP53,18 CREBBP,19,20 and NT5C222,23 genes are associated with relapse. Strikingly, CREBBP mutations are present in ∼ 20% of relapsed BCP-ALL and specifically in > 60% of high-hyperdiploid patients at relapse.20 Several CREBBP and NT5C2 mutations were detected in small subclones at diagnosis and were highly predictive of relapse. These observations suggested clonal evolution and expansion of the minor subclones under the selective pressure of cytotoxic treatment that conferred resistance to therapy.19,21
Associations between abnormalities
We are beginning to understand the associations between these spectra of abnormalities. For example, we have shown the differing nature and distribution of copy number changes according to cytogenetic subtype in a large cohort of childhood ALL.22 The most striking findings were the well-known association of ETV6 deletions and ETV6-RUNX1, as well as the higher incidence of copy number changes overall in this subgroup compared with the other good-risk group: high hyperdiploidy. Among the poor-risk subgroups, MLL-rearranged patients showed a small number of changes compared with the BCR-ABL1 subgroup, with frequent deletions of CDKN2A/B, PAX5, and IKZF1, specifically exons 4 to 7. These findings indicate the more aggressive nature of the MLL translocation in ALL, requiring fewer additional genetic alterations for induction of leukemogenesis than the BCR-ABL1 and ETV6-RUNX1 subtypes. The iAMP21 patients demonstrated a characteristic profile, including a high proportion of P2RY8-CRLF2 and deletions of RB1.
It is also known that Ras/receptor tyrosine kinase (RTK) pathway mutations are common among patients with high hyperdiploid, MLL-rearranged, and near-haploid karyotypes23-26 ; low hypodiploidy is characterized by alterations of TP53,26 whereas JAK2 mutations are strongly associated with CRLF2 rearrangements and IKZF1 deletions in high-risk patients.27
BCR-ABL1–like ALL
An unusual yet specific genetic subgroup accounting for ∼ 15% of BCP-ALL has been identified from gene expression profiling. It is known as BCR-ABL1–like or Ph-like ALL, so called because it shows a similar gene expression signature to BCR-ABL1–positive ALL and shares the same high risk of relapse and poor outcome.14,28 A characteristic genomic landscape is emerging to facilitate the identification BCR-ABL1–like ALL in the many study groups for which gene expression profiling is not available. They are generally mutually exclusive of the common established cytogenetic subgroups and have a high incidence of IKZF1 deletions, deregulated CRLF2, and JAK mutations.29 Recently, using mRNA-seq and a range of next-generation sequencing and molecular techniques, patients with this expression signature have been shown to be characterized by genetic alterations, including rearrangements, mutations, and deletions of a range of kinase and cytokine receptors. These include ABL1 translocations (eg, the NUP214-ABL1 fusion, previously shown to be amplified in T-ALL); translocations involving EPOR, JAK2, PDGFRB, and EBF1; and mutations of FLT3, IL7R, and SH2B3 (a negative regulator of JAK2 signaling).30 Among high-risk cohorts from the Children's Oncology Group, 15% to 20% of patients were classified as BCR-ABL1–like according to their gene expression signature. In total, ∼ 50% had CRLF2 high expression, of which ∼ 30% had JAK2 mutations. The remainder had ABL1, JAK2, PDGFRB, and other kinase rearrangements and sequence mutations. Largely, these genes facilitate leukemic transformation by inducing constitutive kinase activation and signaling through the activation of the ABL1 and/or JAK-STAT pathways.
Treatment beyond BCR-ABL1
Conventional therapy
Childhood patients with high-risk chromosomal abnormalities, whether they are NCI high (> 10 years old or WBC count > 50 × 109/L) or NCI standard risk (< 10 years old and WBC count < 50 × 109/L) are currently recommended for intensive treatment on contemporary protocols. All high-risk adult patients with a suitable donor are typically recommended for allogeneic BM transplantation in first remission.
BCR-ABL1 and other ABL1 translocations
BCR-ABL1–positive patients are now treated with tyrosine kinase inhibitors (TKIs) in addition to conventional chemotherapy in an attempt, particularly for childhood patients, to avoid BM transplantation. BCR-ABL1–positive leukemia represents the paradigm of how understanding the molecular pathogenesis led to major treatment improvements. The formation of the BCR-ABL tyrosine kinase is the critical pathogenetic event in chronic myeloid leukemia, which proved to be an ideal target for therapy in clinical trials of TKI. It was shown that imatinib significantly improved the long-term survival of patients with chronic myeloid leukemia. The advent of next-generation TKIs (eg, dasatinib and nilotinib) has revolutionized the treatment of all BCR-ABL1–positive diseases.31 Reports from pediatric and adult ALL trials using TKIs show promising results, with outcomes substantially higher than those achieved with traditional chemotherapy and stem cell transplantation, potentially reducing the number of patients requiring transplantation.32,33 Because resistance is a clinically significant issue for these patients, many groups are working on third- and fourth-generation TKIs to overcome or minimize the development of resistance; alternative clinical trial designs such as the sequential use of different TKIs, may reduce this problem.
In the United States, a collaborative project involving clinical and research investigators known as TARGET (Therapeutically Applicable Research to Generate Effective Treatments; www.target.gov) has focused on applying high-throughput genomics and sequencing to identify genes and pathways that are consistently altered in high-risk ALL, which may act as novel targets for therapy; for example the BCR-ABL1-like group described on the previous page. Evidence has emerged from such studies that patients with activated kinase signatures among the BCR-ABL1–like patient group may also benefit from treatment with TKIs. For example, 3 cases of the NUP214-ABL1 fusion with active ABL1 signaling as seen in T-ALL have now been reported in BCP-ALL.30,34 An exciting observation was that the transformation induced by these alterations was attenuated by several TKIs in primary samples from 2 of these patients, demonstrated by a reduction in the basal level of substrate phosphorylation determined by phosphoflow cytometry.30 In addition, a NUP214-ABL1 ALL xenograft model of one of these samples responded to dasatinib. However, primary cells from the third case showed no change in viability or levels of apoptosis upon treatment with a range of TKIs.34 It is noteworthy that there are also case reports documenting mixed clinical responses to TKIs in NUP214-ABL1 T-ALL. Differences in response in both B- and T-ALL may result from variable NUP214-ABL1 copy number or the nature of cooperating mutations. We have reported a cohort of patients with ZMIZ1-ABL1 fusion with a good response to conventional therapy.5 It is interesting to speculate whether TKI treatment for these patients would have further improved outcome.
PDGFRB rearrangements
Patients with myeloproliferative disorders and PDGFRB rearrangements show complete hematological and molecular response to imatinib. Therefore, based on the evidence that TKI treatment is effective for all BCR-ABL1–positive diseases, it is likely that patients with rearrangements involving PDGFRB, found among the BCR-ABL1–like cohort, will respond to TKI therapy. In our experience, some patients with PDGFRB rearrangements were refractory to conventional therapy, and a recent report and 2 as yet unpublished cases have shown a complete response to TKIs in a BCP-ALL patient with a PDGFRB rearrangement (EBF1-PDGFRB) who was refractory to conventional induction chemotherapy.35 These observations indicate that screening at diagnosis to identify those BCR-ABL1–like patients harboring kinase-activating alterations who may benefit from treatment with TKIs is a feasible proposition for improving the outcome for this high-risk group of patients.
JAK2 and CRLF2 rearrangements
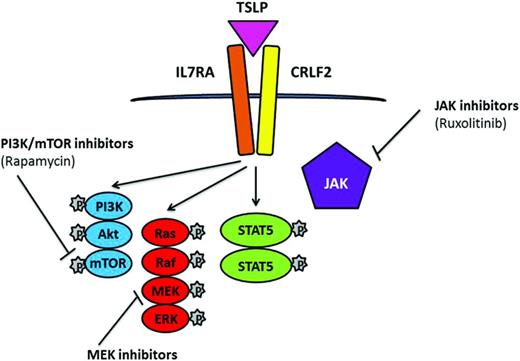
CRLF2 rearrangements are strongly associated with JAK2 mutations. JAK2 translocations, an activating mutation of the cytokine receptor IL7 (IL7R), deletion of SH2B3, and CRLF2 rearrangements have been reported within the BCR-ABL1–like cohort.30 Similar to the NUP214-ABL1 cases, successful response was achieved from direct treatment of cells and xenograft models of these cases with JAK2 translocations by the JAK2 inhibitor ruxolitinib,30 a drug currently undergoing clinical trials. In addition to JAK-STAT, there is evidence of aberrant mTOR/PI3K signaling in ALL with rearranged CRLF2; therefore, combinations of the JAK and PI3K inhibitors ruxolitinib and rapamycin have potential in CRLF2-rearranged and JAK-mutated disease36 (Figure 4).
Simplified representation of signaling pathways associated with CRLF2 rearrangements and JAK mutations in which TSLP induces phosphorylation of STAT5, PI3K, and ERK. Potential therapeutic inhibitors and their targets are indicated.
Simplified representation of signaling pathways associated with CRLF2 rearrangements and JAK mutations in which TSLP induces phosphorylation of STAT5, PI3K, and ERK. Potential therapeutic inhibitors and their targets are indicated.
MLL rearrangements
The histone methyltransferase DOT1L is required for the development and maintenance of MLL-rearranged leukemia.37 A selective inhibitor of DOT1L, EPZ004777, demonstrated selective killing of MLL-rearranged cells in vitro and in a mouse MLL xenograft model. These results provided compelling support for DOT1L inhibition as a basis for targeted therapy against MLL translocations, for which clinical trials with the DOT1L inhibitor EPZ5676 are currently ongoing (www.clinicaltrials.gov identifier NCT01684150).
There is extensive evidence demonstrating the overexpression of wild-type FLT3 in MLL-rearranged infant ALL.38 Because FLT3 inhibitors such as lestaurtinib (CEP-701) and PKC412 selectively kill MLL-rearranged cells, they have shown potential value in the treatment of these infants and have recently been incorporated into clinical trials on the backbone of intensive therapy by the Children's Oncology Group.
CREBBP mutations
CREBBP mutations usually occur within the histone acetyltransferase domain of CREBBP, they may respond to treatment with histone deacetylase inhibitors, particularly at relapse.
Hypodiploidy
Near-haploid and low-hypodiploid patient subgroups demonstrate activation of RAS- and PI3K-signaling pathways and are sensitive to PI3K inhibitors,26 indicating that these drugs may represent a potential therapeutic strategy for this aggressive form of leukemia, as well as other patient subgroups with a high incidence of Ras/RTK pathway mutations, including MLL-rearranged and hyperdiploidy. The use of MAPK-ERK kinase (MEK) inhibitors has been indicated as potential targets for the Ras/Raf/MEK/ERK signaling pathway, which occurs as a consequence of RAS mutations in ALL24 (Figure 4).
Candidate genes identified by deletions
Deletion of genetic material is usually regarded as an impossible target for therapy due to loss rather than gain of function. Understanding the downstream pathways and the respective genes that are deregulated by the loss of function may provide the missing targets for therapeutic intervention. For example, recent insights into IKZF1 biology indicate that IKZF1 deletions may represent good candidates because they occur in ∼ 14% of BCP-ALL cases and are linked to poor outcome. However, one confounding factor is that patients may harbor multiple unique IKZF1 deletions in small subclones,39 which in turn increase the number of potential downstream targets, making elimination of all abnormal populations more difficult.
Important considerations
Although it is evident that much progress is being made in the development of novel targeted therapies for the treatment of high-risk and resistant ALL, as summarized in Table 1, the solutions are not always straightforward. One major consideration resides in the level of clonal heterogeneity and complex diversity of genetic changes, which have been identified within the leukemic cells of patients with ALL.40 To be effective, the driver aberrations need to be druggable, which does not apply to many newly discovered aberrations. They also need to be targeted, which, based on evidence from studies of diagnostic and relapse samples from the same patients, may not always represent the major clone at diagnosis with abnormalities present at a low level at diagnosis conferring resistance emerging at relapse. Therefore, there is an increasing need for highly sensitive detection of these relapse driving mutations at the time of diagnosis, as well as a complete understanding of the interplay between cooccurring genetic abnormalities. The continued development of next-generation sequencing technologies, and their implementation into routine diagnosis and follow-up to identify the genomic abnormalities within the treatment-resistant minimal residual disease populations, is likely to address this need. At the same time, these approaches, while adding a further level of complexity to the leukemia genome, are likely to identify additional novel targets among the heterogeneous subtypes, such as the BCR-ABL1–like group.
The treatment of BCR-ABL1–positive ALL has been revolutionized by TKIs. It has been clearly demonstrated that application of novel agents in the appropriate biological arena to a suitable target can dramatically improve outcome. Evolving studies, such as those carried out on the TARGET project and the handful mentioned herein, are revealing other potential candidates with promise for future therapies. There remain many challenges ahead before these novel drugs become integrated into routine clinical use. As more new agents emerge, the protocols for their integration will become more streamlined toward understanding the relevance of the recent discoveries underlying the pathogenesis of BCP-ALL. The discovery of germline mutations, such as the finding of a high incidence of germline TP53 mutations in patients with hypodiploid ALL,26 has highlighted the role of genetic predisposition to certain subtypes of disease, which are clearly more widespread than previously envisaged. We should continue to search for novel targets that will surely emerge from the detailed analysis of accumulating data from state-of-the-art next-generation sequencing technologies in parallel with expression, proteomic, and epigenetic studies. The goal of 100% cure for ALL may not be too far away.
Disclosures
Conflict-of-interest disclosure: The author declares no competing financial interests. Off-label drug use: None disclosed.
Correspondence
Christine J. Harrison, Northern Institute for Cancer Research, Newcastle University, Sir James Spence Institute, Queen Victoria Road, Newcastle upon Tyne NE1 4LP, United Kingdom; Phone: 44-191-2821320; Fax: 44-191-2821326; e-mail: christine.harrison@newcastle.ac.uk.