Abstract
Throughout its history, chronic myeloid leukemia (CML) has set precedents for cancer research and therapy. These range from the identification of the first specific chromosomal abnormality associated with cancer to the development of imatinib as a specific, targeted therapy for the disease. The successful development of imatinib as a therapeutic agent for CML can be attributed directly to decades of scientific discoveries. These discoveries determined that the BCR-ABL tyrosine kinase is the critical pathogenetic event in CML and an ideal target for therapy. This was confirmed in clinical trials of imatinib, with imatinib significantly improving the long-term survival of patients with CML. Continuing in this tradition of scientific discoveries leading to improved therapies, the understanding of resistance to imatinib has rapidly led to strategies to circumvent resistance. Continued studies of hematologic malignancies will allow this paradigm of targeting molecular pathogenetic events to be applied to many additional hematologic cancers.
Introduction
The hematologic malignancies have frequently been at the forefront of the development of cancer therapies. Many chemotherapeutic agents were first used in hematologic malignancies and were subsequently found to have activity in solid tumor. Thus, the first chemotherapeutic agent, nitrogen mustard, was initially used in patients with Hodgkin disease and lymphocytic leukemias. Methotrexate and adriamycin were first developed in childhood leukemia. Continuing in this tradition, imatinib, the first tyrosine kinase inhibitor, was developed for chronic myeloid leukemia (CML) and has also been found to be active in gastrointestinal stromal tumors and some rare myeloproliferative neoplasms. Imatinib has also established the paradigm of targeting molecular pathogenetic events in cancer. The discoveries that led to the development of imatinib have paralleled much of the 50-year history of the American Society of Hematology (ASH). This began with the discovery of the Philadelphia chromosome, leading to the identification of the BCR-ABL tyrosine kinase as the causative molecular event of CML, which allowed the development of imatinib as a specific inhibitor of this kinase. Scientific discoveries have continued to be integrated into the clinical trials of imatinib, allowing a rapid understanding of mechanisms of resistance to imatinib, development of new drugs to circumvent resistance, and the successful use of imatinib in other malignancies driven by imatinib-sensitive kinases
Identification of the BCR-ABL tyrosine kinase as a therapeutic target
Chronic myeloid leukemia: clinical features
CML was first described by two pathologists, Dr Rudolf Virchow and Dr John Hughes Bennett, in 1845.1,2 Although a debate ensued as to whose description was first, in a public lecture, Virchow acknowledged that Bennett, “observed a case of indubitable leukemia some months before I saw my first case.”3(p2) Of note, these first accounts of CML occurred prior to staining methods for blood, which were not developed until the late 1800s. We now know that CML is a clonal hematopoietic stem cell disorder with an annual incidence of 1 to 2 cases per 100 000 per year.
The chronic, or stable phase of CML is characterized by excess numbers of myeloid cells that differentiate and function normally. Between 90% and 95% of patients will be diagnosed in this phase of the disease. Historically, within an average of 4 to 6 years, the disease transformed through an “accelerated phase” to an invariably fatal acute leukemia, also known as blast crisis. Disease progression is likely due to the accumulation of molecular abnormalities that lead to a progressive loss of the capacity for terminal differentiation of the leukemic clone.4 With currently available therapies, however, median survival might extend to 30 years or more and some patients may never transform and may never die of their leukemia.
Historically, therapy for CML was empirically based. During the late 1800s, the mainstay of therapy for CML was Fowler solution, which was developed by Dr Thomas Fowler in the mid- 1700s. The active ingredient in Fowler solution was probably potassium arsenite and there has been a resurgence of interest in the use of arsenic preparations for CML.3,5,6 During the 1900s, radiation, busulfan, hydroxyurea, interferon-α (IFN-α), and stem cell transplantation were developed for other indications, tried broadly, and found to have activity in CML.3 Allogeneic stem cell transplantation is the only proven curative therapy, but is associated with significant morbidity and mortality.7
Molecular pathogenesis of CML
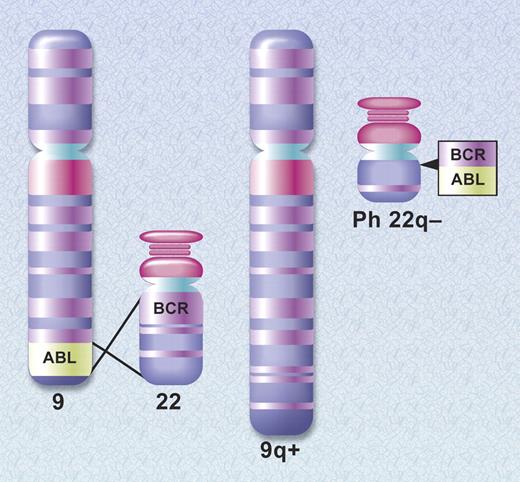
To move from empiric therapy to rationally designed therapy requires a precise understanding of the molecular pathogenesis of a disease, and the unraveling of the molecular pathogenesis of CML began shortly after ASH was formed. In 1960, Peter Nowell and David Hungerford, working in Philadelphia, described a consistent chromosomal abnormality in patients with CML, an acrocentric chromosome that was thought to be a chromosomal deletion.8 This was the first example of a chromosomal abnormality linked to a specific malignancy. In their seminal article, the authors stated, “the findings suggest a causal relationship between the chromosomal abnormality observed and chronic granulocytic leukemia.”8 This prescient statement was met with skepticism as it was felt that the chromosome abnormality was an associated rather than causative phenomenon. As chromosomal banding techniques improved, it became apparent that the chromosome abnormality was a shortened chromosome 22. Then, in 1973, Dr Janet Rowley determined that the shortened chromosome 22, the so-called Philadelphia (Ph) chromosome, was the product of a reciprocal translocation between the long arms of chromosomes 9 and 22, t(9:22)(q34;q11)9 (Figure 1).
Schematic diagram of the translocation that creates the Philadelphia chromosome. The ABL and BCR genes reside on the long arms of chromosomes 9 and 22, respectively. As a result of the (9;22) translocation, a BCR-ABL gene is formed on the derivative chromosome 22 (Philadelphia chromosome). Illustration by A. Y. Chen.
Schematic diagram of the translocation that creates the Philadelphia chromosome. The ABL and BCR genes reside on the long arms of chromosomes 9 and 22, respectively. As a result of the (9;22) translocation, a BCR-ABL gene is formed on the derivative chromosome 22 (Philadelphia chromosome). Illustration by A. Y. Chen.
In the 1970s and 1980s, the investigation of transforming retroviruses led to major breakthroughs in our understanding of human malignancies. These studies revealed that mutations in normal cellular genes could be oncogenic. For example, the Abelson murine leukemia virus, initially described in 1970,10 led to the identification of its transforming gene, v-ABL, and to the cloning of its normal cellular homolog, c-ABL.11-13 By mapping oncogenes to specific chromosomal locations, it was recognized that c-ABL, which normally resides on the long arm of chromosome 9, had been translocated to chromosome 22 in patients with CML.14 As the breakpoints on chromosome 22 clustered in a relatively small region that spanned 5.3 kilobases, this region was named the breakpoint cluster region, or BCR.15,16
This confluence of the analysis of chromosomal abnormalities and transforming retoviruses allowed investigators to determine the exact molecular consequences of the (9;22) chromosomal translocation (Figure 1). Northern blots using ABL probes demonstrated a larger than normal ABL mRNA in patients with CML.17,18 This was subsequently shown to be a chimeric mRNA that was a fusion of BCR and ABL sequences.19 Similarly, a larger than normal ABL protein with tyrosine kinase activity was detected in CML cells and was identified as the product of the BCR-ABL mRNA.20-22 These discoveries provided an important link between the field of oncogenes and the biochemistry of protein kinases, as it had previously been recognized that v-ABL possessed a novel kinase activity, the ability to phosphorylate tyrosine residues.23,24 BCR-ABL is now known to have elevated tyrosine kinase activity compared with c-ABL, and the kinase activity of BCR-ABL is essential for its ability to transform cells.25,26 The leukemogenicity of BCR-ABL was confirmed in 1990 by John Groffen's laboratory and George Daley and Richard van Etten in David Baltimore's laboratory. These investigators expressed BCR-ABL in animal models and demonstrated that BCR-ABL, as the sole oncogenic event, induced leukemia,27,28 thus establishing BCR-ABL as a leukemic oncogene.
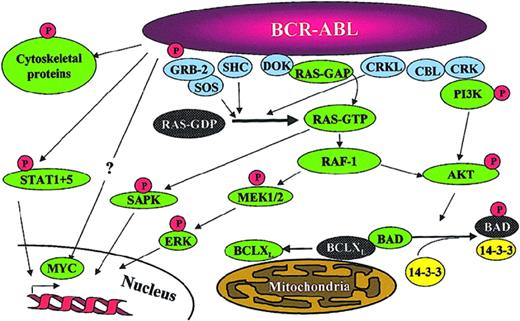
Once identified as the molecular pathogenetic event in CML, significant effort has been placed into understanding the molecular mechanism of action of BCR-ABL through the identification of signaling pathways that are impacted by BCR-ABL tyrosine kinase activity (Figure 2). Numerous substrates and binding partners of BCR-ABL have been identified and current efforts are directed at linking these pathways to the specific pathologic defects that characterize CML.29 These defects include increased proliferation or decreased apoptosis of a hematopoietic stem cell or progenitor cell leading to a massive increase in myeloid cell numbers; premature release of immature myeloid cells into the circulation, postulated to be due to a defect in adherence of myeloid progenitors to marrow stroma; and genetic instability resulting in disease progression. An example of a cellular pathway that links to an increased proliferative rate is activation of the RAS pathway. Protection from programmed cell death may be mediated in part through STAT5 up-regulation of the antiapoptotic molecule BCLXL and phosphorylation of and inactivation of the proapoptotic molecule BAD by AKT.29 CML cells also exhibit reduced adhesion to fibronectin, possibly as a downstream effect of CRKL phosphorylation. Despite the seemingly endless expansion of the list of pathways activated by BCR-ABL and the increasing complexity that is being revealed in these pathways, all of the transforming functions of BCR-ABL are dependent on its tyrosine kinase activity.26
Signaling pathways impacted by BCR-ABL expression. Note that this is a simplified diagram and that many more associations between BCR-ABL and signaling proteins have been reported. Reprinted from Deininger et al.15
Signaling pathways impacted by BCR-ABL expression. Note that this is a simplified diagram and that many more associations between BCR-ABL and signaling proteins have been reported. Reprinted from Deininger et al.15
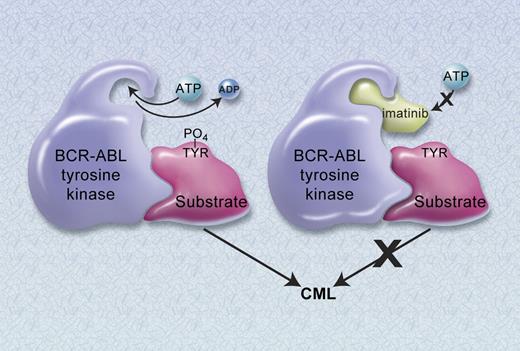
Thus, the period from 1960 to 1990 identified BCR-ABL as an ideal therapeutic target in CML. It is expressed in all patients with CML and it has been shown to be the cause of CML. BCR-ABL functions as a constitutively activated tyrosine kinase and mutagenic analysis has shown that this activity is essential for the transforming function of the protein. For these reasons, an inhibitor of the BCR-ABL kinase would be predicted to be an effective and selective therapeutic agent for CML (Figure 3).
Mechanism of action of imatinib. (A) The constitutively active BCR-ABL tyrosine kinase functions by transferring phosphate from ATP to tyrosine residues on various substrates to cause excess proliferation of myeloid cells characteristic of CML. (B) Imatinib blocks the binding of ATP to the BCR-ABL tyrosine kinase, thus inhibiting kinase activity. Illustration by A. Y. Chen.
Mechanism of action of imatinib. (A) The constitutively active BCR-ABL tyrosine kinase functions by transferring phosphate from ATP to tyrosine residues on various substrates to cause excess proliferation of myeloid cells characteristic of CML. (B) Imatinib blocks the binding of ATP to the BCR-ABL tyrosine kinase, thus inhibiting kinase activity. Illustration by A. Y. Chen.
Development of an ABL-specific tyrosine kinase
Given the success of imatinib and the enormous interest in protein kinase inhibitors, it is easy to forget the degree of skepticism that kinase inhibitors faced from the scientific community and the pharmaceutical industry in the 1980s and 1990s. Much of this skepticism was due to the prevailing thought that inhibitors of ATP binding would lack sufficient target specificity to be clinically useful. Further, some of the early tyrosine kinase–null animals had an embryonic lethal phenotype that led to the belief that tyrosine kinase inhibitors would be extremely toxic. Lastly, it was viewed that targeting of a single molecular defect would not suffice to treat highly heterogeneous cancers. The concern regarding specificity was addressed in 1988, when Yaish et al published a series of compounds, known as tyrphostins, that demonstrated that specific tyrosine kinase inhibitors could be developed.30 Working independently, scientists at Ciba-Geigy (now Novartis), under the direction of Nicholas (Nick) Lydon and Alex Matter, began a kinase inhibitor program.
The group at Ciba-Geigy established collaborations with two investigators at Dana-Farber Cancer Institute from 1987 to 1990 to assist their project: Dr Thomas (Tom) Roberts, a kinase expert, and Dr Charles Stiles, an expert on the platelet-derived growth factor receptor (PDGFR), which was one of the targets selected for drug development. Critical to the success of the project was the ability to generate sufficient quantities of kinases to analyze their chemical libraries. Helen Piwnica-Worms, working in Dr Roberts' laboratory, expressed kinases in the baculovirus expression system,31 and this technology was transferred to Ciba-Geigy. A second reagent was the phosphotyrosine antibody, 4G10, that I had developed with Deborah Morrison while working in Tom's lab.32 This antibody allowed the detection of the activity of tyrosine kinases, tyrosine phosphorylation, and was essential to detecting the ability of chemical compounds to inhibit tyrosine kinases. In addition, Nick Lydon and I established a working relationship as I was the only oncologist working in Tom's lab. At the time, I was working on a variety of tyrosine kinase projects, but not CML or BCR-ABL. Nevertheless, I advised Nick that BCR-ABL would likely be the first and the best cancer in which to validate the paradigm of kinase inhibition. As Nick had a background working on v-ABL, he immediately recognized the importance of including the ABL kinase in their profiling of compounds undergoing optimization. While the Ciba-Geigy group was working to identify kinase inhibitors, I decided to combine my emerging expertise in tyrosine kinase signaling with my background in oncology and established a collaboration with Dr James Griffin to work on BCR-ABL signaling.
With essential reagents in hand, N. Lydon's group performed high-throughput screens of chemical libraries searching for compounds with kinase inhibitory activity. From this time-consuming approach, a lead compound of the 2-phenylaminopyrimidine class was identified. The activity of the 2-phenylaminopyrimidine series was optimized for various kinases by an extremely talented chemist, Joerg Zimmerman, who synthesized chemically related compounds and analyzed the relationship between their structure and activity.33-35 One compound series, optimized for activity against the PDGFR was also found by Elisabeth Buchdunger at Ciba-Geigy to possess inhibitory activity toward ABL.36
In 1993, having moved to Oregon Health & Science University and having established various BCR-ABL–driven models, I reconnected with Nick Lydon as I was interested in finding a company with a BCR-ABL kinase inhibitor. By this point, his group had generated the 2-phenylaminopyrimidine series already noted and he sent me a variety of compounds for testing. From our testing, STI571 (Gleevec, Glivec, imatinib) was identified as the compound that was the most specific at killing CML cells.37 Imatinib ultimately emerged as the lead compound for preclinical development based on its selectivity against CML cells in vitro and its druglike attributes, including pharmacokinetic and formulation properties.38
Preclinical studies
Our initial laboratory studies showed that imatinib had inhibitory activity against ABL and its activated derivatives v-ABL, BCR-ABL, and TEL-ABL.36,37,39 As previously discussed, imatinib was known to inhibit the PDGFR. We also demonstrated potent activity against KIT,40 with minimal activity against a broad range of other tyrosine and serine/threonine kinases, demonstrating that imatinib had a high level of selectivity.41 In cellular experiments, imatinib was shown to selectively suppress the proliferation of BCR-ABL–expressing cells in vitro.37 This selectivity was confirmed using patient cells in colony-forming assays. In these studies, imatinib caused a 92% to 98% decrease in the number of BCR-ABL–positive colonies formed, with minimal inhibition of normal colony formation.37 Lastly, animal studies showed that imatinib treatment led to a dose-dependent inhibition of BCR-ABL–expressing cells with no effects against v-SRC–expressing tumors.37 The first major presentation of this work was at the ASH meeting in December 1995. This was one of the last meetings with Tuesday morning scientific sessions, which were notorious for poor attendance. Despite the typical low turnout, several interested investigators were in attendance and they confirmed and extended our results.39,42,43 This was also a critical meeting for me as I was able to assemble a team of investigators to assist in the planning for clinical trials. This team included Dr John Goldman from the Hammersmith, Dr Charles Sawyers at UCLA, and Dr Moshe Talpaz at M. D. Anderson. All except Dr Goldman participated in the phase 1 study as it was decided to conduct this study only in the United States.
Despite the promising preclinical data, there were still hurdles to overcome before clinical trials commenced. These included concerns about toxicity, whether targeting a single kinase would be an effective anticancer strategy, and most importantly to a large pharmaceutical company, whether they would realize a return on their investment for a small market disease such as CML. At the time, I had patients in my clinic with CML with no effective treatment options remaining and this connection was a crucial turning point. I became their advocate by lobbying my remaining contacts at Novartis, including Alex Matter and Elisabeth Buchdunger, to move this project forward. Ultimately, we prevailed.
Phase 1 clinical trials
A standard dose-escalation phase 1 study of imatinib began in June 1998. The study population consisted of patients with CML in chronic phase, refractory or resistant to IFN-α–based therapy, or intolerant of this drug.44 At later stages of the study, patients with CML in blast crisis and patients with Ph chromosome–positive acute lymphoblastic leukemia were also enrolled.45 Imatinib was well tolerated with the most common side effects including occasional nausea, periorbital edema, and muscle cramps. Despite dose escalation from 25 mg to 1000 mg in 14 cohorts of patients, a maximally tolerated dose could not be defined. Imatinib was administered once daily and pharmacokinetics showed a half-life of 13 to 16 hours.44 At doses of 300 mg and above, significant therapeutic benefits were observed. In chronic phase patients who had failed therapy with IFN-α, 53 of 54 (98%) patients treated at 300 mg and above achieved a complete hematologic response, and with one year of follow-up, only one of these patients relapsed.44 In myeloid blast crisis patients, 21 of 38 (55%) patients treated at doses of 300 mg or more per day responded, with 18% having responses lasting beyond one year.45 The first major international presentation of these phase 1 results were at the plenary session of ASH in December 1999.
Phase 2 studies
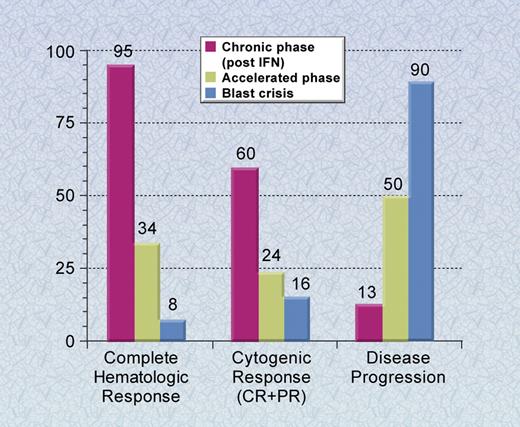
These remarkable phase 1 data led to rapidly accruing phase 2 clinical trials. These clinical trials confirmed the results seen in the phase 1 studies and led to FDA approval of imatinib in May 2001, less than 3 years after the start of the phase 1 study (Figure 4). In chronic phase patients who had failed IFN-α therapy, 95% of patients achieved a complete hematologic response and 60%, a major cytogenetic response, defined as a reduction in the percentage of Ph chromosome–positive metaphases to less than 35.46 The estimated rates of freedom from progression to accelerated phase and blastic phase and overall survival at 6 years were 61% and 76%, respectively.49 In accelerated phase and blast crisis patients, the response rates were also quite high, but relapses have been much more common, with the majority of blast crisis patients relapsing during the first year of therapy.47,48
Phase 2 clinical trials of imatinib for CML. The results of the phase 2 studies are shown in chronic phase patients who previously received interferon therapy, and accelerated phase and blast crisis patients. Results shown are with a median follow-up of up to 30 months and the rate of disease progression is at 24 months.46-48 Complete response (CR) and partial response (PR) for cytogenetic responses include patients with Ph chromosome–positive metaphases of less than or equal to 35%. Illustration by A. Y. Chen.
Phase 2 clinical trials of imatinib for CML. The results of the phase 2 studies are shown in chronic phase patients who previously received interferon therapy, and accelerated phase and blast crisis patients. Results shown are with a median follow-up of up to 30 months and the rate of disease progression is at 24 months.46-48 Complete response (CR) and partial response (PR) for cytogenetic responses include patients with Ph chromosome–positive metaphases of less than or equal to 35%. Illustration by A. Y. Chen.
Cumulative experience with imatinib
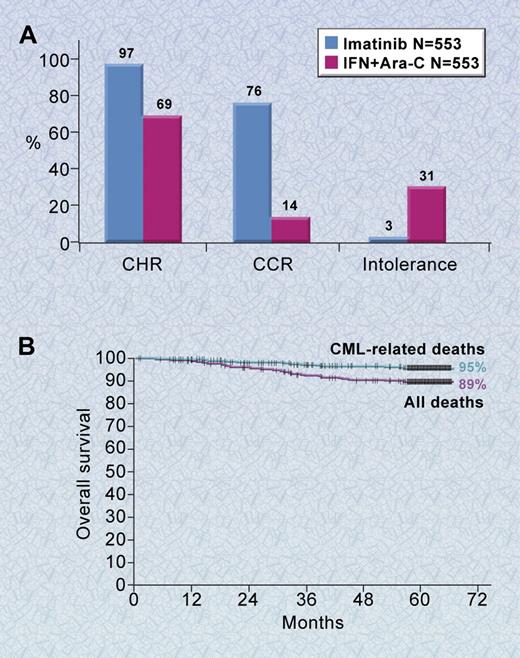
The experience with imatinib has yielded a wealth of information, including data from a randomized clinical trial with 5 years of follow-up (the International Randomized Study of Interferon and STI571 [IRIS] study),50 a crystal structure of the ABL kinase domain in complex with imatinib,51 and important insights into the mechanisms of imatinib resistance.52,53 The IRIS trial was initiated in June 2000 for patients newly diagnosed with CML in the chronic phase. It reached its accrual goal of more than 1000 patients in less than 7 months.54 Five hundred fifty-three patients were randomized to each of the two treatments, imatinib at 400 mg per day or interferon-α plus Ara-C. There were no significant differences in prognostic features on the two arms. With a median follow-up of 19 months, patients randomized to imatinib had significantly better results than patients treated with interferon-α plus Ara-C in all parameters measured, including rates of complete hematologic response (97% vs 56%, P < .001), major and complete cytogenetic responses (85% and 74% vs 22% and 8%, respectively, P < .001), discontinuation of assigned therapy due to intolerance (3% vs 31%), and progression to accelerated phase or blast crisis (3% vs 8%, P < .001; Figure 5A).54 The substantial superiority of imatinib resulted in study results being disclosed early and most patients being crossed over to the imatinib arm. Accordingly, this study is now a long-term follow-up study of patients who received imatinib as initial therapy.
Phase 3 results of imatinib for newly diagnosed patients with CML. (A) The results shown are for newly diagnosed chronic phase patients with a median follow-up of 18 months. (B) The 5-year survival curve for newly diagnosed patients with chronic-phase CML treated with imatinib. Shown is the overall survival among patients treated with imatinib based on an intention-to-treat analysis.50 Illustration by A. Y. Chen.
Phase 3 results of imatinib for newly diagnosed patients with CML. (A) The results shown are for newly diagnosed chronic phase patients with a median follow-up of 18 months. (B) The 5-year survival curve for newly diagnosed patients with chronic-phase CML treated with imatinib. Shown is the overall survival among patients treated with imatinib based on an intention-to-treat analysis.50 Illustration by A. Y. Chen.
In the most recent update of the IRIS study, the overall survival for newly diagnosed chronic phase patients treated with imatinib at 5 years is 89% (Figure 5B).50 An estimated 93% of imatinib-treated patients remain free from disease progression to the accelerated phase or blast crisis.50 An additional 6% of patients have shown some evidence of loss of response to imatinib, but their disease has not progressed to the accelerated phase or blast crisis.50 Most of the side effects of imatinib are mild to moderate, with the most common being edema, muscle cramps, diarrhea, nausea, skin rashes, and myelosuppression. Thus, imatinib therapy has substantially increased survival for patients with CML while simultaneously offering a generally well-tolerated, oral therapy. Although most patients randomized to imatinib achieve a complete cytogenetic response, the majority of these patients have detectable leukemia as analyzed by reverse transcriptase–polymerase chain reaction (RT-PCR) for BCR-ABL.55 Thus, most patients treated with imatinib have persistent disease at the molecular level and most have relapsed if imatinib was discontinued. Interestingly, over the past 4 years, there has been a trend downward in the risk of relapse; if this trend holds, this suggests that CML could be a long-term, controllable illness for the majority of patients.
Mechanisms of relapse
Although most patients with chronic phase CML treated with imatinib have well-controlled disease, some patients have relapsed and/or progressed to accelerated phase or blast crisis.50 Furthermore, most patients with advanced phases of CML initially respond to imatinib but subsequently relapse.44,47,48 Therefore, the mechanisms of relapse emerged as a major question.
The first insights into relapse mechanisms came from assays that evaluated BCR-ABL kinase inhibition. This is where an understanding of BCR-ABL signaling and incorporation of this knowledge into the clinical trials was incredibly useful. In 1994, my lab and two others published the finding that the major tyrosine phosphorylated protein in CML patients was a 39-kDa adaptor protein CRKL.56-58 With this knowledge, we developed an assay that evaluated the phosphorylation status of CRKL in all patients enrolled in the imatinib clinical trials. Using this assay, Charles Sawyers' laboratory determined that the majority of patients who respond to imatinib and then relapse have reactivation of the BCR-ABL tyrosine kinase.59 Reactivation of the kinase would mean that resistance could be due to mechanisms that either prevent imatinib from reaching the target or render the target insensitive to imatinib. In the former category are mechanisms such as drug efflux or protein binding of imatinib. In the latter category would be mutations of the BCR-ABL kinase that render BCR-ABL insensitive to imatinib or amplification of the BCR-ABL protein.
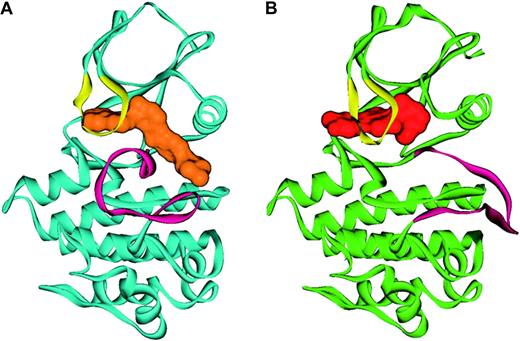
In the background of this, we had been examining a puzzling finding. Imatinib is a potent inhibitor of ABL but not its most closely related kinase, SRC. We generated numerous single amino acid substitutions in residues in the kinase domains of these two proteins that we thought might allow SRC to be inhibited by imatinib or prevent imatinib from inhibiting ABL, with no success. Our conclusions were that either we needed combinations of amino acid changes or that despite their amino acid homology, the 3-dimensional structures of SRC and ABL differed sufficiently to account for these findings. This latter conclusion was confirmed when John Kuriyan's laboratory crystallized the ABL kinase in complex with imatinib (Figure 6).51 This structure demonstrated that imatinib binds to a unique inactive form of ABL and despite their homology, there were significant structural differences between the inactive form of ABL and SRC.
Ribbon representation of ABL in complex with imatinib and PD180970. Shown is (A) the conformation of ABL (blue) in complex with imatinib (orange), with the activation loop (magenta) in the inactive conformation, and (B) ABL (green) in complex with a dual SRC/ABL inhibitor, PD180970 (red), with the activation loop (magenta) in the active conformation. Figure prepared by Sandra W. Cowan-Jacob based on reported structures.60,61 Reprinted from Deininger et al.41
Ribbon representation of ABL in complex with imatinib and PD180970. Shown is (A) the conformation of ABL (blue) in complex with imatinib (orange), with the activation loop (magenta) in the inactive conformation, and (B) ABL (green) in complex with a dual SRC/ABL inhibitor, PD180970 (red), with the activation loop (magenta) in the active conformation. Figure prepared by Sandra W. Cowan-Jacob based on reported structures.60,61 Reprinted from Deininger et al.41
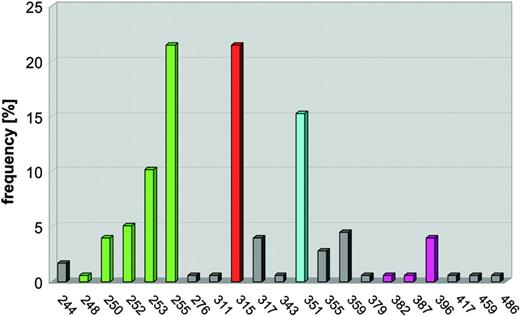
The crystal structural data allowed critical contacts points between the ABL kinase and imatinib to be identified. With these data, we made mutations in all of the contact points and found that substitutions at amino acid 315 resulted in an ABL kinase that could not be inhibited by imatinib.62 This work was originally presented at the ASH meeting in December 2000.63 Subsequently, Charles Sawyers' laboratory reported that T315I mutations were present in patients who relapsed on imatinib therapy.59 We now know that at least 50% of patients who relapse on imatinib therapy have BCR-ABL point mutations in at least 40 different amino acids scattered throughout the ABL kinase domain (Figure 7).52,53 These studies of imatinib relapse mechanisms highlight the importance of incorporating scientific studies into clinical trials.
Schematic of point mutations in the ABL kinase domain. Mutations in the ABL kinase domain (amino acids 240 to 500) cluster in 4 distinct regions, the ATP-binding domain (amino acid 248-255, green), mutations of T315 (red), which form a hydrogen bond with imatinib, M351 (turquoise), which interacts with the SH2 domain and participates in autoregulation of kinase activity, and the activation loop (amino acids 379-398, magenta). The vertical lines represent the frequency each amino acid has been found to be mutated as compiled from the literature. Reprinted from Deininger et al.41
Schematic of point mutations in the ABL kinase domain. Mutations in the ABL kinase domain (amino acids 240 to 500) cluster in 4 distinct regions, the ATP-binding domain (amino acid 248-255, green), mutations of T315 (red), which form a hydrogen bond with imatinib, M351 (turquoise), which interacts with the SH2 domain and participates in autoregulation of kinase activity, and the activation loop (amino acids 379-398, magenta). The vertical lines represent the frequency each amino acid has been found to be mutated as compiled from the literature. Reprinted from Deininger et al.41
In examining the structural basis for imatinib resistance, two main mechanisms have been identified. One obvious set of mutations are those at contact points between the ABL kinase domain and imatinib. Examples of this include T315I and F359V.62 The second category of kinase domain mutations is hypothesized to have conformational effects on the kinase, either favoring the active conformation to which imatinib cannot bind or decreasing the flexibility of the P-loop such that the conformational changes required for imatinib to bind this region cannot be adopted.64 These mutations include those in the P-loop, which bridges the ATP-binding pocket of the kinase domain (M244V, G250E, Q252H, Y253F/H, and E255K/V), and the activation loop (H396R/P).
The second generation of BCR-ABL inhibitors
Much like the original paradigm, whereby an understanding of the molecular pathogenesis of CML led to the development to imatinib, the understanding of the mechanism of resistance to imatinib led to the rapid development of new drugs to circumvent resistance. The information from the crystal structure suggested two approaches for drug design to circumvent resistance. One approach would be to modify imatinib such that it bound more tightly to the ABL kinase. This is exactly what was accomplished by chemists at Novartis in the design of nilotinib.65 In preclinical testing, nilotinib showed 10- to 30-fold increased potency over imatinib against the major resistant mutants, except T315I.65 Nilotinib retains the kinase specificity profile of imatinib, with inhibition confined to ABL kinases, KIT, and PDGFR kinases. As with imatinib, nilotinib binds the kinase domain of ABL in the inactive conformation.65
A second approach to circumventing resistance relates to a comparison of the structure of ABL and SRC in their active and inactive conformations. Each kinase has a unique inactive conformation but similar active configurations. In 2000, Richard Jove published findings that a compound, PD180970, is capable of inhibiting SRC and ABL.66 We reasoned that such as inhibitor would likely bind the active conformation of ABL and that this inhibitor might be capable of inhibiting several of the imatinib-resistant mutations. A crystal structure of ABL with PD180970 from John Kuriyan's laboratory confirmed the first of these predictions,60 and in vitro studies performed in our lab confirmed the second.67 Although PD180970 was unable to be formulated for clinical utility, scientists at Bristol-Myers-Squibb examined dasatinib, originally synthesized as a SRC family inhibitor, and found that it was also a significantly more potent ABL inhibitor than imatinib.68 Dasatinib was subsequently shown to inhibit all imatinib-resistant mutants except T315I.69 A similar approach was taken by scientists at Wyeth in developing bosutinib.70
Dasatinib and nilotinib have progressed rapidly through clinical trials, and both are FDA-approved for patients with resistance or intolerance to imatinib. Both show significant activity and good durability of responses in patients with relapsed, chronic-phase disease.71-74 Both dasatinib and nilotinib have been well tolerated, although each has unique side effects. For example, pleural effusions occur in some patients treated with dasatinib, and elevated liver enzymes and prolongation of QT interval corrected for rate (QTc) occur in some patients treated with nilotinib.71-75
In the clinical trials of dasatinib and nilotinib, one of the most common mechanisms of resistance has been the emergence of clones with the T315I mutation in BCR-ABL.72,74,76-79 Neither nilotinib nor dasatinib inhibit T315I due to direct contacts between this residue and these molecules.65,80 Therefore, T315I remains a recurring problem for all the current ABL kinase inhibitors. Despite this challenge, several preclinical T315I inhibitors have been reported and are advancing to clinical trials.81,82 Preclinical studies have also suggested that combinations of inhibitors could completely circumvent resistance due to mutations.83 Whether combinations are necessary for the majority of patients presenting in chronic phase and the tolerability of combinations remain open questions. As studies define patients with a higher risk of resistance, these patients might be candidates for combination studies. Regardless, persistence of hematopoietic stem cells expressing BCR-ABL is likely to remain an issue,84,85 and studies to define and circumvent persistence will be required to cure patients with CML.
Activity of imatinib in other hematologic malignancies
In addition to inhibiting the ABL tyrosine kinase, imatinib inhibits the PDGFR and KIT tyrosine kinases. There are now several other cancers where imatinib has shown clinical benefits that are based on the profile of kinases inhibited by imatinib and an understanding of the genetic defects causing various malignancies. One such disease is gastrointestinal stromal tumor (GIST), which is driven mainly by KIT mutations.86 Data on the use of imatinib in GIST are reviewed elsewhere.87 Imatinib has also shown significant activity in patients with acute lymphoblastic leukemia (ALL) who are BCR-ABL positive, but responses to single-agent therapy are generally transient.88 Recent data from pediatric clinical trials have shown, however, that combinations of imatinib with standard chemotherapy reverse the negative prognosis conveyed by the presence of the Philadelphia chromosome.89
Translocations involving the PDGFRB gene have been identified in several myeloproliferative and myelodysplastic syndromes. The most common of these translocations, t(5;12)(q33;p13), is seen in a subset of patients with chronic myelomonocytic leukemia (CMML) and results in fusion of the EVT6 (TEL) and PDGFRB genes.90 Patients with CMML containing the (5;12) translocation have been treated with imatinib and significant responses have been observed.91,92
Another disease caused by KIT mutations is systemic mastocytosis. The majority of tumors have a mutation of aspartic acid 816 to valine (D816V) in the kinase domain of KIT, resulting in activation of KIT.86 Unfortunately, the kinase activity of the D816V mutant isoform is resistant to imatinib,85,86,92-94 probably because this mutation induces conformational changes in the activation loop of KIT that prevent the binding of imatinib. Thus, imatinib is unlikely to be useful in this disorder, but an inhibitor of this KIT mutation would be predicted to be an effective therapy.
An interesting example has been the activity of imatinib in hypereosinophilic syndrome (HES). Imatinib was tried empirically in this disorder and dramatic results were observed.95 This prompted investigations of the molecular basis for imatinib's activity in this disease. Two groups arrived at the conclusion—that an intrachromosomal deletion on chromosome 4 resulted in a fusion between a gene of unknown function, FIP1L1, and a truncated PDGFRA in a large percentage of patients with this disorder.96,97 The resulting FIP1L1-PRGFRA fusion protein is a constitutively activated tyrosine kinase that is imatinib sensitive, thus accounting for the responsiveness of this disease to imatinib. The important message from this example is that careful study of responding patients can yield significant insights into disease pathogenesis.
Translating the success of imatinib to other malignancies
The clinical trials with imatinib are a dramatic demonstration of the potential of targeting molecular pathogenetic events in a malignancy. As this paradigm is applied to other malignancies, it is worth remembering that BCR-ABL and CML have several features that were critical to the success of this agent. One of these is that BCR-ABL tyrosine kinase activity has clearly been demonstrated to be critical to the pathogenesis of CML. Thus, not only was the target of imatinib known, but the target is a critical factor required for the development of CML. Another important feature is that as with most malignancies, treatment earlier in the course of the disease yields better results. Specifically, the response rate and durability of responses have been greater in chronic phase patients as opposed to blast phase patients. Thus, for maximal utility as a single agent, the identification of crucial, early events in malignant progression is the first step in reproducing the success with imatinib in other malignancies. An equally as important issue is the selection of patients for clinical trials based on the presence of an appropriate target. Again, in the CML experience, patients with activation of BCR-ABL were easily identifiable by the presence of the Ph chromosome. In this regard, as reagents to analyze molecular end points are developed, these same reagents should be useful in identifying appropriate candidates for treatment with a specific agent. When all of these elements are put together, a critical pathogenetic target that is easily identifiable early in the course of the disease, remarkable results with an agent that targets this abnormality can be achieved. The obvious goal is to identify these early pathogenetic events in each malignancy and to develop agents that specifically target these abnormalities.
As we look back over the past 50 years, it is truly remarkable what has been accomplished by the members of ASH in their pursuit of improved treatments for their patients. In my clinic, it is now common for me to see patients with a CML duration of 10 years or more, a finding that previously would have made me question the diagnosis. It is particularly fitting that this commitment to support of basic discoveries into the molecular pathogenesis of hematologic diseases has been translated into such a successful cancer therapy. With the tools we currently have in hand to understand disease pathogenesis, it will be truly exciting to see what will be accomplished in the next several decades.
Acknowledgments
I gratefully acknowledge the hundreds of people who have participated in this project and assisted me in my career. This includes all of my current and former laboratory staff, my colleagues and mentors at OHSU, the clinical faculty, and our nurses and data managers. I thank my mentors from Dana-Farber Cancer Institute and the dedicated scientific and clinical staff at Novartis, who shepherded imatinib through clinical trials. I also thank the numerous investigators who enrolled patients on our clinical trials. During this time, I have been supported by various funding agencies including the National Cancer Institute, The Leukemia & Lymphoma Society, the Burroughs Wellcome Fund, the T. J. Martell Foundation, the Doris Duke Charitable Foundation, and the Howard Hughes Medical Institute. I am grateful for this support. Lastly, I thank my patients who have gone on this incredible journey with me.
Authorship
Contribution: B.J.D. wrote the paper.
Conflict-of-interest disclosure: Molecular MD–OHSU and B.J.D. have a financial interest in MolecularMD. Technology used in this research has been licensed to MolecularMD. This potential conflict of interest has been reviewed and managed by the OHSU Conflict of Interest in Research Committee and the Integrity Program Oversight Council.
Novartis & BMS: I (B.J.D.) currently am the principal investigator on several Novartis and Bristol-Myers-Squibb clinical trials. My institution has contracts with these companies to pay for patient costs, nurse and data manager salaries, and institutional overhead. I do not derive salary, nor does my lab receive funds from these contracts.
Correspondence: Brian J. Druker, Howard Hughes Medical Institute, JELD-WEN Chair of Leukemia Research, Oregon Health & Science University Cancer Institute, L592, 3181 SW Sam Jackson Park Road, Portland, OR; e-mail: drukerb@ohsu.edu.