Abstract
Most bleeding disorders encountered in clinical practice will be diagnosed, at least initially, by phenotypic assays. However, since the characterization of the genes that encode coagulation factors in the 1980s, significant progress has been made in translating this knowledge for diagnostic and therapeutic purposes. For hemophilia A and B, molecular genetic testing to determine carrier status, prenatal diagnosis, and likelihood of inhibitor development or anaphylaxis to infused coagulation factor concentrates is an established component of comprehensive clinical management. In contrast, although significant recent advances in our understanding of the molecular genetic basis of von Willebrand disease (VWD) have allowed for the development of rational approaches to genetic diagnostics, questions remain about this complex genetic disorder and how to incorporate emerging knowledge into diagnostic strategies. This article will review the state-of-the-art for molecular diagnostics for both hemophilia and VWD.
Learning Objectives
To understand current strategies for molecular diagnostics of hemophilia and von Willebrand disease
To understand how molecular diagnostics can be utilized to improve patient care
The role of genetic testing for hemophilia A and B
Hemophilia A and B are X-linked recessive bleeding disorders that result from deficiency of factor VIII or factor IX, respectively, both of which are required for the propagation of the coagulation cascade and thrombin generation. They represent the most common of the severe inherited bleeding disorders with a combined prevalence of ∼0.01%.1 The clinical presentation of the hemophilias is dependent on the residual coagulant activity: severe disease (factor level <0.01 IU/mL) is characterized by spontaneous bleeding into muscles and joints, moderate (0.01–0.05 IU/mL) and mild hemophilia (>0.05 IU/mL) are characterized by excessive bleeding after trauma or surgery.2,3 Over one-third of hemophilia cases will be caused by de novo mutations and will therefore not have a family history of the disease.4 Given the X-linked inheritance, hemophilia predominantly affects males, however, female carriers may be affected with low levels of FVIII or FIX and a bleeding diathesis that can result from skewed X inactivation. In rare cases, females can also manifest hemophilia as the result of homozygous or compound heterozygous inheritance of F8 or F9 mutations. Additionally, loss of heterozygosity, Turner's syndrome, and Testicular Feminization syndrome need to be considered.5 Studies evaluating the pathogenesis of abnormal bleeding (which is not uncommon) in hemophilia carriers are ongoing. Although the diagnosis of hemophilia is typically based on the much more rapid and easily accessible clotting factor levels rather than genetic analysis (except for prenatal cases), identification of the underlying F8 or F9 mutation is important for carrier and prenatal diagnosis, as well as prediction of risk for inhibitor formation or anaphylactic reaction following infused replacement therapy.6 There may also be a role for genotyping in predicting the outcome of immune tolerance induction and for future gene-based hemophilia therapies.7 The high degree of mutational heterogeneity complicates mutation testing and approaches to genetic diagnosis in both conditions is reviewed.
Hemophilia A
Hemophilia A is an inherited bleeding disorder characterized by reduced or absent levels of the coagulation cofactor factor VIII (FVIII). It occurs in approximately 1/5000 male births. FVIII is encoded by F8, a large gene (186 kb) that is found in the most distal band (Xq28) of the long arm of the X chromosome.8 It is comprised of 26 exons which vary in length from 3.1 kb (exon 14) to 69 bp (exon 26) that encode a 9 kb mRNA transcript.
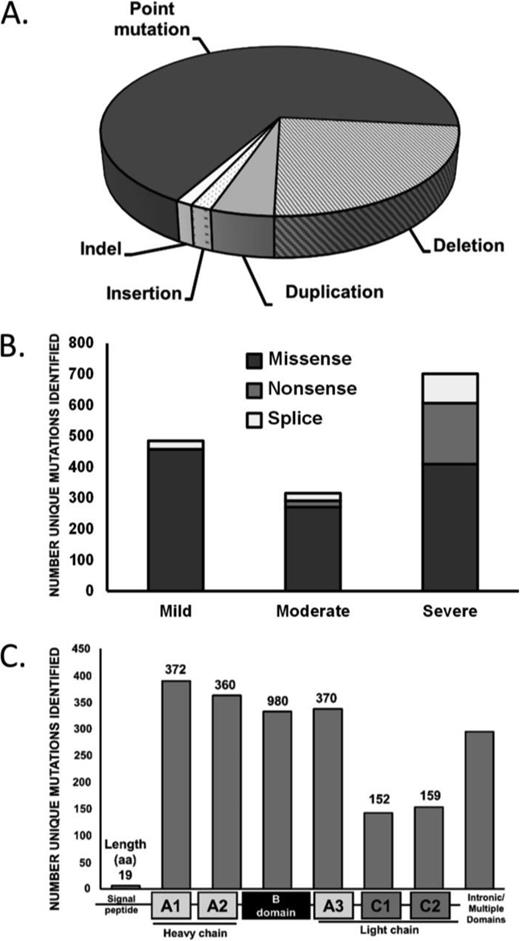
There is a large degree of allelic heterogeneity in the hemophilia A population (Figure 1A) with mutations identified in all F8 exons and intron/exon junctions. The most frequently observed hemophilia A mutation is an intrachromosomal inversion involving intron 22,9 which occurs in ∼45% of severe hemophilia A patients. The inversion event can be detected by a number of approaches including Southern blot analysis, inverse or long range PCR.10,11 Similarly, an inversion at intron 1 accounts for 1%-2% of severe hemophilia A cases, and can be identified by PCR. In addition to these well characterized events, more than 2000 unique mutations have been catalogued in the FVIII Variant Database (www.factorviii-db.org) maintained by the Structural Immunology Group at University College London, United Kingdom.12 Although deletions (23%), duplications (4.8%), insertions (1.6%), and indels (1.3%) are associated with hemophilia A, the majority of unique variants involve point mutations (66.5%; Figure 1A). Of these single nucleotide variants, missense mutations are most frequent, regardless of disease severity, with nonsense mutations accounting for a greater proportion of severe cases (Figure 1B). Mutations occur most often in the A domains of FVIII (Figure 1C).
Mutation type and frequency in hemophilia A. The frequency of mutation types in hemophilia A (A). The association of point mutation types with hemophilia A disease severity (B). The frequency of mutations by FVIII protein domain (C). Data from: http://www.factorviii-db.org, accessed April 20, 2015.
Mutation type and frequency in hemophilia A. The frequency of mutation types in hemophilia A (A). The association of point mutation types with hemophilia A disease severity (B). The frequency of mutations by FVIII protein domain (C). Data from: http://www.factorviii-db.org, accessed April 20, 2015.
The method of mutation detection can vary, with next generation sequencing, linkage analysis or direct sequencing of all 26 exons, intron/exon boundaries, 3′ and 5′ UTR, and promoter proximal regions being utilized.13 MLPA (multiplex ligation-dependent probe amplification) or aCGH (array comparative genomic hybridization) may also be used to identify copy number variants (CNVs) in individuals where no candidate mutations are found.14 The molecular diagnostic algorithm for hemophilia A is largely dependent on clinical presentation as patients with severe hemophilia A (<1% FVIII:C) generally undergo intron 22 and intron 1 inversion testing prior to sequencing. Currently, the causative mutation in ∼5% of hemophilia A (and hemophilia B) cases is not identified. This discrepancy is most likely because of phenotypic misdiagnosis and mutations occurring in noncoding regions of the F8 and F9 genes.15
Hemophilia B
Hemophilia B involves a partial or complete deficiency of the zymogen coagulation factor IX (FIX). FIX is encoded by the F9 gene located on the long arm of the X chromosome at Xq27. The F9 gene is 34 kb in length, comprised of 8 exons, and encodes an mRNA of 2.8 Kb.16 Hemophilia B is less common than hemophilia A, occurring in 1/30 000 male births due to the smaller size of the F9 gene relative to F8 as well as to the absence of high-frequency inversions associated with hemophilia A. Mutation analysis for hemophilia B typically involves direct sequencing of coding and regulatory regions,13 although strategies include CNV analysis, mutation scanning, and next generation sequencing.
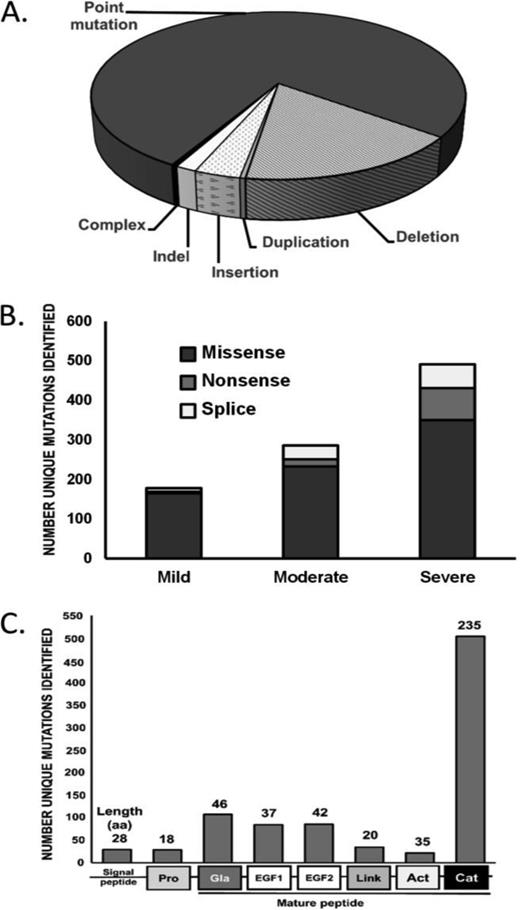
The FIX Variant database, a recently updated catalogue of >1000 unique hemophilia B mutations can be found at: www.factorix.org.17 In this population, mutations have been found throughout the F9 gene including the promoter and 3′ UTR. Although the majority of unique mutations cluster within the coding region for the serine protease domain (56%), this is proportional to its relative size (51%; (Figure 2). The majority of F9 variants are point mutations (73%), followed by deletions (16.3%), insertions (3.3%), indels (1.5%), and duplications (0.4%), whereas no inversions have been identified (Figure 2). Missense mutations account for the majority of single-nucleotide variants across all disease severity types (∼65%), with an increase in splice site and nonsense mutations for cases of severe hemophilia B (Figure 2).
Mutation type and frequency in hemophilia B. The frequency of mutation types in hemophilia B (A). The association of point mutation types with hemophilia B disease severity (B). The frequency of mutations by FIX protein domain (C). Data from: http://www.factorix.org/. Accessed April 20, 2015.
Mutation type and frequency in hemophilia B. The frequency of mutation types in hemophilia B (A). The association of point mutation types with hemophilia B disease severity (B). The frequency of mutations by FIX protein domain (C). Data from: http://www.factorix.org/. Accessed April 20, 2015.
A subset of hemophilia B patients (1.9%)18 present with a severe disease phenotype, and then experience spontaneous normalization of plasma FIX levels post-puberty.19 Termed hemophilia B Leyden, this condition is caused by point mutations in the F9 promoter that disrupt 1 of 3 transcription factor binding sites.20 Phenotypic recovery is related to increased activation of an androgen response element and growth factor activity. Sequencing of cis-regulatory elements associated with hemophilia B Leyden is routinely incorporated into molecular diagnostic protocols and is highly informative with respect to prognosis for these patients, although rare cases in which phenotypic recovery is not documented have been reported.21
Inhibitors and anaphylaxis in hemophilia
The risk for developing inhibitory antibodies in hemophilia A and B patients is strongly influenced by the severity of the causative mutation. Hemophilia A patients with mild and moderate disease have a lower risk for inhibitor development (3%-13%) than patients with severe disease (25%-30%). Accordingly, missense mutations have a lower risk of developing inhibitors (<10%), although there are some identified missense mutations that predispose to inhibitor development in nonsevere hemophilia A.22 In contrast, severe hemophilia A patients with large deletions (68%-88%) and nonsense mutations have a higher risk for inhibitor development.23 Interestingly, individuals with intron 22 inversions have a lower incidence of inhibitor development (∼20%), presumably due to synthesis but not secretion of the FVIII amino acid sequence.24
There is a lower risk of inhibitor development in hemophilia B than in hemophilia A, with 2%-4% of patients affected, the majority of whom have severe disease.18 One potential explanation is that it is more common in hemophilia B for mutant FIX protein to be expressed (CRM+). Although anaphylaxis is rare in hemophilia A, it can occur in up to 60% of hemophilia B patients with inhibitors. Both anaphylaxis and inhibitor development in hemophilia B patients are most frequently associated with large deletions and nonsense mutations.18,25
Hemophilia carrier and prenatal testing
Females with a family history of hemophilia may decide to learn their carrier status once they reach reproductive age, begin family planning, or become pregnant, however most pediatricians will test FVIII or FIX levels in potential carriers who display abnormal bleeding or prior to hemostatic challenge. Knowledge of carrier status may impact the need for neonatal testing, and approach to management of pregnancy. Prenatal diagnosis of hemophilia by genetic analysis can be performed by either chorionic villus sampling, as early as 10 weeks gestation, or amniocentesis at around 14 weeks gestation. Knowledge of the mutation in an affected male relative is of benefit in terms of streamlining testing. Knowledge of mutation status of both mother and newborn is important for planning of care during delivery and the neonatal period. A list of laboratories offering genetic testing services in North America can be found in Table 1.
The role of genetic testing for von Willebrand disease
Von Willebrand disease is the most common inherited bleeding disorder known in humans, with prevalence estimates ranging from 0.1% to 1%.26-28 Although an objective personal history of excessive mucocutaneous bleeding can usually be obtained from the patient, incomplete penetrance and variable expressivity can make the documentation of a family history of the disease difficult, and laboratory tests of hemostasis can be variable in their ability to reveal either a quantitative or qualitative defect of von Willebrand factor (VWF), making the diagnosis of VWD challenging in some situations. Furthermore, there is a lack of international consensus about diagnostic criteria for type 1 VWD. With these issues as background, this review will consider the role of molecular genetic analysis as a complementary diagnostic modality, particularly where existing clinical and laboratory approaches to diagnosis have failed to provide a definitive answer.
The VWF Gene
The VWF gene was cloned in 1985 by 4 groups in the US and Europe. The gene is located on the short arm of chromosome 12 at the locus 12p13.3.29-32 It spans ∼178 kb and contains 52 exons that range in size from 1.3 kb (exon 28) to 40 bp (exon 50).33 Analysis of the VWF gene is complicated by at least 2 other factors in addition to size: (1) there is a partial pseudogene on chromosome 22 with 97% sequence homology to exons 23-34 that necessitates the use of carefully selected chromosome 12 gene-specific PCR amplification primers for this region, and (2) the VWF locus is highly polymorphic.34 This makes direct VWF sequencing an important methodology for genetic analysis, given that mutation screening approaches such as conformation sensitive gel electrophoresis and denaturing high performance liquid chromatography will be complicated by the frequent sequence variants. However, studies have shown that strategies to identify gene deletions or duplications, such as MLPA or aCGH are also important to include in a genetic diagnostic algorithm.35,36
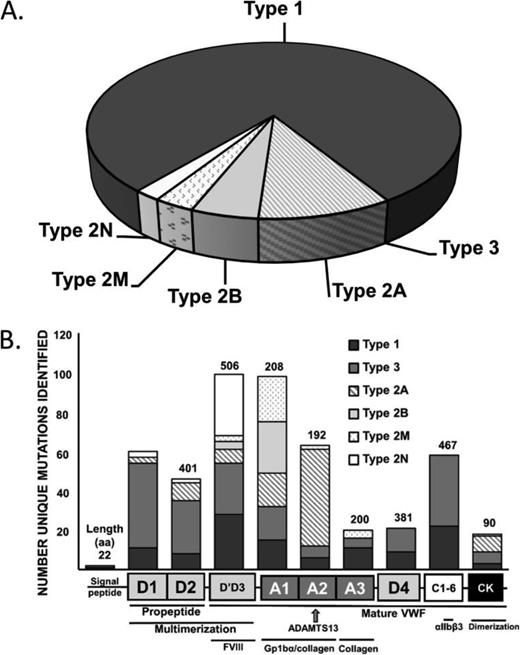
The role of genetic testing for each of the current International Society on Thrombosis and Haemostasis (ISTH) established VWD subtypes (Types 1, 2A, 2B, 2M, 2N, and 3 VWD) will be reviewed (Figure 3A).37 VWD exhibits autosomal inheritance, with Types 1, 2A, 2B, and 2M being primarily dominant and 2N and Type 3 VWD being recessive. The location of VWF mutations by VWD type can be found in Figure 3B. A database of mutations can be found at: http://www.vwf.group.shef.ac.uk/vwd.html.
Diagnosis and mutation identification by VWD subtype. The frequency of diagnosis of VWD subtypes (A). The protein domains of the pre-pro-VWF molecule with functional description and associated frequency of identified VWD mutations by disease type (B). Data from: http://www.vwf.group.shef.ac.uk/vwd.html. Accessed April 20, 2015.
Diagnosis and mutation identification by VWD subtype. The frequency of diagnosis of VWD subtypes (A). The protein domains of the pre-pro-VWF molecule with functional description and associated frequency of identified VWD mutations by disease type (B). Data from: http://www.vwf.group.shef.ac.uk/vwd.html. Accessed April 20, 2015.
Type 1 VWD
Type 1 VWD, a partial deficiency of qualitatively normal VWF, represents the most common form of the disease (∼65%-75% of all VWD cases) and is the most problematic in terms of its diagnosis. The genetic basis of Type 1 VWD has been the focus of much recent investigation and 4 large multicenter studies have reported consistent results on ∼330 families.36,38-40 Mutations in the VWF gene coding region, promoter and splice sites (predominantly missense mutations) were identified in ∼65% of index cases and were found more frequently, and with higher penetrance, in cases with lower VWF levels. The most frequently reported genetic variation (10%-20% of index cases) identified in all studies was a missense mutation resulting in an amino acid substitution of tyrosine to cysteine at codon 1584 (Y1584C).41 Investigation is ongoing as to the pathogenicity and underlying mechanism of this variant. In an ongoing large US study, the frequency of nonpathogenic VWF sequence variation was highlighted, with particular variants showing a racial predisposition, potentially contributing to false positive diagnoses in certain populations.42-44 In contrast, ∼35% of Type 1 VWD patients had no obvious VWF mutation identified and in these (often milder) cases the genetic determinants are likely to be more complex and could involve either intronic sequence variants and/or variants at other genetic loci.45 These studies have therefore confirmed prior suspicions that Type 1 VWD represents a complex genetic trait, manifesting both allelic and locus heterogeneity. This genetic complexity precludes the use of molecular genetic testing as a complementary diagnostic aid in the majority of Type 1 VWD cases at the present time.
Type 2 VWD
Type 2A VWD accounts for ∼10% of all VWD cases and is characterized by the loss of high and intermediate molecular weight multimers that result in fewer GpIb binding sites and less effective platelet binding. Type 2A VWD has been associated with >50 different missense mutations that act through a number of mechanisms, often in combination: (1) impaired dimer and multimer assembly, (2) enhanced susceptibility to ADAMTS13–mediated proteolysis, (3) loss of regulated storage, and (4) intracellular retention.46-48 In addition to providing further insights into VWF structure/function, genetic testing for type 2A VWD can be used when phenotypic uncertainty exists. Type 2A VWD mutations occur most frequently in the A2 domain but are also found in other domains involved in multimerization including D′D3, CK, and the VWF propeptide (D1 and D2).
Type 2B VWD is the result of “gain-of-function” missense mutations within the GpIb binding site on VWF. This leads to spontaneous VWF-platelet interactions that result in the selective depletion of high molecular weight (HMW) multimers and subsequent thrombocytopenia. VWF binding to platelets can also lead to enhanced ADAMTS13–mediated proteolysis, which contributes to the loss of HMW multimers.49,50 The diagnosis of Type 2B VWD is of therapeutic importance given the relative contraindication of desmopressin in managing these patients, and genetic testing can be helpful in this regard, particularly if interpretation of phenotypic assays is difficult. Additionally, genetic testing can definitively distinguish Type 2B VWD and platelet-type (PT)-VWD, which is caused by “gain-of-function” mutations in the GPIBA gene.51 This is a critical distinction as the mainstay of treatment in Type 2B VWD is VWF replacement, whereas in PT-VWD it is platelet transfusion therapy.
Type 2M VWD is classically characterized by decreased VWF–platelet interactions not caused by abnormal multimers. Causative mutations have been localized to the platelet GPIb binding site, in the A1 domain of VWF, although at distinct locations from Type 2B mutations.52 Type 2M VWD mutations have also been reported in the A3 and A1 domains, resulting in decreased VWF binding to sub-endothelial collagen.53,54 Genetic testing can be helpful, although the main therapeutic consequence of Type 2M is a poor clinical response to desmopressin.55
Type 2N VWD was first described as an autosomal form of hemophilia A and is an important differential in the investigation of all individuals (male and female) presenting with a low FVIII level.56 The ease of analysis of exons 17-25 of the VWF gene and the relative lack of availability of FVIII binding assays has increased interest in using genetic testing to confirm this diagnosis. With the different pattern of inheritance and different treatments (Type 2N VWD may respond to desmopressin or will require infusion of a FVIII/VWF concentrate), the distinction between Type 2N VWD and mild hemophilia A is important, and is one that can be definitively resolved with genetic analysis. Whereas types 2A, 2B, and 2M VWD are almost always inherited in an autosomal dominant manner, type 2N VWD is inherited in a homozygous or compound heterozygous manner, involving either two 2N alleles or a 2N and a VWF null allele.
Type 3 VWD
In most instances, the severe clinical phenotype, absent plasma VWF, and very low factor VIII:C (<0.10 U/mL) levels make the diagnosis of Type 3 VWD straightforward. Despite this, type 3 VWD individuals may be interested in genetic testing/counseling for future family planning purposes and mutation detection can provide definitive information that can be utilized for prenatal testing. Recent studies indicate that in addition to the classical recessive pattern of inheritance, some type 3 VWD kindreds demonstrate codominant inheritance.32 Type 3 VWD has a heterogeneous mutational basis with more than 80 different mutations having been described to date including VWF gene insertions, nonsense and missense mutations as well as partial and total VWF gene deletions.57-59 In addition to its use in the setting of family counseling, especially for prenatal diagnosis, VWF genotyping may also be of value with regard to predicting the likelihood of anti-VWF alloantibody development following exposure to therapeutic concentrates.60
Interpretation of synonymous and non-synonymous variations
Despite the abundance of VWF, F8, and F9 gene variant data now available, ascribing a disease phenotype to silent and missense mutations remains challenging. There is a substantial value in the development and maintenance of disease registries to demonstrate the association of these genetic variants with a clinical presentation in unrelated patients, as well as their absence in the normal population (resources include ExAC, dbSNP, and NHLBI exome sequencing project). Consideration of ancestry of the population reported is essential in interpretation such as is available in ExAC. Functional analysis of mutations using in vitro and in vivo methodologies allows for phenotypic characterization of variations in a controlled setting. In silico analysis of missense mutations may predict the relative ability of missense variants to influence the structural or biochemical properties of their associated proteins,61 as well as the influence of promoter and splice site mutations on mRNA synthesis and processing, respectively.
Additionally, synonymous F9 mutations have been described in multiple unrelated hemophilia B families that have no other identified mutations.62 Little is known about how these silent substitutions can influence the function of the encoded protein in the context of inherited bleeding disorders, although it is suspected that in addition to influencing splicing, they may alter the secondary structure and stability of the mRNA, rate of translation, and/or protein folding.63
Future of molecular diagnosis for inherited bleeding disorders
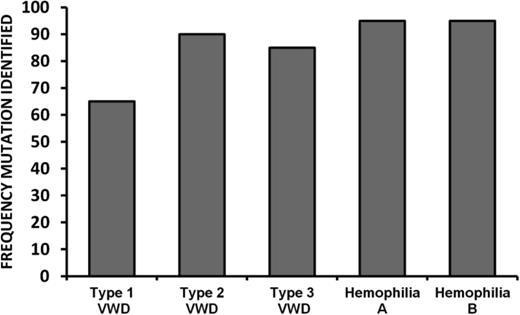
The success rate in the identification of causative mutations in inherited bleeding disorders is related to both the type of molecular analysis performed as well as genetic heterogeneity of the disease. For hemophilia A and B, between 91%-99% of causative mutations can be detected by current diagnostic approaches (Figure 4). Although slightly lower success rates (in terms of mutation identification) are observed for Types 2 and 3 VWD, it is likely that for the majority of mutation negative cases, the causative variations are located within introns or other regulatory sequences that are not routinely analyzed. Next generation sequencing of the whole F8, F9, or VWF genes will eventually lead to decreased costs associated with molecular diagnostics and increase the frequency of mutation identification.64,65
For type 1 VWD, the rate of mutation detection is significantly lower at ∼65%. Although whole-gene sequencing may increase the rate of mutation detection in Type 1 VWD, additional genetic modifiers may also influence this quantitative phenotype. Genome-wide association studies (GWAS) have identified novel loci that contribute to the regulation of VWF plasma levels,66 but the extent to which variants at these loci influence the likelihood of a Type 1 VWD diagnosis has yet to be evaluated.45 Currently, genetic analysis of non-VWF loci in the context of Type 1 VWD should be confined to research studies, but there is a possibility that these assessments could eventually be integrated into a diagnostic algorithm for this complex trait.
Conclusion
Although molecular testing for hemostatic disorders requires expertise not typically available in routine clinical hemostasis laboratories, access to these facilities is becoming more widely available. The current state of knowledge of the molecular genetic aspects of hemophilia and VWD makes the inclusion of this knowledge appropriate in routine clinical care. Additionally, the application of novel advances in genomic analysis including genome-wide linkage analysis and whole-genome/exome sequencing will allow for the detection of loci responsible for bleeding in individuals without diagnosis, ultimately improving the care of patients with inherited bleeding disorders.
Acknowledgments
The authors thank A. Goodeve and D. Lillicrap for critical review of this paper.
Support for the National Inherited Bleeding Disorder Genotyping Laboratory in Kingston is provided by the Association of Hemophilia Clinic Directors of Canada, and L. Swystun is supported by a Canadian Institutes of Health Research postdoctoral fellowship.
Correspondence
Dr Paula James, Department of Medicine, Rm 2025, Etherington Hall, Queen's University, Kingston, ON, Canada K7L 3N6; Phone: 613-533-6329; Fax: 613-533-6855; e-mail: jamesp@queensu.ca.
References
Competing Interests
Conflict-of-interest disclosures: L.L.S. declares no competing financial interests. P.J. has received research funding from CSL Behring, Bayer, and Octapharma, and has received honoraria from Baxter, Biogen Idec, CSL Behring, Bayer, and Octapharma.
Author notes
Off-label drug use: None disclosed.