Key Points
Although common risk alleles for multiple myeloma have been identified, their contribution to familial MM is unknown.
We demonstrate an enrichment of common MM risk alleles in familial cases, providing the first direct evidence for a polygenic contribution.
Abstract
Although common risk alleles for multiple myeloma (MM) were recently identified, their contribution to familial MM is unknown. Analyzing 38 familial cases identified primarily by linking Swedish nationwide registries, we demonstrate an enrichment of common MM risk alleles in familial compared with 1530 sporadic cases (P = 4.8 × 10−2 and 6.0 × 10−2, respectively, for 2 different polygenic risk scores) and 10 171 population-based controls (P = 1.5 × 10−4 and 1.3 × 10−4, respectively). Using mixture modeling, we estimate that about one-third of familial cases result from such enrichments. Our results provide the first direct evidence for a polygenic etiology in a familial hematologic malignancy.
Introduction
Multiple myeloma (MM) is the second most common hematologic malignancy. It is characterized by an uninhibited, clonal growth of plasma cells in the bone marrow, producing a monoclonal immunoglobulin (“M protein”) or light chains that can be detected in peripheral blood.1 It is preceded by monoclonal gammopathy of unknown significance (MGUS),2,3 a common condition (3% of ≥ 50-year-olds) defined as a clonal growth of plasma cells that does not satisfy the criteria for MM, but progresses to MM at a rate of about 1% per year.4,5
Since the 1920s, several authors have reported families with multiple cases of MM, including families both with Mendelian and non-Mendelian pedigrees.6,7 In the 2000s, systematic family studies, including in population-based registries, confirmed that first-degree relatives of patients with MM and its precursor condition, monoclonal gammopathy of unknown significance, have 2 to 4 times higher risk for MM,8-13 and a higher risk for lymphomas and certain solid tumors.14,15 According to estimates based on Swedish nationwide registries, about 2.4% of MM cases have a first-degree relative with MM.14
Recently, genome-wide association studies have identified single-nucleotide polymorphisms (SNPs) at 16 independent loci associated with MM risk.16-19 Although the discovery of risk alleles proves the existence of inherited susceptibility, the genetic background of familial MM remains unclear. The identified risk alleles are common, have modest effects, and have been identified by comparing unselected cases with controls. Although statistical predictions suggest that they account for somewhere on the order of 20% of the familial risk,18 no direct evidence for a polygenic etiology has been presented. It is still an open question whether familial MM is caused by aggregation of unusually high numbers of common risk alleles or by other factors (eg, rare variants with strong effects or exposure of family members to a common environmental factor).
Materials and methods
The study design is described in detail in the supplemental Methods. In essence, we carried out a nationwide search for patients with familial MM in Sweden. For this, we used the Swedish Cancer Registry and the Swedish Multi-generation Registry, which record cancer cases and familial relationships, respectively. Linking the registries, we identified 237 patients with MM diagnosed between 1958 and 2013 who had at least 1 first- or second-degree relative with MM. From the Swedish National Myeloma Biobank, we obtained samples from 36 of these patients, representing 24 families. We also obtained samples from 2 second-degree relatives with MM from Norway. In terms of clinical characteristics, the identified familial cases were comparable to sporadic cases (supplemental Tables 1 and 2).
The samples were analyzed using Illumina OmniExpress SNP microarrays. To increase the genotyping resolution, variants identified by whole-genome sequencing by the UK10K and 1000 Genomes consortia20,21 were imputed into the microarray data. We then extracted the genotypes of tag SNPs for the 16 risk loci that have been robustly associated with MM (supplemental Table 3) and quantified risk allele burden using 2 different scores: 1 calculated as the total number of risk alleles and 1 calculated by summarizing the number of risk alleles weighted by their log-transformed odds ratios (ORs) across all loci. For this, we used the ORs reported in Mitchell et al18 (supplemental Table 3). To avoid statistical inflation resulting from relatedness, gene scores from individuals from the same family were averaged. The corresponding scores were calculated for 2 preexisting sets of SNP microarray data: 1 representing 1530 sporadic MM cases from Sweden and Norway,19 and 1 representing 10 171 population-based controls from Sweden.22 All individuals included in our analyses were of non-Finnish Scandinavian ancestry, as determined using the 2 top principal components calculated using the PLINK software23 (supplemental Figure 1).
Results
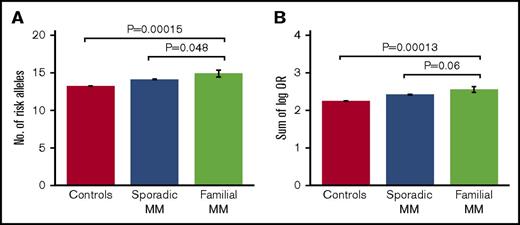
Comparing the different study groups, we observed a borderline significant enrichment of risk alleles in familial cases compared with both sporadic cases (average number of risk alleles, 14.9 vs 14.1 [1-sided Student t test, P = 4.8 × 10−2]; sum of log OR, 2.55 vs 2.42 [P = 6.0×10−2]) and population-based controls (average number of risk alleles, 14.9 vs 13.2 [P = 1.5 × 10−4]; sum of log OR, 2.55 vs 2.25 [P = 1.3 × 10−4]; Figure 1; Table 1). In addition, the majority of the individual risk alleles occurred at higher frequencies in familial cases than in sporadic cases, although only 1 of the individual alleles reached statistical significance after correction for multiple testing (supplemental Table 4). The most overrepresented risk alleles were the ones at CCAT1 (risk allele frequency, 0.55 vs 0.38; uncorrected Cochran-Armitage trend test P = .002; Bonferroni-corrected P = .033) and ELL2 (risk allele frequency, 0.89 vs 0.77; uncorrected P = .011; corrected P = .16; supplemental Table 4). No difference in age of onset was observed between familial and sporadic cases (66.0 vs 68.4 years; t test P = .22). No differences in polygenic risk scores were observed between men and women within the 3 study groups (t test P = .21-.75). These results support that at least a subset of familial MM cases are characterized by an enrichment of known, common MM risk alleles.
Risk allele burdens for familial MM, sporadic MM, and controls. Risk allele burdens for the 3 study groups, quantified as (A) number of risk alleles, and (B) sum of number of risk alleles per locus weighted by their log-transformed ORs. We detected an enrichment of MM risk alleles in familial cases compared with both sporadic cases and population-based controls. The indicated P values were obtained with 1-sided Student t test. Error bars indicate standard error of the mean. Of the 38 familial MM cases, 30 had a first-degree relative with MM and 8 had a second-degree relative. No further increase in average risk score was observed when excluding the cases with second-degree relatives (not shown).
Risk allele burdens for familial MM, sporadic MM, and controls. Risk allele burdens for the 3 study groups, quantified as (A) number of risk alleles, and (B) sum of number of risk alleles per locus weighted by their log-transformed ORs. We detected an enrichment of MM risk alleles in familial cases compared with both sporadic cases and population-based controls. The indicated P values were obtained with 1-sided Student t test. Error bars indicate standard error of the mean. Of the 38 familial MM cases, 30 had a first-degree relative with MM and 8 had a second-degree relative. No further increase in average risk score was observed when excluding the cases with second-degree relatives (not shown).
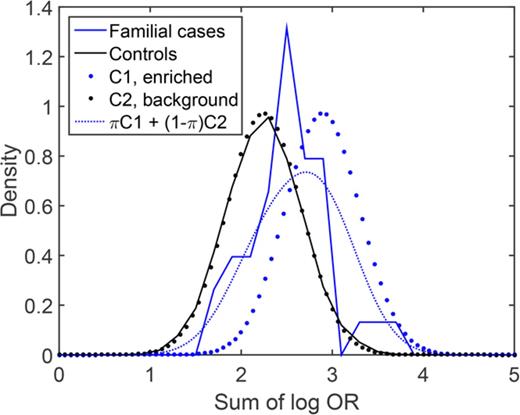
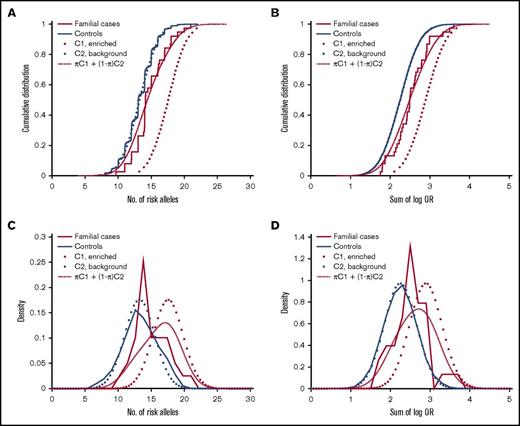
We next sought to estimate the proportion of familial MM cases that associate with enrichment of common risk alleles. Theoretically, a polygenic risk score can be drawn from either of 2 distributions, depending on the etiology: cases that are caused by enrichment of common MM risk alleles will follow a right-shifted risk score distribution, and cases that have other causes (not reflected in our risk scores) will follow the same distribution as the background population. To estimate the proportions of these 2 underlying distributions, we fit a 2-component Gaussian mixture model to the observed risk scores of familial MM cases (Figure 2). With the number-of-risk-alleles score, the proportion of the component representing cases caused by a high risk allele burden was estimated at 32% (90% confidence interval, 19%-53%), with a mean of 17.6 (compared with 13.2 for controls; Figure 2A). With the sum-of-log-OR score, the corresponding proportion was estimated at 37% (90% confidence interval, 21%-65%) with a mean of 2.91 (compared with 2.25 for controls; Figure 2B). These results indicate that about one third of familial cases are a result of enrichments of common MM risk alleles.
Estimation of the proportion of familial MM cases associated with enrichment of common MM risk alleles and other etiologies, respectively. Theoretically, the risk score of an individual with familial MM can be drawn from either of 2 underlying distributions, depending on the etiology: cases of familial MM associated with enrichment of common risk alleles are expected to follow a right-shifted distribution, whereas cases of MM on the basis of etiologies that are not quantified by our polygenic risk scores will follow the same distribution as population controls (solid blue). The risk score distribution for the group as a whole will be a mixture of these 2 components (C1 and C2; dotted) with proportions π and 1-π, respectively. To estimate π, we fit this 2-component Gaussian mixture distribution (dashed red) to the observed risk scores (solid red): (A) with the number-of-risk-alleles score, the corresponding proportion was 30%, and mean of the enriched component was 17.6; (B) with the sum-of-log-OR score, we estimated π at 37%, and the mean of the enriched component was 2.91. These results support that about one-third of familial MM cases are caused by polygenic inheritance of a high risk allele burden. (C-D) Corresponding density distribution plots for the same data.
Estimation of the proportion of familial MM cases associated with enrichment of common MM risk alleles and other etiologies, respectively. Theoretically, the risk score of an individual with familial MM can be drawn from either of 2 underlying distributions, depending on the etiology: cases of familial MM associated with enrichment of common risk alleles are expected to follow a right-shifted distribution, whereas cases of MM on the basis of etiologies that are not quantified by our polygenic risk scores will follow the same distribution as population controls (solid blue). The risk score distribution for the group as a whole will be a mixture of these 2 components (C1 and C2; dotted) with proportions π and 1-π, respectively. To estimate π, we fit this 2-component Gaussian mixture distribution (dashed red) to the observed risk scores (solid red): (A) with the number-of-risk-alleles score, the corresponding proportion was 30%, and mean of the enriched component was 17.6; (B) with the sum-of-log-OR score, we estimated π at 37%, and the mean of the enriched component was 2.91. These results support that about one-third of familial MM cases are caused by polygenic inheritance of a high risk allele burden. (C-D) Corresponding density distribution plots for the same data.
Hypothetically, familial MM could be caused by rare mutations with strong effects that are not represented in the single-nucleotide polymorphism microarray data. For completeness, we therefore analyzed 29 of the Swedish cases plus 6 cases (from 3 families) from Germany by whole-exome sequencing. As expected from the small sample size, we did not detect any variants with exome-wide significance. Looking specifically for rare variants in genes known to be associated with MM16-19,24 (supplemental Table 3) and genes frequently somatically mutated in MM plasma cells25,26 (supplemental Methods), we detected 10 nonsynonymous variants with minor allele frequency less than 1%. Only 2 of these were found in more than 1 case, and none of them were obviously pathogenic (supplemental Table 5).
Discussion
In conclusion, we describe the first direct association between a polygenic feature and familial MM. No similar evidence has been put forward for any hematologic malignancy. Although the enrichment of common MM risk alleles among familial cases is of modest magnitude when calculated across the familial MM study group as a whole, our data indicate that polygenic risk scores for individuals with familial MM follow a bimodal distribution. We estimate that about one third of familial MM cases are caused by enrichments of risk alleles (Figure 2), whereas the rest follow the background distribution and likely have other causes. For example, the remaining two-thirds of cases could be caused by as yet undiscovered rare mutations or exposure to common environmental factors. Unfortunately, our sample size did not allow exome-wide screening for rare mutations. Another limitation of our study is that the Swedish registries do not record monoclonal gammopathy of unknown significance cases. We anticipate that future studies, including large-scale sequencing efforts, will provide a more detailed understanding of the genetic basis of familial MM and set the stage for clinical applications.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank Anna Collin, Adam Kiezun, and Magnus Jöud for their assistance. They are indebted to the clinicians and patients who participated in the project.
This work was supported by research grants from the Swedish Foundation for Strategic Research (KF10-0009), the Marianne and Marcus Wallenberg Foundation (2010.0112), the Knut and Alice Wallenberg Foundation (2012.0193), the Swedish Research Council (2012-1753), the Royal Swedish Academy of Science, ALF grants to the University and Regional Laboratories (Labmedicin Skåne), the Medical Faculty at Lund University, and the Swedish Society of Medicine. The German parts were supported by International Myeloma Foundation, Multiple Myeloma Research Foundation, the German Ministry of Education and Science (01ZX1309B), the Harald Huppert Foundation, and Deutsche Krebshilfe.
Authorship
Contribution: B.-M.H., B.N., and M.H. designed the research; I.T., B.N., M.H., U.-H.M., E.A., H.N., A.-K.W., E.J., K.L., K.H., A.W., N.W., H.G., C.L., M.P., A.F., U.G., and T.G. organized and carried out the collection of samples; B.-M.H., B.N., E.J., C.C., A.N., and M.A. carried out genetic analyses; B.-M.H. and B.N. analyzed data and drafted the manuscript; and all authors contributed to data interpretation and the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Björn Nilsson, Hematology and Transfusion Medicine, Department of Laboratory Medicine, BMC B13, 221 84 Lund, Sweden; e-mail: bjorn.nilsson@med.lu.se.
References
Author notes
M.H. and B.N. contributed equally to this study.