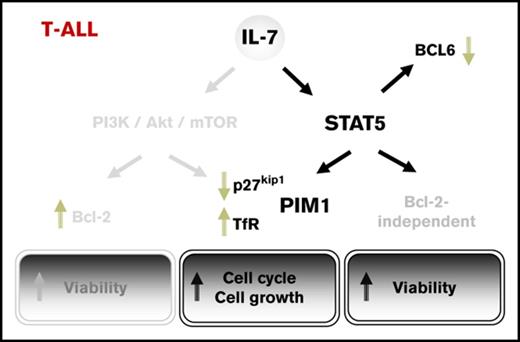
Key Points
STAT5 is required for IL-7–mediated proliferation and viability, but it does not regulate Bcl-2 downstream from IL-7 in T-ALL cells.
PIM1 is required for IL-7-induced leukemia cell cycle progression and proliferation and may be a therapeutic target for IL-7-reliant T-ALLs.
Abstract
T-cell acute lymphoblastic leukemia (T-ALL) constitutes an aggressive subset of ALL, the most frequent childhood malignancy. Whereas interleukin-7 (IL-7) is essential for normal T-cell development, it can also accelerate T-ALL development in vivo and leukemia cell survival and proliferation by activating phosphatidylinositol 3-kinase/protein kinase B/mechanistic target of rapamycin signaling. Here, we investigated whether STAT5 could also mediate IL-7 T-ALL-promoting effects. We show that IL-7 induces STAT pathway activation in T-ALL cells and that STAT5 inactivation prevents IL-7–mediated T-ALL cell viability, growth, and proliferation. At the molecular level, STAT5 is required for IL-7-induced downregulation of p27kip1 and upregulation of the transferrin receptor, CD71. Surprisingly, STAT5 inhibition does not significantly affect IL-7–mediated Bcl-2 upregulation, suggesting that, contrary to normal T-cells, STAT5 promotes leukemia cell survival through a Bcl-2-independent mechanism. STAT5 chromatin immunoprecipitation sequencing and RNA sequencing reveal a diverse IL-7-driven STAT5-dependent transcriptional program in T-ALL cells, which includes BCL6 inactivation by alternative transcription and upregulation of the oncogenic serine/threonine kinase PIM1. Pharmacological inhibition of PIM1 abrogates IL-7–mediated proliferation on T-ALL cells, indicating that strategies involving the use of PIM kinase small-molecule inhibitors may have therapeutic potential against a majority of leukemias that rely on IL-7 receptor (IL-7R) signaling. Overall, our results demonstrate that STAT5, in part by upregulating PIM1 activity, plays a major role in mediating the leukemia-promoting effects of IL-7/IL-7R.
Introduction
Interleukin-7 (IL-7), which is produced by stromal cells in the thymus and bone marrow, and by vascular endothelial cells, intestinal epithelium, keratinocytes, and follicular dendritic cells,1 is essential for normal lymphoid development.2-5 However, IL-7–mediated signaling may also contribute to T-cell leukemogenesis. IL-7 transgenic mice develop B- and T-cell lymphomas6 and spontaneous leukemias that originate in AKR/J mice associate with overexpression of the IL-7 receptor (IL-7R) α chain in developing thymocytes.7 IL-7 is present in the microenvironments where the malignant T cells arise and develop,8 and most human primary T-cell acute lymphoblastic leukemia (T-ALL) samples express IL-7Rα9 and proliferate in vitro in response to IL-7.10-12 Accordingly, we have shown that IL-7 accelerates human T-ALL development in vivo.13 However, the exact molecular mechanisms by which this microenvironmental factor contributes to T-ALL cell viability and proliferation remain only partially characterized.
In developing thymocytes, IL-7 activates phosphatidylinositol-3-kinase/protein kinase B (PKB/Akt) (PI3K/Akt) and signal transducer and activator of transcription (STAT) pathways.14-16 Both pathways are triggered by IL-7 in T-ALL cells as well.17,18 We previously showed that IL-7 promotes T-ALL cell proliferation and viability through activation of PI3K/Akt pathway17 and consequent downregulation of p27kip1 and upregulation of Bcl-2.11 On the other hand, STAT5 appears to be fundamental for IL-7-dependent murine lymphomagenesis,19 and T-ALL patient samples displaying IL-7Rα gain-of-function driver mutations are sensitive to JAK inhibitors.20-22 However, JAKs (namely JAK1 and JAK3) are the most upstream kinases in IL-7–mediated signal transduction, not only required for STAT activation but also for the activation of parallel pathways, such as PI3K/Akt.1,18,23,24 No studies have yet rigorously evaluated the impact of STAT signaling in IL-7 stimulation of human T-ALL cells.
Here, we confirm that IL-7 activates JAK1, JAK3, and STAT5 in T-ALL cells and show that STAT5 is required for increased cell viability, growth, and cell cycle progression induced by IL-7. However, in contrast to what has been previously proposed for normal T cells, IL-7–mediated upregulation of Bcl-2 in T-ALL is independent of STAT5 activity. This suggests that, in the context of IL-7 signaling, STAT5 promotes the viability of malignant T cells by alternative, Bcl-2-independent mechanisms. We further demonstrate that STAT5 directly downregulates BCL6 and promotes the expression of PIM1 in response to IL-7 stimulation and provide evidence that PIM1 plays a role in mediating IL-7 proliferative effects on T-ALL cells.
Methods
T-ALL cell cultures
Primary T-ALL cells collected from pediatric patients at diagnosis (Table 1) were isolated as previously described.13 In all cases, informed consent was obtained in accordance with the Declaration of Helsinki and under institutional ethical review board approval. TAIL7, an IL-7-dependent cell line that was established from the peripheral blood of a pediatric T-ALL patient,18 was cultured in RPMI 1640 medium (Life Technologies) supplemented with 5% fetal bovine serum (FBS; Biowest), 2 mM glutamine, penicillin/streptomycin (100 U/mL; Life Technologies), and 20 ng/mL of recombinant human IL-7 (Peprotech). HPB-ALL, an IL-7-responsive T-ALL cell line,25 was cultured in RPMI 1640 medium supplemented with 10% FBS, 2 mM glutamine, and penicillin/streptomycin. Primary T-ALL and patient-derived xenograft (PDX) samples were cultured in conditions similar to those of TAIL7. Before each experiment, TAIL7 cells were deprived of IL-7 for 24 hours; HPB-ALL cells were cultured in medium with 1% FBS for 24 hours prior to and during the course of the experiment. Unless otherwise indicated, IL-7 was used at 20 ng/mL in culture experiments and at 50 ng/mL for short-term stimulation (up to 120 minutes). In some experiments, we used the STAT5 small-molecule inhibitor N′-((4-oxo-4H-chromen-3-yl)methylene)nicotinohydrazide (100 µM, TAIL7 cells; 150 μM, primary T-ALL cells),26 the PIM1 inhibitor Smi-4a (90 μM; Merck/Calbiochem),27 the pan-PIM inhibitor AZD1208 (1 µM; Selleckchem),28 and the STAT inhibitor parthenolide (5 µM; Sigma-Aldrich).
Immunoblotting and antibodies
Whole cell lysates were prepared as described elsewhere,17 resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, transferred onto nitrocellulose membranes, and immunoblotted with antibodies against p-JAK1/3 (Y1022-Y1023/Y980) (Sigma), p-Akt (S473), Akt, p-STAT1 (Y701), p-STAT3 (Y705), p-STAT5a/b (Y694/Y699), Bcl-xL, Mcl-1, PIM1 (Cell Signaling Technology), p27kip1 (BD Transduction Labs), STAT5, BCL6, PIM1, ZAP-70, and actin (Santa Cruz Biotechnology). Immunodetection was performed by incubation with horseradish-peroxidase–conjugated appropriate secondary antibodies and developed by chemiluminescence.
STAT5 transcriptional activity and DNA binding
TAIL7 cells were nucleofected in a Nucleofector 2b using Solution V (Lonza) with pGL3-β-casein-firefly luciferase and pGL4-SV40-Renilla luciferase as control. Upon nucleofection, cells were left to recover in RPMI with 1% FBS for 12 hours. Cells were then stimulated or not with IL-7 for 24 hours and harvested. Luminescence was determined on an Infinite F500 luminometer (Tecan). The firefly or Renilla luciferase values in nonnucleofected cells were subtracted from the corresponding luciferase values in nucleofected cells. The ratio between firefly and Renilla was determined for the stimulated condition and normalized to the control (medium).
Viral transductions
For STAT5 knockdown, lentiviral plasmids expressing short hairpin RNAs (shRNAs) for STAT5A or scramble control hairpin were obtained from the RNAi Consortium.29 HPB-ALL cells were transduced by spin infection with Polybrene plus lentivirus. Similarly, for myristoylated-Akt (myr-Akt) overexpression, retroviral plasmids overexpressing myr-Akt-IRES-GFP or empty-IRES-GFP control (pLZRS) were used for retrovirus production. HPB-ALL cells were transduced by spin infection with Polybrene plus retrovirus. Upon transductions, viability was monitored daily thereafter.
Proliferation assays
Cells were cultured in triplicates in flat-bottom 96-well plates in the appropriate experimental conditions. Cells were incubated with 3H-thymidine (1 μCi per well) for the last 16 hours of culture before harvest. DNA synthesis, measured by 3H-thymidine incorporation, was assessed using a liquid scintillation counter. Results were expressed as average and standard error of triplicates.
Flow cytometry analyses of viability, cell size, and protein expression
Viability was determined using annexin V–based apoptosis detection kits and the manufacturers’ instructions (R&D Systems or eBioscience). Cell size was assessed by flow cytometry analysis of forward scatter vs side scatter physical parameters gated on the live cell population. Surface analysis of CD71 was done using phycoerythrin-conjugated CD71 antibodies (eBioscience). Intracellular staining of Bcl-2 was performed using a fluorescein isothiocyanate–conjugated Bcl-2 antibody (Dako). Briefly, cells were fixed using a formaldehyde-based fixation buffer and the manufacturer’s instructions (eBioscience), washed in phosphate-buffered saline, resuspended in 1× Perm/Wash Solution (BD Biosciences), and stained with the Bcl-2 antibody. Flow cytometry acquisition was performed in a FACS Calibur or an LSR Fortessa (BD Biosciences). Data analysis was done using FlowJo software (TreeStar). Results are expressed as percentage of positive cells and/or as mean fluorescence intensity.
Cell cycle analysis
Cells (1 × 106 to 2 × 106) were resuspended in phosphate-buffered saline and fixed and permeabilized with an equal volume of ice-cold 80% ethanol. Ribonuclease A was added at 50 μg/mL, and samples were incubated for 30 minutes at 37°C. Propidium iodide was added at a final concentration of 2.5 μg/mL, and samples were analyzed by flow cytometry. Cell cycle distribution was determined using ModFit LT software (Verity).
Reverse transcription quantitative polymerase chain reaction (qPCR)
RNA was extracted from 0.5 × 106 to 1 × 106 cells using TRIzol and the manufacturer’s instructions (Life Technologies). Total RNA (400 ng) was reverse-transcribed using Superscript II reverse transcriptase and random hexamers, according to the manufacturer’s instructions (Invitrogen). Complementary DNA and relevant primers were mixed with SYBR green master mix (Applied Biosystems) according to the manufacturer’s protocol. Reactions were performed in triplicates in ViiA7 system instruments (Applied Biosystems). Relative expression of the messenger RNAs (mRNAs) was estimated using the ddCt method, taking 18S expression as internal control. Primers used are described in supplemental Table 1.
Chromatin immunoprecipitation (ChIP)
RNA sequencing, ChIP sequencing (ChIP-seq), and data analysis
RNA sequencing (RNA-seq) library preparation was done to enrich for mRNAs using a 2-step protocol. Libraries were constructed and sequenced on a SOLiD/AB sequencer (Applied Biosystems). Details can be found in the supplemental Methods. Data were deposited in the Sequence Read Archive from the National Center for Biotechnology Information (accession number SRP139928).
Statistics
Data were analyzed using GraphPad Prism software (San Diego, CA). Differences between conditions were calculated using 2-tailed Student t test and 1-way analysis of variance, as appropriate, and considered significant for P < .05.
Results
IL-7 induces JAK/STAT pathway activation in T-ALL cells
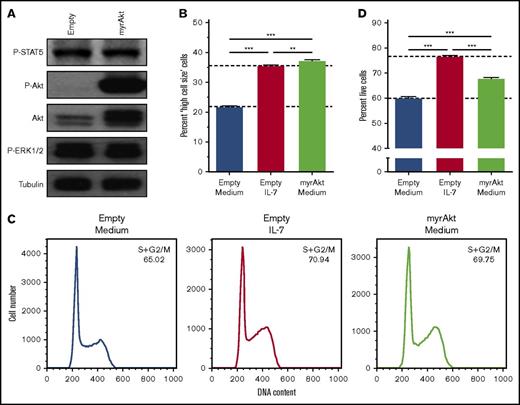
We previously showed that PI3K/Akt signaling pathway is mandatory for IL-7–mediated T-ALL cell survival, growth, and proliferation.17,32 Here, we first questioned whether activation of PI3K/Akt signaling is sufficient to reproduce the effects elicited by IL-7 on T-ALL cells. IL-7-responsive HPB-ALL cells were transduced with myristoylated Akt (myr-Akt), which displays constitutive kinase activity33 (Figure 1A). Akt activation alone is sufficient to fully mimic IL-7-induced cell growth (Figure 1B; supplemental Figure 1A), and, in accordance with the association between cell size and proliferation in lymphocytes,17,34 acceleration of cell cycle progression from G0/G1 to S and G2/M (Figure 1C). In contrast, myr-Akt only recapitulates partially the ability of IL-7 to promote T-ALL cell viability (Figure 1D; supplemental Figure 1B).
Akt activation fully mimics IL-7-induced T-ALL cell growth and cell cycle progression but does not completely recapitulate the effects on cell viability. HPB-ALL cells were stably transduced with retroviral vectors driving the expression of constitutively active, myristoylated-Akt (myrAkt), or empty control (Empty). (A) Transduced, regularly cultured HPB-ALL cells collected for immunoblot analysis of Akt expression (Akt) and PI3K/Akt, JAK/STAT5, and MEK/extracellular signal-regulated kinase (ERK) pathway activation (P-Akt, P-STAT5, and P-ERK1/2, respectively). Starved transduced HPB-ALL cells were stimulated or not with IL-7 (20 ng/mL) and were assessed by flow cytometry analysis of cell growth at 96 hours (B), cell cycle at 72 hours (C) and viability at 96 hours (D).
Akt activation fully mimics IL-7-induced T-ALL cell growth and cell cycle progression but does not completely recapitulate the effects on cell viability. HPB-ALL cells were stably transduced with retroviral vectors driving the expression of constitutively active, myristoylated-Akt (myrAkt), or empty control (Empty). (A) Transduced, regularly cultured HPB-ALL cells collected for immunoblot analysis of Akt expression (Akt) and PI3K/Akt, JAK/STAT5, and MEK/extracellular signal-regulated kinase (ERK) pathway activation (P-Akt, P-STAT5, and P-ERK1/2, respectively). Starved transduced HPB-ALL cells were stimulated or not with IL-7 (20 ng/mL) and were assessed by flow cytometry analysis of cell growth at 96 hours (B), cell cycle at 72 hours (C) and viability at 96 hours (D).
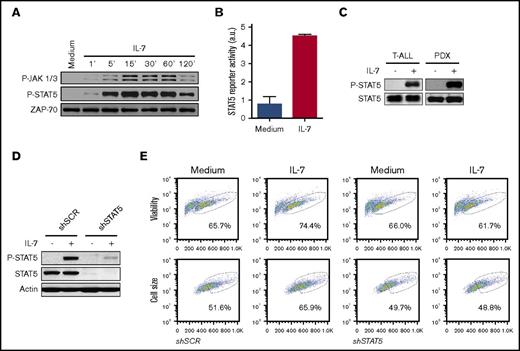
Because PI3K/Akt pathway activation is not sufficient to replace IL-7, we sought to identify additional functionally relevant signaling pathways that can operate concomitantly with Akt activation. MEK/ERK pathway inhibition does not prevent the effects triggered by IL-717 or, to a large extent, by mutational activation of the IL-7R.35 We then focused on JAK/STAT, the canonical pathway activated by cytokine receptor-mediated signaling, including IL-7/IL-7R.20,36,37 Using TAIL7 cells, which are appropriate surrogates for IL-7-dependent primary T-ALL cells,18,38 we confirmed that IL-7 stimulation induces JAK1, JAK3, and STAT5 phosphorylation18 (Figure 2A), leading to increased STAT5 transcriptional activity (Figure 2B). In agreement, IL-7 induces STAT5 activation in primary cells collected from pediatric T-ALL patients at diagnosis and in PDX samples (Figure 2C).
JAK/STAT5 pathway activation is required for IL-7-induced T-ALL cell viability and growth. (A) IL-7-starved TAIL7 cells were incubated with IL-7 for the indicated periods of time, and JAK-STAT5 pathway activation was analyzed by immunoblot. In the P-JAK1/3 panel, the upper bands correspond to P-JAK1 and the lower bands to P-JAK3. Results are representative of at least 2 independent experiments. (B) TAIL7 cells were nucleofected with pGL3-β-casein-firefly luciferase vector and pGL4-SV40-Renilla luciferase, followed by IL-7 stimulation for 24 hours. Luciferase activity from cell extracts was measured in a luminometer. STAT5 transcriptional activity was calculated as described in “Methods.” (C) Primary T-ALL or PDX cells were stimulated with IL-7 for 15 minutes or 2 hours, respectively, followed by immunoblot analysis of STAT5 activation. Data representative of 2 patients and 3 PDX analyzed. (D) HPB-ALL cells, stably transduced with STAT5A shRNA (shSTAT5) or scramble control (shSCR), were stimulated with IL-7 for 15 minutes and evaluated for STAT5 activation (P-STAT5) and total STAT5 expression. (E) Stably transduced HPB-ALL cells cultured for 72 hours in the indicated conditions were assessed for viability and cell growth. Results are representative of 3 independent experiments. Graphics represent average of triplicates ± standard error of the mean (SEM).
JAK/STAT5 pathway activation is required for IL-7-induced T-ALL cell viability and growth. (A) IL-7-starved TAIL7 cells were incubated with IL-7 for the indicated periods of time, and JAK-STAT5 pathway activation was analyzed by immunoblot. In the P-JAK1/3 panel, the upper bands correspond to P-JAK1 and the lower bands to P-JAK3. Results are representative of at least 2 independent experiments. (B) TAIL7 cells were nucleofected with pGL3-β-casein-firefly luciferase vector and pGL4-SV40-Renilla luciferase, followed by IL-7 stimulation for 24 hours. Luciferase activity from cell extracts was measured in a luminometer. STAT5 transcriptional activity was calculated as described in “Methods.” (C) Primary T-ALL or PDX cells were stimulated with IL-7 for 15 minutes or 2 hours, respectively, followed by immunoblot analysis of STAT5 activation. Data representative of 2 patients and 3 PDX analyzed. (D) HPB-ALL cells, stably transduced with STAT5A shRNA (shSTAT5) or scramble control (shSCR), were stimulated with IL-7 for 15 minutes and evaluated for STAT5 activation (P-STAT5) and total STAT5 expression. (E) Stably transduced HPB-ALL cells cultured for 72 hours in the indicated conditions were assessed for viability and cell growth. Results are representative of 3 independent experiments. Graphics represent average of triplicates ± standard error of the mean (SEM).
STAT5 is required for IL-7–mediated T-ALL cell survival, growth, and proliferation
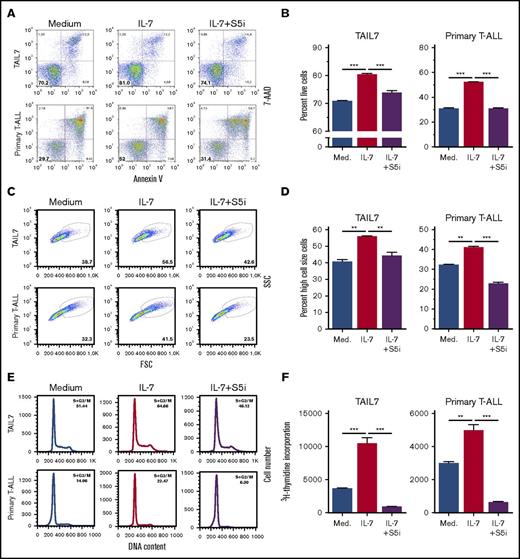
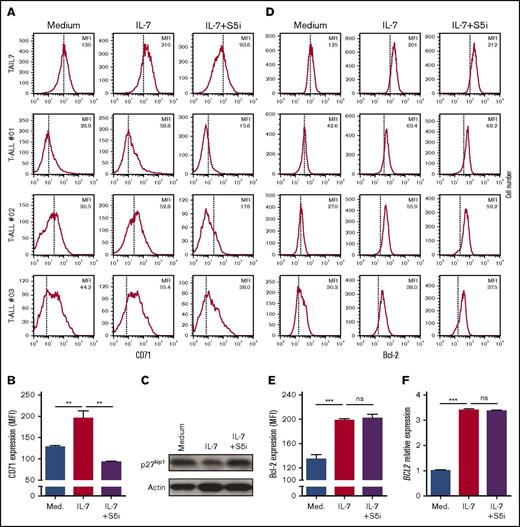
To determine the functional involvement of STAT5 in IL-7–mediated T-ALL cell stimulation, we transduced HPB-ALL cells with either STAT5A shRNA or scrambled control (Figure 2D). We observed that STAT5 silencing abrogates IL-7–mediated increase in T-ALL cell viability and cell size (Figure 2E). To further dissect the functional consequences of, and identify molecular mechanisms associated with, STAT5 inactivation, we cultured TAIL7 and primary T-ALL cells with IL-7 in the presence or absence of a STAT5-specific small-molecule inhibitor [S5i; N′-((4-oxo-4H-chromen-3-yl)methylene) nicotinohydrazide].20,26 STAT5 pharmacological inhibition completely prevents IL-7-induced T-ALL cell viability (Figure 3A-B), growth (Figure 3C-D), cell cycle progression (Figure 3E), and proliferation (Figure 3F). Taken together, these results demonstrate that STAT5 is required for the impact of IL-7 on T-ALL cells. In agreement, S5i prevents IL-7–mediated upregulation of the transferrin receptor, CD71 (Figure 4A-B), a marker of cell growth and proliferation,39 while reversing IL-7-induced downregulation of the cell cycle inhibitor p27kip1 (Figure 4C). These results are in line with those we obtained using the pan-STAT/NF-κB inhibitor parthenolide (supplemental Figure 2).
STAT5 inhibition abrogates IL-7–mediated T-ALL cell viability, cell growth, cell cycle progression, and proliferation. IL-7-deprived TAIL7 or primary T-ALL cells were cultured in medium, IL-7, or IL-7 plus S5i (100 µM and 150 µM, respectively). Cell viability (A-B), cell growth (C-D), cell cycle (E), and proliferation (F) were determined at 72 hours. Results are representative of at least 2 independent experiments or patients. Graphics represent average of triplicates ± SEM. **P < .01; ***P < .001.
STAT5 inhibition abrogates IL-7–mediated T-ALL cell viability, cell growth, cell cycle progression, and proliferation. IL-7-deprived TAIL7 or primary T-ALL cells were cultured in medium, IL-7, or IL-7 plus S5i (100 µM and 150 µM, respectively). Cell viability (A-B), cell growth (C-D), cell cycle (E), and proliferation (F) were determined at 72 hours. Results are representative of at least 2 independent experiments or patients. Graphics represent average of triplicates ± SEM. **P < .01; ***P < .001.
STAT5 inhibition abrogates IL-7–mediated upregulation of CD71, and downregulation of p27kip1, but not upregulation of Bcl-2 in T-ALL cells. IL-7-deprived TAIL7 or primary T-ALL cells were cultured for 72 hours in medium, IL-7, or IL-7 plus S5i, and analyzed for CD71 surface expression (A-B), expression of p27kip1 (C), and Bcl-2 levels (D-E). Mean fluorescence intensity (MFI) values are indicated for each flow cytometry histogram. Vertical lines indicate peak value in medium alone. (F) Under the same experimental conditions, TAIL7 cells were collected at 24 hours, and BCL2 transcript levels determined by qPCR. Fold induction is normalized to the medium alone. Results are representative of at least 2 independent experiments or 3 patients. Graphics represent average of triplicates ± SEM. **P < .01; ***P < .001. ns, not significant (P ≥ .05).
STAT5 inhibition abrogates IL-7–mediated upregulation of CD71, and downregulation of p27kip1, but not upregulation of Bcl-2 in T-ALL cells. IL-7-deprived TAIL7 or primary T-ALL cells were cultured for 72 hours in medium, IL-7, or IL-7 plus S5i, and analyzed for CD71 surface expression (A-B), expression of p27kip1 (C), and Bcl-2 levels (D-E). Mean fluorescence intensity (MFI) values are indicated for each flow cytometry histogram. Vertical lines indicate peak value in medium alone. (F) Under the same experimental conditions, TAIL7 cells were collected at 24 hours, and BCL2 transcript levels determined by qPCR. Fold induction is normalized to the medium alone. Results are representative of at least 2 independent experiments or 3 patients. Graphics represent average of triplicates ± SEM. **P < .01; ***P < .001. ns, not significant (P ≥ .05).
IL-7–mediated upregulation of Bcl-2 in T-ALL cells does not require STAT5 activity
Surprisingly, STAT5 inhibition does not block IL-7–mediated upregulation of Bcl-2 protein expression in T-ALL cells (Figure 4D-E). Likewise, BCL2 transcript levels are not negatively impacted by STAT5 pharmacological inhibition (Figure 4F) or STAT5 silencing (supplemental Figure 3). These results are in contradiction with what occurs in normal T cells, where Bcl-2 is a reported STAT5 target gene implicated in IL-7–mediated STAT5 function in developing thymocytes and mature T cells.40-42 However, in malignant T cells IL-7 was shown to upregulate Bcl-2 via PI3K/Akt pathway,17 which suggests that STAT5 may regulate leukemia T-cell survival by an alternative, Bcl-2-independent mechanism. Notably, this mechanism does not appear to rely on Bcl-2 family members Mcl-1 or Bcl-xL. IL-7 does not upregulate MCL1 transcript levels in TAIL7 cells (supplemental Figure 4), whereas IL-7–mediated upregulation of BCL2L1 (coding for Bcl-xL) is not reversed by STAT5 inhibition (supplemental Figure 5). In HPB-ALL cells, IL-7 upregulates both BCL2L1 and MCL1 by twofold to threefold, but STAT5 silencing has only a mild ability to reverse this effect. This is in sharp contrast to the major upregulation of CISH by IL-7 and the complete abrogation of this effect by STAT5 silencing (supplemental Figure 3).
IL-7-stimulated, STAT5-dependent transcriptional network analysis in T-ALL
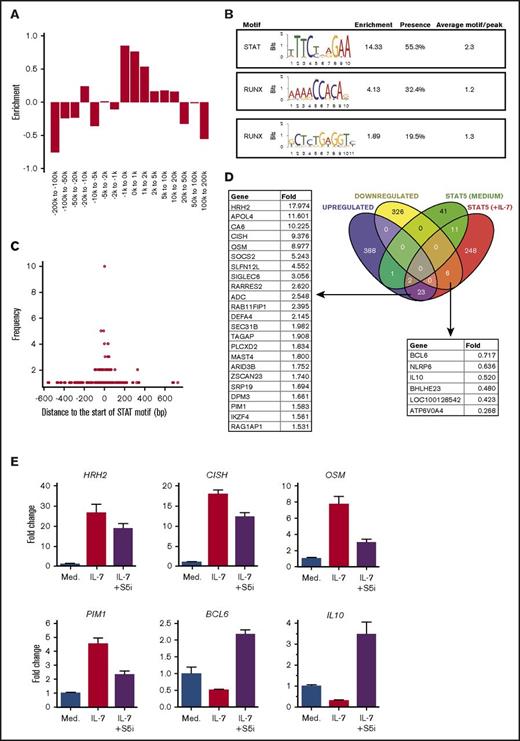
To gain insight into the STAT5-dependent transcriptional events associated with IL-7 stimulation in T-ALL cells, we performed STAT5 ChIP-seq and RNA-seq in the presence or absence of IL-7 with TAIL7 cells. ChIP-seq data analysis revealed that STAT5 binding is mainly enriched at core promoters but can be also detected at distal enhancers (Figure 5A). De novo motif analysis shows, as expected, a preferential enrichment for STAT DNA binding motifs upon IL-7 stimulation (Figure 5B). Interestingly, there is also enrichment for Runt-related transcription factor (RUNX) binding motifs (Figure 5B). We did not find motif enrichment in the IL-7-depleted samples. Activated STAT5 may bind DNA in a dimeric (requiring 1 binding site) or tetrameric (requiring 2 binding sites) form.43 Notably, peaks usually contain >1 STAT motif, with an average of 2.3 STAT motifs per peak (Figure 5B). This suggests that STAT5 may favor DNA binding as a tetrameric complex, a feature that has been associated with leukemia.44 Noteworthy, the STAT and the RUNX motifs (taken together) are both present in >50% of the peaks. A more detailed analysis revealed that a RUNX motif typically occurs close to a STAT motif and is particularly enriched for 1-bp distance (Figure 5C), indicating potential transcription factor interaction/competition.
Cross-analysis of STAT5 ChIP-seq and RNA-seq data on IL-7-stimulated TAIL7 cells. IL-7-deprived TAIL7 cells were cultured with or without IL-7 for 24 hours, and then collected for STAT5 ChIP-seq or RNA-seq enriched for mRNA. (A) Relative STAT5 peak enrichment/depletion in IL-7-stimulated cells. The relative enrichment of each STAT5 peak interval was calculated as described in “Methods.” Value of 0 equals random binding, negative values indicate STAT5 depletion, and positive values indicate STAT5 enrichment. (B) De novo motif discovery and identification on STAT5 ChiP-seq peaks from IL-7-stimulated cells. Enrichment cutoff at 1.5. The “Presence” column denotes the relative presence of the motif on all peaks. “Average motif/peak” denotes the number of times a motif appears on the peak. (C) Graph showing the distance, in base pairs (bp), of RUNX motifs to the STAT motif (horizontal axis) within each peak found in panel B, plotted against the frequency of each occurrence (vertical axis). (D) Venn diagram showing the overlap of genes found in the RNA-seq analysis (purple and yellow sets) and ChIP-seq analysis (green and red sets). Analysis was performed with genes with a STAT5a peak within 20 kb from the transcription start site. The genes with an IL-7-induced STAT5 peak whose expression was up- or downregulated by IL-7 are indicated on the left and right lists, respectively. (E) qPCR validation of ChIP-seq and RNA-seq data. IL-7-deprived TAIL7 cells cultured in medium, IL-7, or IL-7 plus S5i were collected for mRNA extraction and qPCR analysis at 24 hours (HRH2, CISH, OSM, and PIM1) or 48 hours (BCL6 and IL10). Fold change is normalized for the medium condition. Validation results are representative of at least 3 independent experiments. Results represent average of triplicates ± SEM.
Cross-analysis of STAT5 ChIP-seq and RNA-seq data on IL-7-stimulated TAIL7 cells. IL-7-deprived TAIL7 cells were cultured with or without IL-7 for 24 hours, and then collected for STAT5 ChIP-seq or RNA-seq enriched for mRNA. (A) Relative STAT5 peak enrichment/depletion in IL-7-stimulated cells. The relative enrichment of each STAT5 peak interval was calculated as described in “Methods.” Value of 0 equals random binding, negative values indicate STAT5 depletion, and positive values indicate STAT5 enrichment. (B) De novo motif discovery and identification on STAT5 ChiP-seq peaks from IL-7-stimulated cells. Enrichment cutoff at 1.5. The “Presence” column denotes the relative presence of the motif on all peaks. “Average motif/peak” denotes the number of times a motif appears on the peak. (C) Graph showing the distance, in base pairs (bp), of RUNX motifs to the STAT motif (horizontal axis) within each peak found in panel B, plotted against the frequency of each occurrence (vertical axis). (D) Venn diagram showing the overlap of genes found in the RNA-seq analysis (purple and yellow sets) and ChIP-seq analysis (green and red sets). Analysis was performed with genes with a STAT5a peak within 20 kb from the transcription start site. The genes with an IL-7-induced STAT5 peak whose expression was up- or downregulated by IL-7 are indicated on the left and right lists, respectively. (E) qPCR validation of ChIP-seq and RNA-seq data. IL-7-deprived TAIL7 cells cultured in medium, IL-7, or IL-7 plus S5i were collected for mRNA extraction and qPCR analysis at 24 hours (HRH2, CISH, OSM, and PIM1) or 48 hours (BCL6 and IL10). Fold change is normalized for the medium condition. Validation results are representative of at least 3 independent experiments. Results represent average of triplicates ± SEM.
Next, we compared the ChIP-seq with RNA-seq data in order to obtain information on the differential expression, upon IL-7 stimulation, of the genes displaying a STAT5 peak within 20 kb from the transcription start site. RNA-seq analysis confirmed that BCL2 transcript levels are upregulated by IL-7 (supplemental Table 2). However, in agreement with our observation that IL-7-dependent BCL2 upregulation is not dependent on STAT5 (Figure 4F), we did not detect any STAT5 peak in the BCL2 locus upon IL-7 stimulation (supplemental Table 3). Interestingly, we found that STAT5 could bind near the MCL1 gene, albeit without leading to its transcriptional activation (supplemental Figure 4), which is in agreement with the fact that IL-7 does not upregulate Mcl-1 protein in these cells (supplemental Figure 6). Genes upregulated by IL-7 in a STAT5-dependent manner in TAIL7 cells include HRH2, APOL4, and CA6, as well as, expectedly, OSM, IKZF4, PIM1, SOCS2, and CISH,45-49 whereas among the top downregulated genes, we found BCL6, NLRP6, and IL10 (Figure 5D; supplemental Figure 4). We validated these results by qPCR analysis of IL-7-treated TAIL7 cells in the presence or absence of S5i. Pharmacological inhibition of STAT5 partially or completely reverses IL-7–mediated gene upregulation (HRH2, CISH, OSM, and PIM1) and restores the expression of genes (BCL6 and IL10) downregulated by IL-7 (Figure 5E).
IL-7 downregulates BCL6 expression in T-ALL in a STAT5-dependent manner
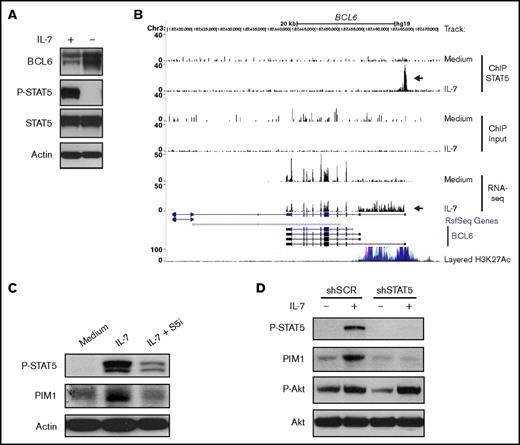
To explore some of the functional consequences of these observations, we focused our analysis on PIM1 and BCL6, genes that were respectively up- and downregulated by STAT5 downstream from IL-7 stimulation and that were potentially relevant mediators of IL-7 effects on T-ALL cells based on their known functions.50-54 BCL6 encodes for B-cell lymphoma 6 protein (BCL6), a transcriptional repressor and an important oncogene in diffuse large B-cell lymphoma.50 Moreover, BCL6 was shown to be an effector of resistance to chemotherapy in adult BCR-ABL–positive ALL,51 again indicating its oncogenic role. However, BCL6 can also act as a tumor suppressor in certain cancers.55 In immature T-cells, IL-7 was shown to repress BCL6.56 In addition, an IL-7/STAT5/BCL6 link was demonstrated in follicular helper T-cell differentiation57 and B-cell development.58 However, whether IL-7 regulates BCL6 in T-ALL has not been reported. In agreement with the downregulation of BCL6 transcript levels (Figure 5D), we observed that culture of TAIL7 cells with IL-7 sustains high STAT5 activation and low BCL6 protein expression, whereas IL-7 withdrawal leads to loss of STAT5 phosphorylation and augmented BCL6 levels (Figure 6A). Interestingly, IL-7–mediated binding of STAT5 in the BCL6 promoter region associates not only with significantly lower BCL6 expression but also with the transcription of an alternate longer variant that included the processing of intron 1 into the mRNA (Figure 6B). In agreement, IL-7 does not induce changes in the repressive mark H3K27me359 in the BCL6 STAT5 binding region (supplemental Figures 7A and 8A). Instead, both TAIL7 and HPB-ALL cells display constitutively high levels of H3K27ac, which associates with active enhancers.60 These levels are further upregulated by IL-7 in HPB-ALL cells (supplemental Figure 8A). The H3K4me3 modification, which is downregulated by IL-7 (supplemental Figures 7A and 8A), is found at high levels near transcription start sites,59 but can also occur at lower levels at enhancer regions.60,61 STAT5 silencing abrogates these IL-7–mediated histone modifications (supplemental Figure 8A).
IL-7 directly modulates PIM1 and BCL6 expression via STAT5. (A) TAIL7 cells were withdrawn or not from IL-7 for 96 hours and collected for immunoblot analysis of BCL6, P-STAT5, and STAT5. (B) Data from ChIP-seq and RNA-seq were uploaded to University of California, Santa Cruz genome browser visualization tool (top 6 tracks). The browser is centered on the human BCL6 gene locus (hg19). Custom tracks are paired as control (Medium) and IL-7. ChIP STAT5 track pair represents peaks found upon STAT5 IP, and the arrow highlights a STAT5 binding peak. Input track represents control input for ChIP. RNA-seq track represents mRNA expression. Peak height is proportional to the expression. The bottom arrow in RNA-seq tracks highlights the decrease in overall BCL6 gene expression and concomitant processing of intron 1 into the mRNA. (C) IL-7-deprived TAIL7 cells were cultured for 72 hours in medium, IL-7, or IL-7 plus S5i and analyzed for P-STAT5 and PIM1. (D) Serum-starved, stably transduced HPB-ALL cells were cultured with or without IL-7 for 24 hours and analyzed for P-STAT5, PIM1, and P-Akt. Results are representative of at least 3 independent experiments.
IL-7 directly modulates PIM1 and BCL6 expression via STAT5. (A) TAIL7 cells were withdrawn or not from IL-7 for 96 hours and collected for immunoblot analysis of BCL6, P-STAT5, and STAT5. (B) Data from ChIP-seq and RNA-seq were uploaded to University of California, Santa Cruz genome browser visualization tool (top 6 tracks). The browser is centered on the human BCL6 gene locus (hg19). Custom tracks are paired as control (Medium) and IL-7. ChIP STAT5 track pair represents peaks found upon STAT5 IP, and the arrow highlights a STAT5 binding peak. Input track represents control input for ChIP. RNA-seq track represents mRNA expression. Peak height is proportional to the expression. The bottom arrow in RNA-seq tracks highlights the decrease in overall BCL6 gene expression and concomitant processing of intron 1 into the mRNA. (C) IL-7-deprived TAIL7 cells were cultured for 72 hours in medium, IL-7, or IL-7 plus S5i and analyzed for P-STAT5 and PIM1. (D) Serum-starved, stably transduced HPB-ALL cells were cultured with or without IL-7 for 24 hours and analyzed for P-STAT5, PIM1, and P-Akt. Results are representative of at least 3 independent experiments.
PIM1 is required for IL-7-dependent increased T-ALL cell proliferation
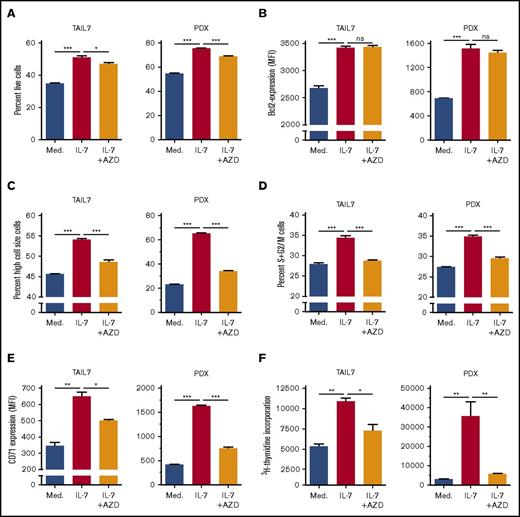
PIM1 kinase, encoded by PIM1, is frequently overexpressed in cancer, including hematological malignancies.52 Moreover, PIM1 is involved in cell cycle regulation53 and apoptosis,54,62 thereby being a possible alternative to Bcl-2-dependent prevention of apoptosis. Although known to be a transcriptional target of either IL-7 or STAT5 in other contexts,52 its involvement on STAT5-dependent IL-7–mediated effects on T-ALL was never evaluated. Treatment with S5i confirmed that IL-7 upregulates PIM1 protein expression in a STAT5-dependent manner (Figure 6C). This was further validated by knockdown of STAT5A, which prevents IL-7-dependent increase in PIM1 expression (Figure 6D). In agreement, IL-7 upregulates H3K27ac in the STAT5 binding region of the PIM1 locus both in TAIL7 and HPB-ALL cells (supplemental Figures 7B and 8B), an effect that is prevented by STAT5 silencing (supplemental Figure 8B). To evaluate the functional consequences of IL-7-dependent PIM1 upregulation, we treated TAIL7 and primary T-ALL cells with a PIM-specific inhibitor (AZD1208), which did not affect IL-7-induced cell viability (Figure 7A; supplemental Figure 9A) or Bcl-2 expression (Figure 7B; supplemental Figure 9B). Likewise, AZD1208 did not affect Mcl-1 or Bcl-xL expression (supplemental Figure 6). In contrast, PIM inhibition reversed IL-7–mediated cell growth (Figure 7C; supplemental Figure 9C), cell cycle progression (Figure 7D; supplemental Figure 9D), and CD71 expression (Figure 7E; supplemental Figure 9E). Accordingly, AZD1208 significantly downregulated IL-7–mediated proliferation (Figure 7F). Similar effects on cell size and proliferation were observed using an unrelated PIM1 inhibitor (supplemental Figure 10).
PIM inhibition abrogates IL-7–mediated T-ALL cell growth, proliferation, cell cycle progression, and CD71 expression but does not impact cell viability or Bcl-2 expression. IL-7-deprived TAIL7 or PDX T-ALL cells were cultured in medium, IL-7, or IL-7 plus the pan-PIM inhibitor AZD1208 (1 µM) and analyzed by flow cytometry for viability (A), intracellular Bcl-2 levels (B), cell size (C), CD71 surface expression (E), cell cycle (D), and proliferation (F). Results are representative of 2 independent experiments or 2 PDX. Results in graphics represent average of triplicates ± SEM. *P < .05; **P < .01; ***P < .001.
PIM inhibition abrogates IL-7–mediated T-ALL cell growth, proliferation, cell cycle progression, and CD71 expression but does not impact cell viability or Bcl-2 expression. IL-7-deprived TAIL7 or PDX T-ALL cells were cultured in medium, IL-7, or IL-7 plus the pan-PIM inhibitor AZD1208 (1 µM) and analyzed by flow cytometry for viability (A), intracellular Bcl-2 levels (B), cell size (C), CD71 surface expression (E), cell cycle (D), and proliferation (F). Results are representative of 2 independent experiments or 2 PDX. Results in graphics represent average of triplicates ± SEM. *P < .05; **P < .01; ***P < .001.
Discussion
IL-7/IL-7R signaling is essential for normal T-cell development, but it can also contribute to T-cell malignancy.63 In T-ALL, IL-7 rapidly activates PI3K/Akt, MEK/ERK, and JAK/STAT pathways,1,17,18,64 which contrasts with the heterogeneous ability of IL-7 to activate MEK/ERK pathway, and the delayed kinetics of Akt activation, in normal T-cells.48,65,66 In fact, all 3 pathways are activated not only by IL-7 binding to its receptor, leading to heterodimerization of IL-7Rα with γc chain,1,64 but also by mutational activation of IL-7Rα, which occurs in 9% to 10% of T-ALL cases, relies on receptor homodimerization, and does not require the ligand.20,67-69
We previously showed that MEK/ERK pathway is dispensable for IL-7-induced viability, growth, or proliferation of T-ALL cells cultured in vitro,17 whereas PI3K/Akt signaling plays a critical role in mediating IL-7 proleukemia effects.11,17,32 Also, JAK3 inhibition prevents IL-7–mediated viability and proliferation of T-ALL cells18 (and data not shown), and ruxolitinib, a clinical-stage JAK1/2 pharmacological inhibitor, displays activity against IL7R mutant T-ALL patient-derived samples.20,22 Ruxolitinib further revealed that IL-7–mediated T-ALL patient sample resistance to in vitro treatment with glucocorticoids depends on JAK1.70,71 However, JAK1 and JAK3 directly bind IL-7Rα and γc, respectively, and are the most upstream kinases in IL-7/IL-7R-mediated signaling, determining the subsequent activation of downstream effectors that include not only the canonical STAT5, as well as STAT1 and STAT315,42 (supplemental Figure 11), but also MEK/ERK and PI3K/Akt pathways.17,18 Consequently, it remains essentially unknown to what extent the JAK-to-STAT5 axis specifically determines the functional outcome of IL-7/IL-7R signaling in T-cell leukemia. Here, we demonstrate for the first time that, similar to PI3K/Akt, signaling via STAT5 is mandatory for IL-7–mediated survival, proliferation, and growth of T-ALL cells.
STAT5 activating mutations, reported in T-ALL,72-74 provide resistance to JAK inhibitors, which can be overcome by the Bcl-2 family inhibitor navitoclax.74 Moreover, STAT5 induces the expression of Bcl-2 or Bcl-xL in different scenarios,75-77 including downstream from IL-7 during normal mouse B-cell development,40,78 and enforced expression of Bcl-2 in Il7r-deficient mice restores thymopoiesis in stages where IL-7 has a major prosurvival role.79 In recent thymic emigrants, IL-7–mediated activation of STAT5 is associated with survival, whereas activation of PI3K/Akt pathway is associated with proliferation.80 These observations suggest that in mature T cells IL-7 upregulates Bcl-2 in a STAT5-dependent manner. However, there is also evidence that IL-7 promotes survival and proliferation of early T-cell progenitors via PI3K/Akt pathway,16 which is compatible with PI3K/Akt, rather than STAT5, regulating Bcl-2 in healthy thymocytes. Notably, we found that IL-7–mediated upregulation of BCL2 was unaffected by STAT5 inhibition or silencing in T-ALL cells. This is in line with studies directly demonstrating that IL-7 upregulates Bcl-2 via a PI3K/Akt-dependent mechanism in T-ALL.17 Whether the uncoupling of STAT5 and Bcl-2 merely reflects the immaturity of T-ALL blasts or is a specific feature of transformed T cells remains to be determined. We should also cautiously note that, although highly consistent, our results were based on the analysis of 2 cell lines (TAIL7 and HPB-ALL), and a limited number of primary or PDX T-ALL cases (Table 1). Thus, it remains possible that T-ALL cells may have heterogeneous wiring of signaling networks and that some cases may depend on STAT5 for regulation of Bcl-2. Nonetheless, the T-ALL samples and cell lines we analyzed are heterogeneous in immunophenotype (ranging from pre-T-ALL to mature T-ALL) and genetic subtype (eg, TAL-LMO, HOXA, or TLX3), suggesting that the inability of STAT5 to upregulate Bcl-2 upon IL-7 stimulation is not restricted to a particular T-ALL subset.
De novo motif analysis of our ChIP-seq data resulted in the identification of RUNX DNA-binding motifs close to STAT motifs, with a great proportion being 1 bp apart. Interaction between STAT5 and RUNX proteins has been reported previously and characterized as mutually inhibitory.81 Runx1−/− mice have a high propensity to develop chemically induced T-cell lymphomas, suggesting a tumor suppressor role for Runx182 that was confirmed by more recent reports.83,84 However, other studies indicate that in TAL1-overexpressing T-ALL, TAL1, GATA3, and RUNX1 form a positive autoregulatory loop promoting expression of MYB and TRIB2 genes and survival of leukemia cells,85 suggesting that RUNX1 may be involved in leukemogenesis in some cases. Notably, expression of IL-7Rα in thymocytes and mature CD4 T cells was shown to be positively regulated by RUNX1.86 More detailed studies are warranted to determine how RUNX1 and STAT5 may crosstalk in the context of IL-7-dependent leukemias.
Our observation that BCL6 was directly downregulated by STAT5 upon IL-7 stimulation is in line with previous studies in normal thymocytes and follicular helper T cells.56,87 Our data further demonstrate that BCL6 repression in IL-7-cultured T-ALL cells is not a simple shutdown of transcription. Instead, it is accompanied by the processing of intronic DNA into mRNA, suggestive of changes in alternative transcript variant expression, which, by mechanisms we did yet not characterize, ultimately results in diminished expression of the canonical BCL6 protein. How BCL6 downregulation contributes to IL-7 leukemogenic effects also remains unexplored. Recently, IL-7 was shown to repress Bcl6 during β-selection thereby contributing to transient self-renewal and delayed differentiation of DN4 thymocytes,56 features required for the massive proliferation of β-selected thymocytes. In this light, one may hypothesize that inhibition of BCL6 by IL-7 in T-ALL cells reflects a normal physiological process that, nonetheless, has obvious potential to contribute to transformation (by diminishing differentiation and dramatically promoting proliferation).
Our functional studies focused on another IL-7-STAT5 target, PIM1, for a number of reasons. Although both IL-7 and STAT5 upregulate PIM1,46 IL-7 regulates PIM1 via JunD/AP-1 in a mouse T-cell line,88 and there was no formal demonstration, to our knowledge, of the actual existence of an IL-7/STAT5/PIM1 axis. Moreover, PIM1 is a well-known oncogene involved in different solid tumors and hematological malignancies, including T-ALL.89-92 Finally, PIM1 is a serine/threonine kinase and thus more easily targetable than STAT5. In fact, a number of PIM kinase small-molecule inhibitors have been developed that hold clinical promise93 and entered clinical trials for acute myeloid leukemia and other cancers (eg, AZD1208; www.clinicaltrials.gov, #NCT01588548). Notably, our results in human T-ALL are in line with very recent studies on mouse T-ALL cells indicating PIM1 as a JAK-STAT pathway target downstream from IL-7.49
Overall, our work unveiled STAT5 as mandatory for IL-7/IL-7R-dependent T-ALL cell survival and proliferation, in large part by activating the proto-oncogene PIM1. Small-molecule inhibitors specific for PIM1 eliminate IL-7–mediated proleukemia effects and therefore may constitute attractive tools for therapeutic intervention in T-ALL.
The data reported in this article have been deposited in the Sequence Read Archive (accession number SRP139928).
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank Luís Moita for providing the shRNAs for STAT5, Sérgio Almeida for valuable advice on ChIP for epigenetic marks, and Luís Vinagre and Afonso Almeida for their help. The authors especially thank the generosity of patients and their families, and the collaboration of the team from the Pediatrics Service of Instituto Português de Oncologia de Lisboa.
This work was supported by LISBOA-01-0145-FEDER-007391, project cofunded by FEDER, through POR Lisboa2020 Programa Operacional Regional de Lisboa, PORTUGAL 2020, and Fundação para a Ciência e a Tecnologia (FCT, Lisbon, Portugal), by the grants PTDC/SAU-OBD/104816, PTDC/SAU-ONC/122428, and FAPESP/20015/2014 grant from FCT, and by the consolidator grant ERC CoG-648455 from the European Research Council (J.T.B.). J.T.B. is an FCT investigator (consolidator). D.R. and A.M. received FCT fellowships.
Authorship
Contribution: D.R. designed and performed experiments, analyzed and interpreted data, and wrote the manuscript; A.M. designed and performed experiments and analyzed and interpreted data; R.v.B. analyzed and interpreted data regarding next generation sequencing experiments; C.I.S., M.C.S., A.S., and B.A.C. performed experiments and analyzed and interpreted data; P.J.C. contributed to the supervision of the next generation sequencing experiments; J.T.B. supervised and coordinated the studies, designed research, analyzed and interpreted data, and wrote the manuscript; and all authors critically read and contributed to the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: João T. Barata, Instituto de Medicina Molecular João Lobo Antunes, Lisbon University Medical School, Av. Prof. Egas Moniz, 1649-028 Lisboa, Portugal; e-mail: joao_barata@medicina.ulisboa.pt.