Key Points
Intronic variants of PKLR are associated with hospitalization rate for acute pain in sickle cell disease.
Allele-specific expression analysis suggests that variants influence PKLR activity, providing a functional basis for the association.
Abstract
Acute pain, the most prominent complication of sickle cell disease (SCD), results from vaso-occlusion triggered by sickling of deoxygenated red blood cells (RBCs). Concentration of 2,3-diphosphoglycerate (2,3-DPG) in RBCs promotes deoxygenation by preferentially binding to the low-affinity T conformation of HbS. 2,3-DPG is an intermediate substrate in the glycolytic pathway in which pyruvate kinase (gene PKLR, protein PKR) is a rate-limiting enzyme; variants in PKLR may affect PKR activity, 2,3-DPG levels in RBCs, RBC sickling, and acute pain episodes (APEs). We performed a candidate gene association study using 2 cohorts: 242 adult SCD-HbSS patients and 977 children with SCD-HbSS or SCD-HbSβ0 thalassemia. Seven of 47 PKLR variants evaluated in the adult cohort were associated with hospitalization: intron 4, rs2071053; intron 2, rs8177970, rs116244351, rs114455416, rs12741350, rs3020781, and rs8177964. All 7 variants showed consistent effect directions in both cohorts and remained significant in weighted Fisher's meta-analyses of the adult and pediatric cohorts using P < .0071 as threshold to correct for multiple testing. Allele-specific expression analyses in an independent cohort of 52 SCD adults showed that the intronic variants are likely to influence APE by affecting expression of PKLR, although the causal variant and mechanism are not defined.
Introduction
Episodic acute pain is the most prominent complication of sickle cell disease (SCD), a result of microvascular vaso-occlusion triggered by hemoglobin S (HbS) polymerization and sickling of red blood cells. HbS polymerizes only when deoxygenated, and a key factor influencing HbS oxygenation is the intracellular concentration of 2,3-diphosphoglycerate (2,3-DPG).1 2,3-DPG is a glycolytic intermediate2 and allosteric effector1 that decreases oxygen affinity by preferentially binding to the low-affinity T conformation. It also stabilizes deoxygenated HbS fibers and further promotes polymerization by decreasing the intraerythrocyte pH.3,4 Pyruvate kinase (gene, PKLR; protein, PKR) is a rate-limiting enzyme in glycolysis2 ; reduced PKR activity leads to accumulation of the upstream enzyme intermediates, including 2,3-DPG, and deficiency of ATP, factors detrimental to SCD.1 Indeed, coinheritance of PKR deficiency and heterozygous HbS may induce sickling, causing an SCD phenotype.5,6
PKR levels comprise a spectrum7 and could represent a quantitative trait that modifies the risk of sickling and frequency of acute pain episodes (APEs). In this study, using annualized hospitalization rates for APEs and allelic imbalance expression analysis, we suggest that PKLR variants are associated with acute sickle pain by perturbing PKLR gene expression.
Patients and methods
The population of the genetic association study consisted of 2 cohorts: (1) 242 adults with HbSS from King’s College Hospital (KCH; London, United Kingdom) with 10-year hospitalization records,8 and (2) 977 children with HbSS or HbSβ0 thalassemia from the Silent Infarct Transfusion (SIT) trial (registered at www.clinicaltrials.gov as #NCT00072761)9,10 with 3-year hospitalization records.10,11 Both studies were approved by the local institutional review boards at KCH (LREC 01-083) and Vanderbilt University Medical Center, respectively. Genome scan for the KCH cohort samples was performed using the lllumina Infinium MEGA chip.8 The SIT cohort DNA samples were genotyped using Illumina HumanHap650Y array 5 or Illumina Infinium HumanOmni1-Quad array. The results were quality controlled followed by genotype imputation based on the 1000 Genomes Project phase 3 data. An annualized hospitalization rate as a measure of acute pain incidence rate was calculated by dividing the number of hospital admissions for acute pain by the number of years of observation (Table 1) and double-loge transformed for statistical analysis.
An independent cohort for evaluation of imbalance in allele expression comprised 52 adults with SCD enrolled under 3 protocols, NCT00011648, NCT00081523, and NCT03685721, approved by the National Heart, Lung, and Blood Institute Review Board (National Institutes of Health). For the allele expression assays, a synonymous variant, rs1052176 (R596R) in exon 11 of PKLR, acted as a marker of relative expression levels of the 2 alleles of the gene using the respective genomic DNA as control. A total of 279 samples were screened to obtain 52 heterozygotes for R596R, of which 29 were also heterozygous for the associated intron 2 variants (test) and 23 did not have the associated intron 2 variants (control). Allele-specific expression analysis was carried using the Bio-Rad droplet digital polymerase chain reaction system (version 1.7.4.0917; Bio-Rad Laboratories, Hercules, CA).
Details on study cohort and analytic methodology are provided in the data supplement.
Results and discussion
Forty-seven variants (supplemental Table 1) were identified in the PKLR locus and evaluated for association with hospitalization rate in the KCH cohort, using a modified significance level of P < .001268 after correction for multiple testing (Cheverud et al12 method taking into account linkage disequilibrium [LD]). Seven variants (1 in intron 4 [rs2071053] and 6 in intron 2 [rs8177970, rs116244351, rs114455416, rs12741350, rs3020781, and rs8177964]) were significantly associated (Table 2; Figure 1A).8 Eighty-three variants were identified in the PKLR gene in the SIT cohort (supplemental Table 2). All 7 associated variants in the KCH cohort showed consistent effect directions in the SIT cohort but did not reach statistical significance (Table 2). Using weighted Fisher’s meta-analyses,13 we summarized the findings for all 7 variants in both cohorts (Table 2).
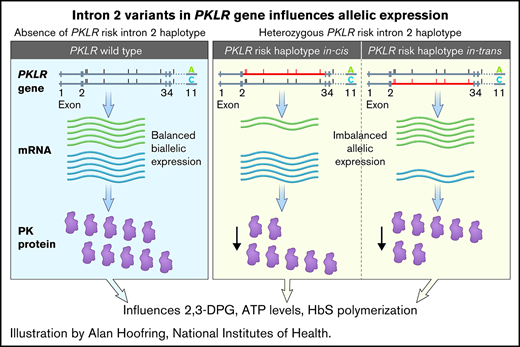
PKLR intronic risk variants and allelic specific expression. (A) PKLR gene with associated variants in intron 2 and coding variant R596R. Line representation of the PKLR gene with the associated intron variants (6 in intron 2 and 1 in intron 4). rs1052176 (R596R) is the synonymous single-nucleotide polymorphism in exon 11 that was used as a marker for allele-specific expression analysis. (B) PKLR allelic-specific expression analysis. Fifty-two heterozygous R569R individuals were selected: 23 without and 29 heterozygous for the associated PKLR intron 2 risk haplotype. Allele-specific expression analysis was carried out using the Bio-Rad droplet digital polymerase chain reaction system (QX200 Droplet Digital PCR System; Bio-Rad Laboratories) and analyzed with QuantSoft software. Ratios of the 2 R596R alleles in complementary DNA (cDNA) and genomic DNA (gDNA), respectively, were derived for each sample. The absolute difference in the ratios between the cDNA and gDNA samples was then calculated for each participant. The average expression ratio in those heterozygous for the variant intron 2 haplotype was 0.2073 (± standard deviation [SD], 0.0135) compared with an average expression ratio of 0.1239 (± SD, 0.0682) in those without the variant intron 2 (homozygous wild type). Wilcoxon rank sum test P = .0297.
PKLR intronic risk variants and allelic specific expression. (A) PKLR gene with associated variants in intron 2 and coding variant R596R. Line representation of the PKLR gene with the associated intron variants (6 in intron 2 and 1 in intron 4). rs1052176 (R596R) is the synonymous single-nucleotide polymorphism in exon 11 that was used as a marker for allele-specific expression analysis. (B) PKLR allelic-specific expression analysis. Fifty-two heterozygous R569R individuals were selected: 23 without and 29 heterozygous for the associated PKLR intron 2 risk haplotype. Allele-specific expression analysis was carried out using the Bio-Rad droplet digital polymerase chain reaction system (QX200 Droplet Digital PCR System; Bio-Rad Laboratories) and analyzed with QuantSoft software. Ratios of the 2 R596R alleles in complementary DNA (cDNA) and genomic DNA (gDNA), respectively, were derived for each sample. The absolute difference in the ratios between the cDNA and gDNA samples was then calculated for each participant. The average expression ratio in those heterozygous for the variant intron 2 haplotype was 0.2073 (± standard deviation [SD], 0.0135) compared with an average expression ratio of 0.1239 (± SD, 0.0682) in those without the variant intron 2 (homozygous wild type). Wilcoxon rank sum test P = .0297.
We hypothesize that the intronic variants alter gene expression in cis, detectable as allelic imbalance in PKLR expression.14,15 Screening of PKLR revealed a synonymous variant (R596R; rs1052176) in exon 11 (Figure 1A); its prevalence in almost 50% of National Institutes of Health patients with SCD makes it a highly suitable marker for differentiating expression of the 2 alleles. We examined the pairwise LD between PKLR variants and found that all 6 intron 2 variants were in tight LD, whereas R596R belonged to another LD block (supplemental Figure 1). Two of the associated intron 2 variants, rs8177964 and rs8177970, were also identified in a child heterozygous for PKLR c.994G>A mutation, and rs8177964 was proposed as causing rapid messenger RNA degradation using in silico analysis.16 Allele-specific sequence analysis (Sanger sequencing) showed minimal amounts of complementary DNA transcribed from the PKLR allele in phase with the risk intron 2 variant rs8177964, supporting results of the in silico analysis.16
Here, we show that rs8177964 and rs8177970 were inherited in an LD block with the other 4 intron 2 variants (supplemental Figure 1), and we defined this LD block as the intron 2 risk haplotype if individuals carried any minor allele from these 2 variants. Fifty-two individuals with SCD were heterozygous for the R596R variant, of whom 29 were also heterozygous for the intron 2 risk haplotype associated with APEs in SCD (test group), whereas 23 individuals did not have the risk haplotype (control) (Table 3; supplemental Table 3). We compared variation in PKLR expression between the 2 alleles in individuals heterozygous for and without the intron 2 risk haplotype, using genomic DNA as internal control for each individual. A Wilcoxon rank sum test revealed significant deviation from the expected expression ratio in those heterozygous for the intron 2 risk haplotype (mean, 0.2073, +/− SD 0.0135) when compared with to those without the “risk” haplotype (mean ± standard deviation, 0.1239 ± 0.0682; P = .0297; Figure 1B; supplemental Table 3). Our data support imbalance in allelic expression in individuals carrying the associated intron 2 risk haplotype, but it is not clear if the risk haplotype leads to an increase or decrease of PKLR gene expression because the heterozygous transcribed variant (R596R) is in imperfect linkage with the associated risk haplotype. It is also not clear which of the 6 intron 2 variants is causal, because they are in tight LD.
The frequency of acute pain varies widely in SCD, even within the same genotypic group, and is a measure of disease severity and a predictive marker of early mortality.11 Numerous genetic variants have been proposed as modifiers of acute pain, but none have been confirmed for the main reason that capturing APE frequency is challenging because of the complexity of the phenotype. A major strength of our study is the use of annualized hospitalization as a robust marker for acute pain. The lack of replication may be related to the many differences in the 2 cohorts, including the different age groups (adults vs pediatric), different health care settings, and different clinical definitions of what constitutes a pain episode requiring hospital admission. It was noted that the frequency of hospitalization was much lower in the adult KCH cohort compared with the SIT pediatric cohort. This difference could have arisen from a number of factors: (1) the observation in the KCH cohort is >10 years compared with the SIT cohort, for which observation is >3 years; (2) the KCH cohort is from a single center that provides both secondary and tertiary care, with hospital records that have been carefully curated over 10 years, whereas the SIT study enrolls participants from multiple sites, and medical records may not be as complete; and (3) there could also be cultural differences with regard to threshold levels of pain for presentation to hospital. Additionally, the KCH cohort is ethnically more homogeneous compared with the SIT cohort, which could explain some differences in the frequencies of the variants and the β coefficient effects, although the latter were in the same direction.
The introduction of genetic testing using next-generation sequencing technologies has revealed an increasing number of PKLR variants,17,18 which include deep intronic variants as encountered here, the pathologic significance of which is often unclear. Allele-specific expression analyses16 or more complex minigene constructs19 are 2 approaches that have been used to evaluate their pathogenicity. The minigene construct approach is set up for detection of alternative splicing20 and will not be practical for detecting RNA stability, which has been suggested as the mechanism underlying the reduced PKLR expression for rs8177964 based on in silico analyses.16
We identified 7 PKLR variants (1 in intron 4 and 6 in intron 2) that were significantly associated with acute sickle pain using annualized hospitalization as a surrogate. The 6 intron 2 variants are in an LD block; 2 (rs8177964 and rs8177970) were also identified in a child heterozygous for PKLR c.994G>A mutation with severe anemia, and rs8177964 was proposed as causing rapid messenger RNA degradation using in silico analysis.16 Allele-specific expression analysis using Sanger sequencing analysis of the proband who was a carrier for rs8177964 showed reduced PKLR expression in cis.16 This report adds to the widening literature that noncoding PKLR variants can contribute to PK deficiency. Of note, the proband inherited rs8177964 and rs8177970 from the mother, who was heterozygous for the variants and who was reported to have low normal hemoglobin accompanied by a 50% reduction in PK activity.
The rs8177964 variant is present in homozygosity in 61 of 4350 people of African or African American ethnicity in the gnomAD database.21 No clinical or laboratory details are available on these individuals who are presumably asymptomatic, bearing in mind that there is wide variability in PK deficiency.22
To circumvent analytic challenges resulting from low expression of PKLR in peripheral red blood cells and potentially small differences in expression associated with the intronic variants, we compared relative expression levels of 2 alleles of the same gene using RNA and genomic DNA from the same individual.23 Allelic variation in human gene expression is common, and population-based studies provide increasing evidence that noncoding variants influence trait variance and, quite likely, response to pharmacotherapy.15,23 Studies have suggested that PK deficiency provides protection from malaria infection, accounting for the high frequency of PKLR variants in sub-Saharan African populations.24,25 Indeed, the PKLR intron 2 variants described here have a minor allele frequency of 14% to 15% in African populations in the 1000 Genomes Project and are almost absent in non-African populations.
This preliminary study highlights that a group of PKLR intron 2 variants in LD affects gene expression, but it is not clear from this study if presence of the intron 2 risk haplotype increases or decreases PKLR expression; it is also not clear which of the 6 intron 2 variants is causative.
Nonetheless, our results support PKLR as a genetic modifier of acute pain in SCD and activating PKR as an antisickling strategy. Two different PKR activators, mitapivat (NCT04000165)26 and FT4202 (NCT03815695),27 are now under clinical development. It should be noted that the intron 2 PKLR variants are frequent and may underlie variation in response to PKR activators, although the influence may be subtle given the myriad of factors that could influence sickling.
Acknowledgments
The authors thank Michael Debaun (Vanderbilt-Meharry-Matthew Walker Center of Excellence in Sickle Cell Disease, Vanderbilt University School of Medicine, Nashville, TN) for providing access to the SIT trial data set, Neal Jeffries (Office of Biostatistics Research, National Heart, Lung, and Blood Institute, National Institutes of Health), Laxminath Tumburu, and members of the Thein laboratory for helpful discussions. The authors are thankful for the excellent support received from the clinical staff of the Sickle Cell Branch.
This work was supported by the Division of Intramural Research, National Heart, Lung, and Blood Institute, National Institutes of Health.
Authorship
Contribution: S.L.T. developed the idea of a candidate gene association study with K.G. and designed the study with X.W., K.G., and M.B.T. S.L.T., X.W., and K.G. wrote the paper; X.W., M.B.T., and C.A. performed genetic and droplet digital polymerase chain reaction experiments and analyses. F.T.S., C.L.D., and M.P. provided bioinformatic support. F.T.S., S.M., H.P., Y.-P.F., M.P., and K.G. performed statistical analyses. S.L.T. supervised the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Swee Lay Thein, National Heart, Lung and Blood Institute/National Institutes of Health, Building 10-CRC, Room 6S241, 10 Center Dr, Bethesda, MD 20892; e-mail: sl.thein@nih.gov.
References
Author notes
X.W., K.G., and M.B.T. contributed equally to this work.
Requests for data sharing may be submitted to Swee Lay Thein (sl.thein@nih.gov).
The full-text version of this article contains a data supplement.

![PKLR intronic risk variants and allelic specific expression. (A) PKLR gene with associated variants in intron 2 and coding variant R596R. Line representation of the PKLR gene with the associated intron variants (6 in intron 2 and 1 in intron 4). rs1052176 (R596R) is the synonymous single-nucleotide polymorphism in exon 11 that was used as a marker for allele-specific expression analysis. (B) PKLR allelic-specific expression analysis. Fifty-two heterozygous R569R individuals were selected: 23 without and 29 heterozygous for the associated PKLR intron 2 risk haplotype. Allele-specific expression analysis was carried out using the Bio-Rad droplet digital polymerase chain reaction system (QX200 Droplet Digital PCR System; Bio-Rad Laboratories) and analyzed with QuantSoft software. Ratios of the 2 R596R alleles in complementary DNA (cDNA) and genomic DNA (gDNA), respectively, were derived for each sample. The absolute difference in the ratios between the cDNA and gDNA samples was then calculated for each participant. The average expression ratio in those heterozygous for the variant intron 2 haplotype was 0.2073 (± standard deviation [SD], 0.0135) compared with an average expression ratio of 0.1239 (± SD, 0.0682) in those without the variant intron 2 (homozygous wild type). Wilcoxon rank sum test P = .0297.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/6/11/10.1182_bloodadvances.2021006668/2/m_advancesadv2021006668f1.png?Expires=1764402921&Signature=XZjqIlgTVHgrRWo93Tfo56fanUe3Ew7EdlJfmLbTz4CJUh-VHWw0eVm~QZ-v3durPMMP1iBwWAS3ivJ1H3DZO4Rf9mBvPE46e4hMlhkE22vfeqmwdTAMNRr930Pej3qnz1L9iYdnDnmhmb8z~b2L6Pm0Y54IBuf~KAuOiXc3y21RC-HIoTXUKU5-1K2xh2XJDoaAgTPJLhP3H7azc4aAz8HmHP0YFB-Yq0OWbGC~z5uzuFh5QJyydmW59YtTL-5IBl2NyTLXFV8LchuL2AxLquNqbGZQKu4uiMj6EI6o3~hsejPZiRgO53WlIBG4eMTWrCGmxNDl2ZAGC4PyGMj4pw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![PKLR intronic risk variants and allelic specific expression. (A) PKLR gene with associated variants in intron 2 and coding variant R596R. Line representation of the PKLR gene with the associated intron variants (6 in intron 2 and 1 in intron 4). rs1052176 (R596R) is the synonymous single-nucleotide polymorphism in exon 11 that was used as a marker for allele-specific expression analysis. (B) PKLR allelic-specific expression analysis. Fifty-two heterozygous R569R individuals were selected: 23 without and 29 heterozygous for the associated PKLR intron 2 risk haplotype. Allele-specific expression analysis was carried out using the Bio-Rad droplet digital polymerase chain reaction system (QX200 Droplet Digital PCR System; Bio-Rad Laboratories) and analyzed with QuantSoft software. Ratios of the 2 R596R alleles in complementary DNA (cDNA) and genomic DNA (gDNA), respectively, were derived for each sample. The absolute difference in the ratios between the cDNA and gDNA samples was then calculated for each participant. The average expression ratio in those heterozygous for the variant intron 2 haplotype was 0.2073 (± standard deviation [SD], 0.0135) compared with an average expression ratio of 0.1239 (± SD, 0.0682) in those without the variant intron 2 (homozygous wild type). Wilcoxon rank sum test P = .0297.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/6/11/10.1182_bloodadvances.2021006668/2/m_advancesadv2021006668f1.png?Expires=1764402922&Signature=Ekrtg9zma2cjptU18NWAD1suX~t2r9Wb3pPgNhVtOvyK9MBxtLJ8jPdE131Pn4771RSsG9pXb5CBS1Z8OIu5sIXFS~9U8bGYzsrcj1uAnraHVrvDLP9bLUyiVsOOR11r3ygKE~A6Cy8nXStuV1S~MOTQJhCQ3KKVY07lA6jH-SSo-nXb0AB~I35GNRaTDGVLKHc1B3LAq1LO7PYXRYq08IkFaODHnVggJeBpvwIxRoP42CzTH1etCllh5ca3Zrk5HNMGYKrMwoozOgbQKdW21D6oMZKh0QsT-2hWBB~b9mjmSzGDveSRBq-I5Jv~p4gr9s2vytGI5LZryqF3H8oZIQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)