Key Points
Polymorphic alleles of either ABCB1 or VEGF genes predicted better outcome in patients with MCL receiving LEN after ASCT.
ABCB1, NCF4, GSTP1, and CRBN polymorphisms were associated with toxicity during induction and with LEN dose reductions.
Abstract
In the Fondazione Italiana Linfomi MCL0208 phase 3 trial, lenalidomide maintenance (LEN) after autologous stem cell transplantation (ASCT) in mantle cell lymphoma (MCL) improved progression-free survival (PFS) vs observation (OBS). The host pharmacogenetic background was analyzed to decipher whether single-nucleotide polymorphisms (SNPs) of genes encoding transmembrane transporters, metabolic enzymes, or cell-surface receptors might predict drug efficacy. Genotypes were obtained via real-time polymerase chain reaction of the peripheral blood germ line DNA. Polymorphisms of ABCB1 and VEGF were found in 69% and 79% of 278 patients, respectively, and predicted favorable PFS vs homozygous wild-type (WT) in the LEN arm was 3-year PFS of 85% vs 70% (P < .05) and 85% vs 60% (P < .01), respectively. Patients carrying both ABCB1 and VEGF WT had the poorest 3-year PFS (46%) and overall survival (76%); in fact, in these patients, LEN did not improve PFS vs OBS (3-year PFS, 44% vs 60%; P = .62). Moreover, the CRBN polymorphism (n = 28) was associated with lenalidomide dose reduction or discontinuation. Finally, ABCB1, NCF4, and GSTP1 polymorphisms predicted lower hematological toxicity during induction, whereas ABCB1 and CRBN polymorphisms predicted lower risk of grade ≥3 infections. This study demonstrates that specific SNPs represent candidate predictive biomarkers of immunochemotherapy toxicity and LEN efficacy after ASCT in MCL.
Introduction
Mantle cell lymphoma (MCL) is an aggressive, mature B-cell non-Hodgkin lymphoma historically known for its poor long-term outcome.1 In recent years, the introduction of novel treatment platforms has led to substantial improvement in patients’ prognosis, almost uniformly including a maintenance phase, which is now considered a critical stem of successful treatment.2-5 Among maintenance regimens, lenalidomide, an oral immunomodulatory drug, has been demonstrated to have significant activity and a manageable safety profile, both alone and in combination with rituximab.3-7 However, treatment response appears nonuniform across patients, and predicting the clinical drug profile remains an unmet need.
Many factors may affect interindividual responsiveness to drugs, and the role of the host genetic background has also been investigated in lymphoma.8-11 In particular, gene polymorphisms consist of variations in the gene DNA sequence occurring in a population with a frequency ≥1% and resulting in changes in the expression, structure, and activity of the proteins encoded by these genes. Interestingly, previous reports have suggested that single-nucleotide polymorphisms (SNPs) involving drug metabolic pathways are predictive of both response and toxicity in different histotypes of non-Hodgkin lymphoma.8-15 This has been proven for several agents, including anthracyclines,12 monoclonal antibodies,13,14 and immunomodulatory drugs, including lenalidomide, which is affected by SNPs in ABCB1, ERCC5, XPA, and GSTP1, as demonstrated in multiple myeloma (MM).14,15
The Fondazione Italiana Linfomi (FIL) MCL0208, a prospective multicenter, randomized phase 3 trial, showed a significant progression-free survival (PFS) benefit of lenalidomide maintenance (LEN) vs observation (OBS) in young (<66 years old), previously untreated patients with MCL.3 The study enrolled 300 patients, and a large number of biological samples were stored centrally at fixed time points to document the minimal residual disease (MRD).16 Moreover, several biological substudies,17,18 including a pharmacogenetic analysis, were planned. In particular, we investigated specific germ line polymorphisms of transmembrane transporters, metabolic enzymes, and cell-surface receptors (ABCB1, ABCG2, VEGFA, FCGR2A, NCF4, GSTP1, and CRBN) that might predict the efficacy and safety of immunochemotherapy and LEN.
Methods
Patients and treatment
This study was performed on biological samples collected from the phase 3 MCL0208 study (NCT02354313) sponsored by FIL. The trial enrolled 300 previously untreated patients with MCL, aged from 18 to 65 years, without clinically significant comorbidities. The planned treatment is shown in supplemental Figure 1. Briefly, patients received 3 Rituximab-Cyclophosphamide, Doxorubicin, Vincristine and Prednisone (R-CHOP21), followed by R-high-dose cyclophosphamide (4 g/m2), 2 cycles of R-high-dose Ara-C (2 g/m2 q12h x 3d), and autologous stem cell transplantation (ASCT) conditioned by using the BEAM or FEAM regimen.3 After ASCT, responding patients (either complete or partial response) were randomly assigned to OBS or LEN; 15 mg on days 1-21 every 28 days) for 24 months. Clinical results of the trial have already been published.3 All patients provided written informed consent for the use of their biological samples for research purposes, in accordance with the institutional review board’s requirements and the Helsinki Declaration. The clinical trial as well as the substudy was approved by the ethics committees of all the enrolling centers. All the samples were centralized for scientific analysis in the hematological laboratory of Torino University.
Biological samples
Bone marrow (BM) and peripheral blood (PB) samples were collected at diagnosis and during follow-up, based on the preplanned time points of MRD analysis16 (supplemental Figure 1). Biological samples were identified with a subject study number that can be traced or linked back to the subject only by the site investigator.
MRD and mutational analysis
MRD monitoring was assessed for BM and PB samples by using an allele-specific oligonucleotide real-time quantitative polymerase chain reaction (ASO RQ-PCR) approach (either on immunoglobulin heavy chain (IGH) gene or B-cell lymphoma-1 (BCL-1)/IGH rearrangements) at the indicated time points and evaluated in accordance with the criteria of the EuroMRD standardization group.16
TP53 disruptions as well as KMT2D mutations were identified via next-generation targeted resequencing and copy-number alteration analysis on CD-19–selected tumoral cells from BM baseline samples or as previously described.17
Pharmacogenetic analyses
For the purposes of the pharmacogenetic study, germinal DNA was extracted by using commercially available kits (QIAamp DNA Blood Mini Kit, Qiagen; Maxwell RSC Blood, Promega; and DNAzol, Invitrogen). gDNA was extracted from different specimens, preferably selecting samples devoid of lymphoma infiltration. A total of 193 patients were studied using a follow-up PB sample, mainly those who were tested to be MRD-negative or had very low levels of MRD (supplemental Table 5). When FU samples were not available, baseline PB was used (n = 60), characterized by a median tumor infiltration of 3% via flow cytometry (range, 0.03%-88%; supplemental Table 5)
Specific germ line polymorphisms of transmembrane transporters, metabolic enzymes, and cell-surface receptors were evaluated. The selected polymorphisms of the ABCB1 gene were rs1128503, rs2032582, and rs1045642. Additional SNPs belonging to other genes were chosen based on the mechanistic role of their encoded proteins in the pharmacokinetics and pharmacodynamics of lenalidomide. They included ABCB2 rs2231142, VEGF-A rs699947, FCGR2A rs1801274, NCF4 rs1883112, GSTP1 rs1695, and CRBN rs1714327 and rs1705814 (Figure 1). The full list of investigated SNPs with their respective putative functions is shown in supplemental Table 2. For all the SNPs, a minor allele frequency of ≥0.3 in the European population was considered as a general criterion for selection.
Depiction of the biological functions of the proteins encoded by the investigated genes and the impact of relative polymorphisms. ROS, reactive oxygen species; GSTP1, glutathione-S-transferase Pi1; ATP, adenosine triphosphate; FCGR, Fc gamma receptor; VEGF-A, vascular endothelial growth factor A, ABCB1, ATP-binding cassette subfamily B member 1; Ub, ubiquitin; eNOS, endothelial nitric oxide synthase.
Depiction of the biological functions of the proteins encoded by the investigated genes and the impact of relative polymorphisms. ROS, reactive oxygen species; GSTP1, glutathione-S-transferase Pi1; ATP, adenosine triphosphate; FCGR, Fc gamma receptor; VEGF-A, vascular endothelial growth factor A, ABCB1, ATP-binding cassette subfamily B member 1; Ub, ubiquitin; eNOS, endothelial nitric oxide synthase.
Patients’ genotypes with respect to the investigated SNPs were obtained by using specific TaqMan SNP Genotyping Assays (ABI, Applied Biosystems, Foster City, CA) on an ABI Prism HT7900 Sequence Detection System instrument, per the manufacturer’s instructions. Approximately 10% of samples were analyzed in duplicates, and the results showed a concordance rate of >99%. The SDS Software (Applied Biosystems) was used to impute patients’ genotypes, and allele frequencies and genotypes were calculated. The Hardy-Weinberg equilibrium was checked using the Pearson χ2 test (threshold value, P = 3.841) for each locus, whereas haplotypes and their frequencies were imputed with Arlequin software.19 Finally, differences in genotype (or haplotype) distributions between groups of the study (ie, the LEN vs OBS arm) were evaluated using the Pearson χ2 test.
Toxicity evaluations and lenalidomide dose intensity
Toxic events were recorded per the Common Terminology Criteria for Adverse Events version 4.0. Only hematological toxicities were available in detail and were then correlated with pharmacogenetic data. In particular, granulocytes, platelets, and hemoglobin reductions were considered together across different treatment phases (induction, consolidation, and LEN/OBS). Infective toxicity was evaluated as a cumulative event per the Common Terminology Criteria for Adverse Events, version 4.0. Lenalidomide dose intensity was calculated as the ratio between the effective received dose and the planned dose.
Statistical analysis
Because of the high number of collected biological and clinical variables, the entire data set underwent systematic post hoc quality control through data warehousing.20
Statistical analysis was carried out using R version 4.0.0. For continuous variables, the Kruskal-Wallis test was used to compare medians between groups; for categorical variables, depending on the number, the χ2 test or Fisher exact test was used.
For survival analysis, PFS and overall survival (OS) were used as clinical end points. PFS was calculated from the date of enrollment in the clinical study to the date of disease progression (event), death from any cause (event), or last follow-up (censoring). OS was measured from the date of enrollment in the clinical study to the date of death from any cause (event) or last follow-up (censoring).21 Survival was estimated with the Kaplan-Meier method, and the log-rank test was applied to compare the survival distributions of the patients. The Cox proportional hazards model was implemented for the univariate and multivariate survival analysis. For all statistical analyses, the level of significance was set at P ≤ .05. The outcome data for the present analysis were updated as of December 2017, as planned in the clinical trial.
Results
Feasibility of the study
Overall, 300 patients were enrolled in the FIL-MCL0208 clinical trial, and 93% of these patients (278 of 300) were included in the pharmacogenetic study because of the availability of adequate biological samples. In particular, 96% (197 of 205) of the randomly assigned population, 97% (101 of 104) of patients randomly assigned to LEN, and 95% (96 of 101) of those randomly assigned to OBS were genotyped (supplemental Figure 2). The main clinical features of these patients are described in supplemental Table 1.
Pharmacogenetic analyses
Allele and genotype frequencies of all the investigated SNPs are detailed in Table 1. The minor allele frequency values did agree with those already calculated in European populations, without significant differences between the LEN and OBS arms. Of note, all SNPs were in Hardy-Weinberg equilibrium, except for the CRBN locus, rs1705814.
The expected linkage between the 3 ABCB1 SNPs was confirmed (supplemental Table 3), with percentage values in the LEN and OBS arms of 18.8% and 16.7%; 51.5% and 50.0%; and 29.7% and 33.3% for wild-type (WT) homozygous (ie, CGC/CGC), heterozygous polymorphic (POL) (ie, CGC/TTT), and POL homozygous (ie, TTT/CTT) patients, respectively. No significant differences in haplotype distribution were observed between the enrolled and randomly assigned populations as well as between the 2 arms (χ2 = 0.264; P = .876).
Impact of SNPs on treatment efficacy
Among the investigated polymorphisms, only 2, namely ABCB1 rs2032582 and VEGF rs699947, were associated with lenalidomide efficacy in terms of both PFS and OS. In the randomly assigned population, 60 (31%), 107 (54%), and 30 (15%) patients were WT homozygotes (HoWT), heterozygotes (HePOL), and POL homozygotes (HoPOL), respectively, for the ABCB1 locus. For the VEGF rs699947 locus, 42 (21%), 96 (49%), and 59 (30%) patients were HoWT, HePOL, and HoPOL, respectively (supplemental Table 4).
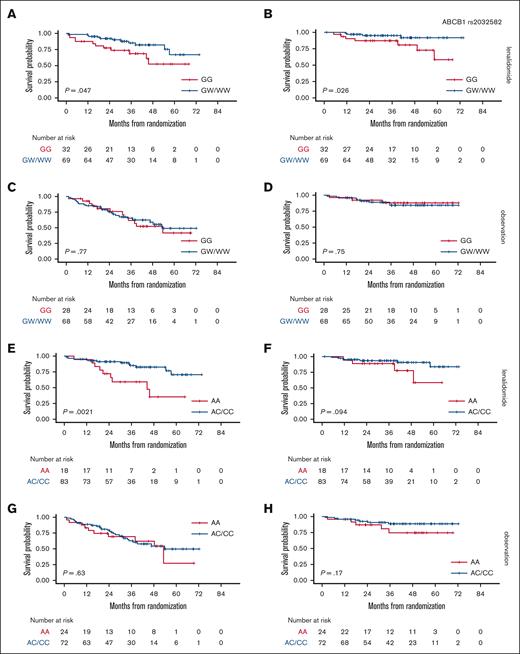
Firstly, survival analysis was performed by separately comparing these 3 groups (supplemental Figure 4), but HoPOL and HePOL patients were then grouped together because these patients showed superimposable outcomes. In the randomly assigned population, 137 patients (70%) carried at least 1 POL allele for the ABCB1 rs2032582 locus. Interestingly, HoPOL (WW) and HePOL (GW) had better outcomes when compared with HoWT (GG) in the LEN arm (3-year PFS, 85% vs 70% [P = .047] and 3-year OS, 98% vs 90% [P = .026]) but not in the OBS arm (Figure 2A-D).
Association between ABCB1 or VEGF-A–grouped genotypes and survival in a randomly assigned population. PFS stratified based on ABCB1 genotypes in the LEN (A) and OBS arms (C); OS stratified based on ABCB1 genotypes in the LEN (B) and OBS arms (D); PFS stratified based on VEGF-A genotypes in the LEN (E) and OBS arms (G); OS stratified based on VEGF-A genotypes in the LEN (F) and OBS arms (H). ABCB1: HoWT = GG, HePOL = GW, HoPOL = WW; VEGF-A: HoWT = AA, HePOL = AC, HoPOL = CC.
Association between ABCB1 or VEGF-A–grouped genotypes and survival in a randomly assigned population. PFS stratified based on ABCB1 genotypes in the LEN (A) and OBS arms (C); OS stratified based on ABCB1 genotypes in the LEN (B) and OBS arms (D); PFS stratified based on VEGF-A genotypes in the LEN (E) and OBS arms (G); OS stratified based on VEGF-A genotypes in the LEN (F) and OBS arms (H). ABCB1: HoWT = GG, HePOL = GW, HoPOL = WW; VEGF-A: HoWT = AA, HePOL = AC, HoPOL = CC.
Similarly, 155 patients (79%) carried at least 1 variant allele of the VEGF-A locus, such that HoPOL (CC) and HePOL (AC) had better outcomes than HoWT in the LEN arm (3-year PFS, 85% vs 60% [P = .0021] and 3-year OS, 90% vs 86.5% [P = .094]) but not in the OBS arm (Figure 2E-H).
Accordingly, when compared with HoWTs, patients carrying variant genotypes of either gene (ABCB1 or VEGF-A) showed a trend toward deeper MRD clearance via RQ-PCR in the BM after 6 months of LEN (supplemental Figure 3). In this population, as well as in the overall series, HoWT did not differ from HoPOL and HePOL based on baseline clinical features, classical prognosticators (including TP53 aberrations), ABCB1, and VEGF-A polymorphisms (supplemental Table 3).
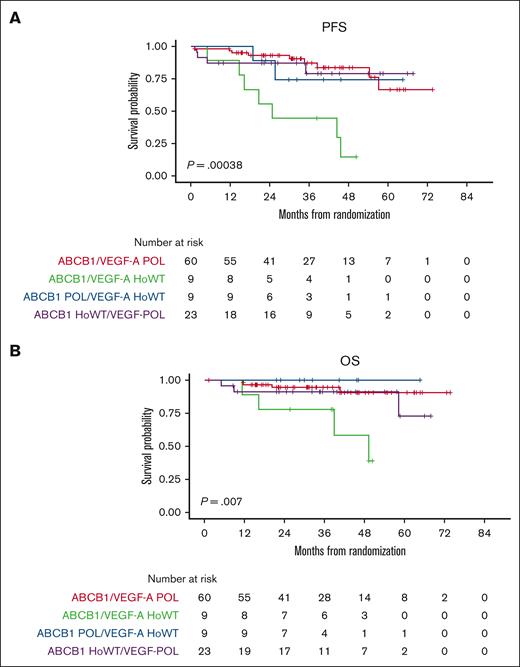
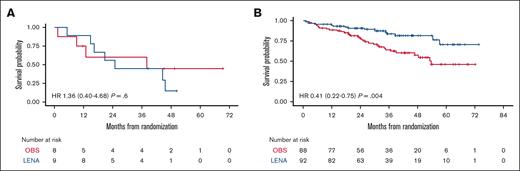
Moreover, by analyzing the combinatorial effect of each polymorphism in the same patients, only the small group of double WT cases (ie, ABCB1 HoWT and VEGF-A HoWT) receiving lenalidomide were actually associated with poor survival, in terms of both PFS and OS (Figure 3). Therefore, we reconsidered the efficacy of lenalidomide maintenance by stratifying the randomly assigned population based on its pharmacogenomic background. Survival analysis showed that patients carrying at least 1 variant allele of ABCB1/VEGFA POL loci experienced the highest benefit from LEN (PFS hazard ratio = 0.41 [95% confidence interval [CI], 0.22-0.75]; P = .004, vs PFS hazard ratio = 0.51 [95% CI, 0.30-0.87]; P = .012) of the entire population.22 In contrast, based on the limited number of patients who were double WT (n = 17), this genotype seemed to confer no benefit at all in the LEN vs the OBS arm (P = .632), as shown in Figure 4.
Association between combined ABCB1/VEGF-A genotypes and survival in the LEN population. PFS (A) and OS (B) stratified based on the combination of ABCB1 and VEGF-A genotypes.
Association between combined ABCB1/VEGF-A genotypes and survival in the LEN population. PFS (A) and OS (B) stratified based on the combination of ABCB1 and VEGF-A genotypes.
PFS by randomization arm stratified on the pharmacogenomic background. PFS stratified by ABCB1 and VEGF-A genotypes in the LEN and OBS arms: ABCB1/VEGF-A HoWT (A) and ABCB1/VEGF-A POL (B). HR, hazard ratio.
PFS by randomization arm stratified on the pharmacogenomic background. PFS stratified by ABCB1 and VEGF-A genotypes in the LEN and OBS arms: ABCB1/VEGF-A HoWT (A) and ABCB1/VEGF-A POL (B). HR, hazard ratio.
Impact of SNPs on treatment toxicity
Lenalidomide dose reduction
Dose reduction of lenalidomide was significantly associated only with the CRBN rs1705814 genotype. In the studied population, 113 patients (41%) were HoWT (TT), 91 (33%) were HePOL (TC), and 73 (26%) were HoPOL (CC) (supplemental Table 4). It is worth noting that 28 patients randomly assigned to LEN (28%) were HoPOL for the CRBN rs1705814 locus and had a higher risk of major lenalidomide dose reduction (more than 66%) or discontinuation during the maintenance phase with respect to HoWT/HePOL (odds ratio [OR], 3.24; CI, 1.69-6.21; P = .013). Nonetheless, in the LEN randomization arm, no statistically significant impact of the CRBN rs1705814 SNP was observed on either hematological toxicities or infections (Table 2).
Hematological toxicity
Three polymorphisms were associated with hematological toxicity in the whole population of patients: ABCB1 rs2032582, NCF4 rs1883112, and GSTP1 rs1695 (see supplemental Table 3 for the distribution of genotypes). Indeed, 46 patients (17%) carrying ABCB1 TT/AT/AA genotypes had a lower risk of hematological toxicity after the first R-CHOP cycle than did patients carrying HePOL/HoWT (OR for G ≥3 toxicity, 0.39; CI, 0.15-0.88; P = .033). Moreover, 162 patients (58%) carried NCF4 AG/GG genotypes and were exposed to an overall lower risk of hematological toxicities during induction (within the R-high-dose cyclophosphamide cycle), when compared with HoWT (OR, 0.56; CI, 0.34-–0.92; P = .024; Table 2). Similarly, 28 patients (10%) carried the GSTP1 GG HoPOL genotype and were exposed to a lower risk of toxicities than HePOL/HoWT (OR, 0.35; CI, 0.15-0.79; P = .014; Table 2). Interestingly, by combining the postulated protective effect of both SNPs, we were able to identify a subgroup of patients (NCF4 and GSTP1 HoWT, n = 48) who were at a higher risk of hematological toxicity during induction (OR, 2.26; CI, 1.09-4.83; P = .031). No impact of these SNPs was observed during later treatment phases (R-high dose Ara-C)or on hematological recovery after ASCT.
Infective toxicity
Two polymorphisms, ABCB1 rs1045642 and CRBN rs1705814, were associated with infections in the whole series of patients during chemoimmunotherapy. In the studied population, the following allele frequencies were found: for ABCB1 c.3435.C>T, 70 patients (25%) were HoWT (CC), 143 (51%) were HePOL (CT), and 65 (23%) were HoPOL (TT). For CRBN rs1705814 T>C, 113 patients (41%) were HoWT (TT), 91 (33%) were HePOL (TC), and 73 (26%) were HoPOL (CC) (supplemental Table 3). The 65 out of 278 patients carrying the ABCB1 TT genotype (supplemental Table 4) were exposed to a lower risk of infections than were CT/CC individuals (OR, 0.53; CI, 0.30-0.95; P = .030). Similarly, 73 patients HoPOL for the CRBN rs1705814 locus had a lower risk of severe infections than those who were HoWT/HePOL (OR for G≥3 toxicity, 0.39; CI, 0.22-0.68; P = .001). On In contrast, none of the SNPs predicted the onset of infections during LEN.
Discussion
The pharmacogenetic study of the FIL MCL0208 phase 3 randomized trial showed that SNPs of genes encoding specific cellular proteins may be associated with clinical outcome in younger patients with MCL receiving LEN after ASCT. Most importantly, we observed that
ABCB1 c.2677 SNP (transmembrane transporter) was significantly associated with better PFS and OS;
VEGF-A c.2055 SNP (angiogenic factor) was significantly associated with better PFS;
by combining ABCB1 and VEGF-A genotypes, a small subgroup of patients with MCL who did not benefit from LEN can be identified;
and CRBN rs1705814 SNP (a lenalidomide molecular target) was associated with a higher risk of lenalidomide dose reduction or discontinuation.
Notably, no significant differences in genotype distribution were observed between the 2 randomization arms, resetting any possible bias due to the unbalanced stratification of patients and reducing the weight of confounding factors in the LEN phase. In addition, some SNPs (namely, ABCB1 c.3435C>T, NCF4 c.368G>A, CRBN rs1705814 T>C, and GSTP1 c.313.A>G) were associated with hematological and infective toxicity during the immunochemotherapy phase preceding the randomization.
In recent years, several studies have underlined the potential role of interindividual genetic differences in shaping treatment response and toxicity in lymphoma.23,24 Nonetheless, to date, little pharmacogenetic knowledge is available in the MCL literature,25-28 and no data relative to lenalidomide treatment in MCL have been published. In contrast, several pharmacogenetic studies have been published, mainly in MM, assessing the relation of lenalidomide activity with polymorphisms of CRBN, one of lenalidomide’s most investigated molecular targets.22,29-31 Thus, to our knowledge, this study provides the first pharmacogenetic data related to lenalidomide treatment in MCL by investigating the effects of candidate genes and polymorphisms on both efficacy and toxicity. Because some of the genes may play a role in immunochemotherapy outcomes and tolerability, the investigated panel of loci was also assessed for these effects in the whole population of patients.
Our main findings suggested a key role of ABCB1 and VEGF-A polymorphisms in enhancing the clinical activity of LEN after ASCT in MCL. Biologically, we might hypothesize that variant alleles of ABCB1, a transmembrane drug transporter, leading either to decreased gene expression in gut cells and renal tubules or to reduced ejection activity of the transporter,32-34 resulted in greater lenalidomide bioavailability, leading to an increased pharmacological effect due to higher plasma and tissue drug concentrations. This hypothesis is in line with published literature on lenalidomide pharmacokinetics in MM,35,36 but in this study, no gut or renal tissue was available to investigate the hypothesis.
Similarly, the POL genotype of VEGF-A might lead to decreased gene expression (as suggested in other scenarios37-39), resulting in decreased stimulation by this key cytokine in the PI3K-Akt pathway.40 Unfortunately, adequate lymphoma tissue for gene expression analysis was available only for a minority of the patients in this trial, so no statistically relevant conclusions might be drawn from these data.41 Anyway, given the direct inhibitory effect of lenalidomide on VEGF-A–mediated Akt phosphorylation, the presence of the VEGF-A C POL allele might additively enhance lenalidomide’s pharmacological effect. Therefore, we hypothesized that the absence of both POL genotypes might lead to lower lenalidomide and higher VEGF-A concentrations and, thus, act as a predictive biomarker of poor response to lenalidomide in MCL. Accordingly, patients carrying neither ABCB1 nor VEGF-A polymorphisms (actually, 9% of the series) did not benefit from LEN when compared with OBS only (Figure 4B). Thus, these results suggest a possible means of selecting patient candidates for lenalidomide to improve its efficacy and concomitantly reduce the risk of toxicity. More cumbersome is the association found between the CRBN rs1705814 POL genotype and lenalidomide dose reduction. We might hypothesize that this genotype could reduce the intracellular expression of cereblon, the molecular target of lenalidomide, and the minimum lenalidomide dose required to obtain the therapeutic effect. However, our analysis was not able to show any statistically significant association between this SNP and a higher risk of hematological or infectious toxicities, possibly explaining the observed lenalidomide dose reduction. Further analyses are required to explain this finding, including the roles of different toxic events not reported in the trial electronic case report form (eCRFs) as well as a wider validation on an external, well-annotated patient population, such as the recently completed EuMCLNet R2 Elderly trial.42 Finally, we cannot exclude the possibility that other polymorphisms might serve as possible biomarkers of lenalidomide efficacy, but none of the analyzed genes other than ABCB1 and VEGF-A showed promising survival trends that deserved to be further investigated (data not shown).
Considering the standard chemotherapeutic regimens administered to patients before ASCT, it was hypothesized that some of the SNPs identified and evaluated in the present work could also act as predictive markers of tolerability because proteins encoded by the corresponding genes are involved in the transmembrane transport (ABCB1) or detoxification (GSTP1 and NCF) of cyclophosphamide, vincristine, and doxorubicin. Indeed, we found that at least 3 loci in different genes were significantly associated with hematological toxicities during the induction phase, with an increased risk for patients carrying WT genotypes. That risk was coupled with an increased incidence of severe infections, at least when considering ABCB1.
Our study has some strengths. Firstly, the robustness and reproducibility of RQ-PCR analysis ensure its theoretical large-scale applicability in clinical practice across different labs because the present analyses can be carried out on several instrumental platforms, including the most recent ones. Easy standardization, a patient-friendly PB source, and overall limited costs also favor this approach. Furthermore, it is intriguing to derive predictive biomarkers by investigating the genetic background of every single patient, in addition to the intrinsic alterations of tumor cells, and, hence, allow wider approaches to obtain somatic DNA (ie, buccal swab). Nevertheless, although promising, our strategy identified only a limited number of cases with an adverse genotype, and we totally lack functional studies; therefore, an external validation on an independent series of patients receiving lenalidomide is needed for validation of these candidate biomarkers. Moreover, we acknowledge that we have investigated, in this cohort, only a finite number of gene polymorphisms, selected based on data already available in the literature, and they probably do not represent all the pharmacogenomics targets potentially involved in lenalidomide’s mechanism of action. In this regard, current genome-wide sequencing tools may be applied in the future as more convenient technical approaches to study, in one-run, larger panels of different gene polymorphisms. Finally, the number of patients with MCL enrolled in this study is the largest so far. However, the sample size of this study might have limited the comprehensive detection of associations with less common alleles. Additional studies on larger MCL populations will help confirm the current findings and may reveal additional variants that influence drug-associated efficacy and toxicity. Overall, pharmacogenomics might play a role in MCL therapy, enabling clinicians to provide tailored treatment based on individual patients’ genetic profiles, possibly complementing classical mutational and expression analysis carried out on tumor cells.17,43 This paradigm might also be scalable to several novel drugs currently used in MCL treatment (such as Bruton tyrosine kinase inhibitors or bispecific antibodies)44,45 as well as in other lymphomas46 or MM, for which lenalidomide is widely used.47,48 In fact, the comprehensive identification of patients who benefit the most from lenalidomide or other therapeutic approaches would not only bring clinical benefits and improved quality of life to our patients but would also be valuable from a pharmacoeconomic perspective.
In conclusion, this study demonstrates that pharmacogenetic analysis of PB samples could easily yield predictive biomarkers of poor response to LEN after ASCT in patients with MCL. Despite some methodological limitations, this approach is shown to be promising and theoretically scalable to different clinical contexts. Thus, it is a perfect example of how precision medicine might present an exceptional opportunity for our patients in the near future.
Acknowledgments
The authors thank all the patients who participated in the study. They are grateful to Daniela Barbero, Pier Paola Fenoglio, Daniela Drandi, and Barbara Mantoan for their scientific advice and to Sonia Perticone, Antonella Ferranti, Daniela Gioia, Antonella Fiorillo, and Giulia Bondielli for their assistance.
This work was supported by Progetto di Rilevante Interesse Nazionale (PRIN2009) from the Ministero Italiano dell'Università e della Ricerca, Roma, Italy (7.07.02.60 AE01); Progetto di Ricerca Sanitaria Finalizzata 2009 (RF-2009-1469205) and 2010 (RF-2010-2307262) (S.C.), AOS Maurizio, Bolzano/Bozen, Italy; Fondi di Ricerca Locale, Università degli Studi di Torino, Italy; Fondazione Neoplasie Del Sangue, Torino, Italy; Fondazione CRT (project codes 2016.0677 and 2018.1284), Torino, Italy; AL-AIL ODV Sezione Interprovinciale di Alessandria-Asti; Associazione DaRosa, Torino, Italy; the Gilead Fellowship Program 2019, Milano, Italy; and Cancer Research UK (C355/A26819) and FC AECC and AIRC under the Accelerator Award Program.
Authorship
Contribution: G.A.P., S.G., S.C., A.D.P., and M.L. conceived and designed the study; S.F., D.G., A.D.R., A.R., V.S., F.C., C.B., M.B., V.Z., F.M., L.A., E.L., F.B., A.J.M.F., B.P., S.C., and M.L. enrolled patients and provided biological samples; E.A., B.A., and E.G. performed experiments; S.F., D.G., M.P., G.M.Z., M.G., and A.D.P. collected and analyzed data; G.D.L. performed statistical analysis; and S.F., D.G., M.P., R.T., G.A.P., S.G., A.D.P., and M.L. wrote the paper; and all the authors approved the final version of the manuscript.
Conflict-of-interest disclosures: S.F. is a consultant for Janssen and EUSA Pharma; is on the advisory board of Janssen, EUSA Pharma, Incyte, and Clinigen; received speaker’s honoraria from Janssen, EUSA Pharma, Servier, and Gentili ; and received research funding from Gilead and MorphoSys G.A.P. received speaker’s fees from AbbVie, Bristol Myers Squibb (BMS), Incyte, and Novartis; reports service on advisory boards for AbbVie Orphan Pharmaceuticals, AstraZeneca, Bristol Myers Squibb, and Novartis; and reports support for attending meetings from AbbVie, Bristol Myers Squibb, Janssen, and Novartis. M.L. received honoraria from AbbVie, Acerta, Amgen, GSKI, Gentili, Sandoz, Gilead/Kite, Novartis, Roche, EUSA Pharma, Takeda, Regeneron, Incyte, and Jazz; received honoraria and research funding from ADC Therapeutics, BeiGene, Celgene, and Janssen. L.A. reports consulting or advisory role in Roche, Janssen-Cilag, Verastem, Incyte, EUSA Pharma, Celgene/Bristol Myers Squibb, Kite/Gilead, and ADC Therapeutics; is on speakers' bureau at EUSA Pharma and Novartis; and received research funding from Gilead Sciences. V.Z. reports consulting or advisory role in Takeda, Janssen, MSD, Roche, Servier, and Kite; received support for travel, accommodations, and expenses from Takeda and Janssen. S.G. reports being speaker at events supported by Jazz, Janssen, Pfizer, Novartis, Incyte, AstraZeneca, and AbbVie. F.C. reports advisory or consulting role for Roche and AstraZeneca; reports being speaker for Servier; and receives travel accommodation from Takeda, AstraZeneca, and Roche. C.B. reports consulting or advisory role in AbbVie and AstraZeneca; and receives support for travel, accommodations, and expenses from Janssen. The remaining authors declare no competing financial interests.
Correspondence: Simone Ferrero, Department of Molecular Biotechnologies and Health Sciences, University of Torino, Italy SC Ematologia 1 U AOU Città della Salute e della Scienza di Torino, via Genova 3, 10126 Torino, Italy; e-mail: simone.ferrero@unito.it.
References
Author notes
∗A.D.P. and M.L. are joint last authors.
All data are available on request from corresponding author, Simone Ferrero (simone.ferrero@unito.it).
The full-text version of this article contains a data supplement.