TO THE EDITOR:
As reported, 40% of patients with diffuse large B-cell lymphoma (DLBCL) still experience refractory disease or relapse after the standard R-CHOP (cyclophosphamide, doxorubicin, vincristine, and prednisone+rituximab) therapy.1,2 Various salvage therapeutic strategies, including autologous hematopoietic stem cell transplantation, have been attempted to overcome the resistance to treatment and improve overall survival (OS). Nevertheless, the disease still relapses in 30% of patients.3,4 Regarding immunochemotherapy, chimeric antigen receptor (CAR) T-cell therapy has been shown to be promising for treating patients with refractory or relapsed DLBCL (rrDLBCL). However, CD19-CAR T-cell therapy has shown only a 40% to 54% complete remission (CR) rate in patients with rrDLBCL,5-8 which may be related to the genetic heterogeneity of the rrDLBCLs. The advent of high-throughput next-generation sequencing technology has rapidly increased our knowledge of genomic alterations of DLBCL.9-11 Several reports have proposed the concept of molecular subtypes.12-16 Distinct classification describes partially overlapping features, suggesting the existence of molecular subtypes and guiding novel-targeted therapy. It is unclear whether the diverse molecular subgroups experience different efficacy of CAR T-cell therapy. To investigate, we performed targeted deep sequencing of 92 hematologic-related genes in a cohort of 105 patients with rrDLBCL, in which most of the patients underwent CAR T-cell immunotherapy after having a poor response to multiple lines of treatment. This trial is registered on the Chinese Clinical Trials Registry as #ChiCTR1900020980.
One hundred five patients with rrDLBCL diagnosed from 2019 through 2020, including 6 patients with transformed follicular lymphoma, 2 with transformed mucosa-associated lymphoid tissue lymphoma, and 2 with transformed chronic lymphocytic leukemia lymphoma, were recruited for the study. Eighty-four patients (86%, 84/105) had been treated with CAR T-cell therapy before enrollment in the study. The patients were followed up until 15 April 2021. The study was approved by the Institutional Review Board of Boren Hospital. Informed consent was obtained from all patients in accordance with the Declaration of Helsinki. The baseline clinical characteristics of the 105 patients included in the study are summarized in Table 1 and supplemental Table 1. The median age of the entire cohort at initial diagnosis was 49 (range, 13-79) years. Twenty-nine patients (27.6%) had a relatively high-risk International Prognostic Index score of 4 to 5. The median time of relapse was 12.7 months. The median number of chemotherapy cycles was 11. Among our cohort of patients, 11 with germinal center B cells (11 of 31, 35.5%) were classified as “double hit” or “triple hit” (DH/TH), harboring translocations of MYC and BCL2/BCL6, whereas only 2 patients without germinal center B-cells (GCB; 2 of 74; 2.7%) were classified as DH/TH. Most of the patients presented with advanced disease. Written informed consent was obtained from each patient, and the study was approved by the Ethics Committee at the Beijing Boren Hospital, according to guidelines of the 1975 Declaration of Helsinki.
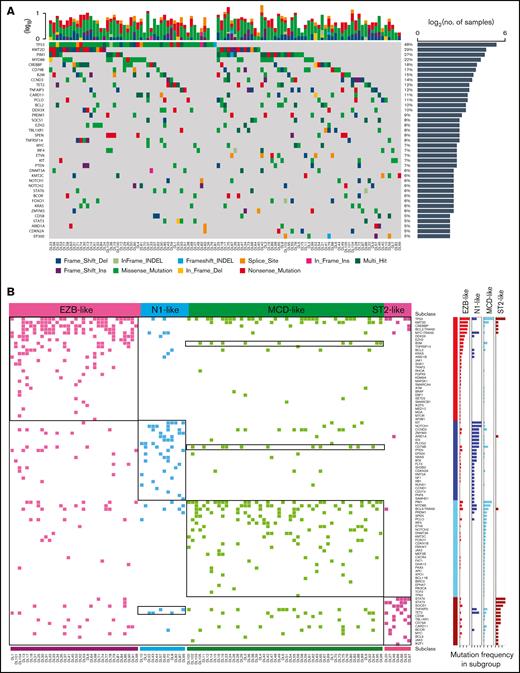
Figure 1A shows the distribution of the mutations of candidate cancer genes found in more than 5% of the patients with rrDLBCL. The patients in this series harbored a median of 6 (range, 1-24) genetic drivers (supplemental Tables 2 and 3). The most frequently mutated genes, found in >15% of cases, were TP53, KMT2D, PIM1, MYD88, CREBBP, CD79B, and B2M (Figure 1A; supplemental Tables 2 and 3). To assess the relationship between gene-reated lesions and the success of CAR T-cell immunotherapy, we determined the genetic classification of patients with rrDLBCL and discovered 4 robust subsets of tumors (clusters; Figure 2). To display the genetic composition of the subtypes, we selected a set of genetic features that were significantly associated with a subtype (P < .05) and were present in >10% of the tumors of each subtype (supplemental Table 5): EZB-like, MCD-like, N1-like, and ST2-like.
Identification of groups of tumors with coordinate genetic signatures in patients with rrDLBCL. (A) A heat map showing individual mutations in 105 patient samples color coded by mutation type. The top plot shows the absolute number of oncogenic mutations in each patient. The graph on the right shows the number of mutations in each gene. Percentages represent the fraction of tumors with at least 1 mutation in the specified gene. Fifty recurrently and significantly mutated genes are sorted according to their mutational frequencies. Genes with a mutation frequency >5% are shown. (B) Nonnegative matrix factorization consensus clustering was performed on mutated genes in the 105 DLBCL samples. Clusters with their associated landmark genetic alterations are shown. Distinct clusters are identified by color. Genetic alterations that were positively associated with each cluster were identified by 1-sided Fisher's exact test and ranked by frequency. P < .05, right.
Identification of groups of tumors with coordinate genetic signatures in patients with rrDLBCL. (A) A heat map showing individual mutations in 105 patient samples color coded by mutation type. The top plot shows the absolute number of oncogenic mutations in each patient. The graph on the right shows the number of mutations in each gene. Percentages represent the fraction of tumors with at least 1 mutation in the specified gene. Fifty recurrently and significantly mutated genes are sorted according to their mutational frequencies. Genes with a mutation frequency >5% are shown. (B) Nonnegative matrix factorization consensus clustering was performed on mutated genes in the 105 DLBCL samples. Clusters with their associated landmark genetic alterations are shown. Distinct clusters are identified by color. Genetic alterations that were positively associated with each cluster were identified by 1-sided Fisher's exact test and ranked by frequency. P < .05, right.
Most of the 34 DLBCL tumors harbored a DH (BCL2 translocation and MYC break) and also exhibited frequent mutations in the chromatin modifiers KMT2D, CREBBP, and EZH2, as well as the mutation of DDX3X with nonsynonymous single-nucleotide variant and stop-gain mutations. Consistent with this finding, the EZB-like subtype was found in most of the cases of transformed follicular lymphoma and was strongly enriched in cases of DH GCB-type DLBCL. The genetic features of the subgroup were similar to those of the EZB-like tumors in Wright et al.16
The 12 DLBCL tumors were characterized by gain-of-function of NOTCH1 mutations, the B-cell differentiation regulator ID3, and the BCR signaling intermediates BTK and PLCG2. A somatic KIT p.M541L somatic mutation in N1-like tumors enhances the ability of KIT to activate PI3K-AKT-MTOR and MAPK signal transduction and to induce sustained proliferation of tumor cells and inactivation of apoptosis signaling pathways.17 This subgroup exhibited features analogous with the N1 tumors in Wright et al.16
The 52 DLBCL tumors exhibited frequent mutations in PIM1, MYD88, and CD79B alterations which had been associated with ABC-type DLBCL tumors.18 In tumors with the MCD-like subtype, 33% (17 of 52) carried mutations in MYD88, and 21% carried CD79B aberrations (mostly the mutation Y196), with 11.5% (6 of 52) bearing both MYD88L265P and abnormalities in CD79B. Additional alterations linked to ABC-DLBCL tumors included alterations in IRF4 and ETV6 and inactivation of PRMD1. This subgroup demonstrated features similar to those of the MCD subtype in Wright et al.16
The 7 DLBCL tumors were characterized by mutations in JAK/STAT pathway members (SOCS1, STAT6, and STAT3) and immune escape (CD58). Half of the cases with this subtype were found to have mediastinal involvement, suggesting a degree of biological similarity to primary mediastinal B-cell lymphoma. The genetic features of the subgroup were similar to the ST2 tumors in Wright et al.16
Tumors in half of all cases were classified as MCD-like, 32.4% as EZB-like, 11.4% as N1-like, and 6.7% as ST2-like (supplemental Figure 1A). The N1-, MCD-, and ST2-like subtypes were dominated by non-GCB cases, and EZB-like included mostly GCB cases (supplemental Figure 1B-C). Most of the patients with DH/TH DLBCL belonged to the GCB subgroup and the EZB-like subtype (supplemental Figure 1D).
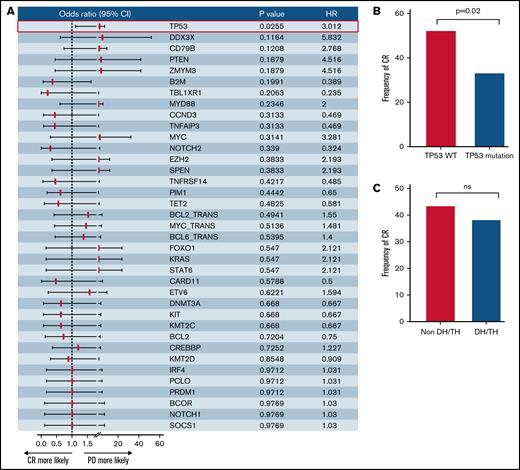
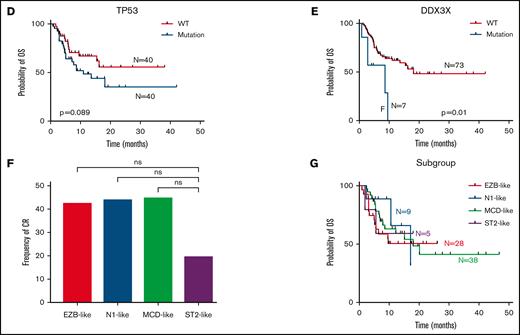
Eighty-four patients (86%; 84 of 105) in our cohort were treated with CAR T-cells. After 3 months of CAR T-cell infusion, the CR rate was 42.8%, and the overall response rate was 58.3%. A total of 39.2% of the patients remained in CR at the cutoff date. The median follow-up time was 13.63 months (95% confidence interval [CI], 10.3-16.1). The median progression-free survival and OS in patients with CAR T-cell treatment was 6.23 months (95% CI, 2.4-not reached) and 16.2 months (95% CI, 8.7-not reached), respectively, with a more favorable progression-free survival and OS than the patients without CAR T-cell treatment (supplemental Figure 2A-B). We next assessed whether the differences in genomic lesions determine the therapeutic benefit after immunotherapy with CAR T cells. Patients reacted differently to CAR T-cell treatment. Therefore, we next assessed whether the differences in genomic lesions determine the therapeutic benefit after immunotherapy with CAR T cells. Among tumor genetic factors, only TP53 alterations were predictive of an inferior rate of CR in univariable logistic regression (Figure 2A-B). No association of other cytogenetic features with CR rate was observed (Figure 2A). The patients carrying mutations in TP53 and DDX3X had inferior OS after CAR T-cell immunotherapy vs the wild-type group (Figure 2D-E). In these patients with TP53 mutation, CAR T-cell immunotherapy can improve the survival (supplemental Figure 2C). The CR rate was similar between the DH/TH and non-DH/TH groups (Figure 2C). Statistically significant differences in CR rates were not found between the molecular subtypes (Figure 2F). We also found no significant differences in OS between any of the molecular subtypes (Figure 2G), but the patients with CAR T-cell treatment in the MCD- and EZB-like subgroups had a more favorable OS than those without CAR T-cell treatment (supplemental Figure 2D-E).
In summary, we identified 4 major genetically distinct rrDLBCL groups, which resembled high-risk primary molecular subgroups reported perviously.16 We found a limited effect of genetic classification on the outcome of CAR T-cell therapy. We also demonstrated the potential prognostic impact of TP53 and DDX3X mutations in patients with rrDLBCL who are treated with CAR T-cell therapy. The patients with rrDLBCL who underwent CAR T-cell treatment had a favorable OS. Nonetheless, further study is needed to demonstrate an improvement in OS for rrDLBCL patients after CAR T-cell therapy.
Acknowledgments: The authors thank the histological processing and analysis services provided by the molecular biology laboratory, genetics laboratory, immunohistochemistry laboratory, cell biology laboratory, and Fluorescent Confocal Microscopy (FCM) Core of Beijing Boren Hospital.
Contribution: K.H. and X.K. conceived the design for the study; H.S. designed the experiments; H.S., P.Z., R. Liu, T.X., F.Y., S.F., Y.G., L.M., H.L., Y. Lei, R. Li, B.D., S.H., Y. Li, Q.Z., K.H., and X.K., performed the experiments; H.S. wrote the manuscript; and all authors discussed the results and commented on the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Xiaoyan Ke, Department of Hematology, Peking University Third Hospital, Beijing Boren Hospital, No. 6, Jitong East Rd, Fengtai District, Beijing, China; e-mail: kexy@gobroadhealthcare.com; and Kai Hu, Department of Hematology and Lymphoma Research Center, Beijing Boren Hospital, No. 6, Jitong East Rd, Fengtai District, Beijing, China; e-mail: huk@gobroadhealthcare.com.
References
Author notes
Sequencing data can be found in the Genomic Sequence Archive for Human, part of the Chinese National Genomic Data Center (accession numbers HRA002670 and GVM000357). Any data issues can be addressed by contacting the corresponding author, Kai Hu (huk@gobroadhealthcare.com).
The full-text version of this article contains a data supplement.