Key Points
BL with granulomatous reaction was characterized by an M1 signature and INFG in CD8+ cells, whereas BL with starry sky showed M2 genes.
Tumor-associated macrophages repolarization and epigenetic regulators may open new therapeutic options for the fight against BL lymphoma.
Visual Abstract
Burkitt lymphoma (BL) is characterized by a tumor microenvironment (TME) in which macrophages represent the main component, determining a distinct histological appearance known as “starry sky” pattern. However, in some instances, BL may exhibit a granulomatous reaction that has been previously linked to favorable prognosis and spontaneous regression. The aim of our study was to deeply characterize the immune landscape of 7 cases of Epstein-Barr virus–positive (EBV+) BL with granulomatous reaction compared with 8 cases of EBV+ BL and 8 EBV-negative (EBV–) BL, both with typical starry sky pattern, by Gene expression profiling performed on the NanoString nCounter platform. Subsequently, the data were validated using multiplex and combined immunostaining. Based on unsupervised clustering of differentially expressed genes, BL samples formed 3 distinct clusters differentially enriched in BL with a diffuse granulomatous reaction (cluster 1), EBV+ BL with typical starry sky pattern (cluster 2), EBV– BL with typical “starry sky” (cluster 3). We observed variations in the immune response signature among BL with granulomatous reaction and BL with typical “starry sky,” both EBV+ and EBV–. The TME signature in BL with diffuse granulomatous reaction showed a proinflammatory response, whereas BLs with “starry sky” were characterized by upregulation of M2 polarization and protumor response. Moreover, the analysis of additional signatures revealed an upregulation of the dark zone signature and epigenetic signature in BL with a typical starry sky. Tumor-associated macrophages and epigenetic regulators may be promising targets for additional therapies for BL lymphoma, opening novel immunotherapeutic strategies.
Introduction
The tumor microenvironment (TME) in B-cell lymphomas is highly variable in terms of spatial arrangement and the type of inflammatory cells, blood and lymphatic vascular networks, and extracellular matrix.1 Growing evidence suggests that TME plays an important role in various processes, including the development and dissemination of B-cell lymphomas.2,3 In aggressive lymphomas, particularly Burkitt lymphoma (BL), owing to their high proliferation rate, intensive chemotherapy is the standard of care. Unfortunately, these treatments are not as effective in the older patients who are immunocompromised and in the equatorial African population, in which BL is the most common childhood cancer.4,5 Having a complete understanding of TME biology in BL and the way in which lymphoma cells interact with TME can truly make all the difference when it comes to developing new target therapies and tumor prognosis prediction. Certainly, research in the field of lymphomas is highly complex and multifaceted, and there is still much to learn. However, with continued exploration and innovative thinking, there is a strong hope for improved outcomes for those affected by BL and other lymphomas. BL is a type of lymphoma that has been historically divided into 3 categories: endemic , sporadic , and immunodeficiency associated. However, these factors are tightly confounded by the presence or absence of Epstein-Barr virus (EBV). Thus, the most recent developments in BL biology have prompted the fifth edition of the World Health Organization classification of hematolymphoid tumors to recommend distinguishing EBV-positive (EBV+) and EBV-negative (EBV–) BL, based on their molecular features regardless of the epidemiological context and geographic location.6,7 At the histological level, BL shows macrophages with abundant cytoplasm dispersed in the background of a monomorphic population of rapidly proliferating B cells with basophilic cytoplasm. Reactive infiltrating lymphocytes are few, whereas scattered phagocytic macrophages give rise to the characteristic histological aspect of BL, known as the “starry sky” appearance.8 These macrophages, along with mesenchymal stem cells, stromal cells, and soluble factors, represent the main components of the TME of BL. The role of macrophages in BL tumor-associated macrophages (TAMs) is not fully understood, but they may contribute to tumor advancement by secreting chemokines and cytokines.9,10 Over the last 2 decades, different macrophage polarizations have been recognized as increasingly relevant for lymphomagenesis, contributing to the effects of the immune microenvironment.11,12 A constructive model inspired by T helper 1 (Th1) vs Th2 nomenclature has been developed to describe the complex mechanism of macrophage activation as polarization toward 2 opposite states: M1 with proinflammatory properties and M2 with protumoral properties. M1 macrophages produce proinflammatory cytokines which boost cancer immunosurveillance and cytotoxicity.13,14 However, these effects are counterbalanced by M2 macrophages with the anti-inflammatory and protumoral effects. Interestingly, there are some cases of BL that are characterized by conspicuous granulomatous reactions. These reactions can make detection of neoplastic proliferation difficult. Usually, these cases are EBV+ and tend to occur in the early stage of the disease, showing at times spontaneous regression without therapy.15-19 Macrophages in granulomas are derived both from circulating monocytes attracted by chemotaxis and from local resident macrophages recruited by T-cell–derived growth factors. CD4+ T cells accumulate in the center of epithelioid granulomas, whereas most CD8+ T cells are found at their periphery. Recent data have shown, using multiplex immunohistochemistry, that the TME of BL with granulomatous reaction is characterized by the prevalence of M1 macrophages and Th1 lymphocytes with a proinflammatory response, which may explain the spontaneous regression of such cases.19 Still, the differences in the composition of the cellular and soluble components of the BL TME in patients with both EBV+ and EBV– remain unclear. Therefore, the aim of this study was to delve deeper into the immune landscape of BL in 7 cases of EBV+ BL with granulomatous reaction compared with 8 cases of EBV+ BL and 8 BL EBV–, both with typical starry sky patterns, by applying NanoString technologies, focusing on the immune gene categories using a large panel of immune-related genes.
Materials and methods
Case selection
Formalin-fixed paraffin-embedded (FFPE) BL samples were obtained from the Department of Medical Biotechnologies (University of Siena, Siena, Italy), the Pathology section of the University of Florence (Florence, Italy), and the University of Nairobi (Nairobi, Kenya). The diagnosis of BL was made by hematopathology experts, who followed the essential criteria reported in the fifth edition of the World Health Organization classification of hematolymphoid tumors. Diagnostic immunohistochemistry was performed using a large panel of antibodies including CD20, CD10, BCL6, and LMO2. BCL2, Myc, and Ki67 on the Ventana BenchUltra (Roche Diagnostics, Monza, Italia), according to the manufacturer’s instructions. To assess the presence of EBV, in situ hybridization for EBV-encoded small RNAs was performed on all FFPE cases, cut at 5 μm, using an automated staining system (Ventana BenchMark ULTRA, Roche Diagnostics), as previously described10. Fluorescence in situ hybridization analysis for MYC rearrangement (Vysis MYC Dual Color Break Apart Rearrangements Probe; Abbott, Wiesbaden, Germany), translocation t (8;14) (q24;q32) (MYC::IgH, ZytoVision GmbH, Bremerhaven, Germany), and MYC::IgL (probes kindly provided by the Institute of Human Genetics, Ulm University Medical Center, Ulm, Germany) was performed for each case. BCL6 and BCL2 rearrangements were investigated in each case using respective break apart probes (Vysis BCL2 or BCL6 Dual Color Break Apart Rearrangements Probe; Abbott, Wiesbaden, Germany) according to the manufacturer’s instructions. Furthermore, aberrations on the long arm of chromosome 11q were evaluated using fluorescence in situ hybridization analysis (ZytoLight SPEC 11q gain/loss Triple Color Probe).19
Immune-related GEP using the NanoString platform
Digital multiplexed gene expression profiling (GEP) of 730 immune-related genes and 40 housekeeping genes was performed using the nCounter PanCancer Immune Profiling Panel (NanoString Technologies, Seattle, WA) on primary diagnostic FFPE tumor tissue.20 Total RNA from 23 FFPE 10 μm thick sections from each diagnostic sample was isolated using the RNeasy FFPE kit (Qiagen, Hilden, Germany). The protocol included deparaffinization, proteinase K digestion, extraction, elution, or hydration procedures, and DNase treatment to obtain DNase-free RNA. RNA concentration was measured using the Qubit RNA HS Assay Kit following the manufacturer’s instructions.
Bioinformatics and data analysis
Raw expression data were analyzed using the NanoStringNorm R package for background correction and between-sample normalization. Normalized data were then used to perform and plot principal component analysis (PCA) with the FactoMine and ggplot2 R packages, respectively. For hierarchical clustering analysis, the Euclidean distance metric across samples was considered for building trees within the pheatmap package. Differential expression analyses were carried out using the Bioconductor package limma and considering pairwise comparisons between different BL subtypes.21,22 We applied the voom21 method to model the mean-variance relationship, after which lmFit was used to fit per-gene linear models, and empirical Bayes moderation was applied with the eBayes function. Upregulated or downregulated genes were selected for subsequent analysis if their expression values were found to exceed an adjusted P value cutoff of <.05, after multiple testing corrections using a moderated t statistic.22 Genes differentially expressed were considered statistically significant when the adjusted P value (false discovery rate [FDR]) was <.05. Gene set enrichment analysis (GSEA) was performed using the gsea23 function of the phenoTest R package24 to test the association between the predefined groups of genes and BL subtypes. The gene lists used for this analysis were derived from KEGG and Reactome databases,25 of which only gene sets represented in the nCounter PanCancer Immune Profiling Panel were included in the computation. The output of GSEA is an enrichment score, a normalized enrichment score that accounts for the size of the gene set being tested, a P value, and an estimated FDR. Computing NES, P values and FDR requires a permutation-based approach, for which we computed 10 000 permutations. Heat maps were used for gene pathway representation. Additional pathways of interest were considered using the NanoString Panel Pro tool (https://nanostring.com/products/ncounter-assays-panels/panel-selection-tool/). The Kruskal-Wallis (nonparametric analysis of variance) test was used to identify pathways whose genes were associated with the BL groups. The heat map vertical bars indicate the significance of the Kruskal-Wallis P values. CIBERSORTx software was used to estimate the immune deconvolution fractions.26 The immune deconvolution fractions were compared between EBV+ BL with granulomatous reaction and BL with typical “starry sky” (both EBV+ and EBV–) using the Mann-Whitney nonparametric rank test, instead of the unpaired t test, because the data could not be assumed to be normally distributed. Bioinformatics and statistical analyses were performed using R statistical software (v4.3.0; http://www.R-project.org)27,28 (a detailed description of bioinformatic analysis is provided in the supplemental Data).
In situ messenger RNA (mRNA) hybridization and immunolocalization analyses
The human Interferon Gamma (IFNG) probe hybridization (Cod. 310501) was performed using the RNAscope 2.5 HD Detection Reagent-BROWN (Advanced Cell Diagnostic) in accordance with the manufacturer’s protocol. For multiple-marker immunostaining, the tissue section was subjected to sequential rounds of single-marker immunostaining, and the binding of the primary antibodies was revealed by fusing specific secondary antibodies conjugated with different enzymes (a detailed description of in situ hybridization is provided in supplemental Data29,30).
Multiplex immunostaining
Four-micrometers-thick human sections were deparaffinized, rehydrated, and unmasked using Novocastra Epitope Retrieval Solutions pH 9 in a thermostatic bath at 98°C for 30 minutes. Subsequently, sections were brought to room temperature and washed with phosphate-buffered saline. After Fc blocking by a specific protein block (Novocastra), the samples were incubated with primary antibodies against CD206 (1:4000 pH 9; Abcam), CD163 (clone 10D6, 1:100 Ph6; Novocastra), and CD68 (clone KP1, 1:50 pH 9; Dako), and the binding of the primary antibodies was revealed using specific secondary antibodies conjugated with different fluorophores anti-mouse and anti-rabbit (Alexa Fluor 488 and 568 conjugates). The slides were analyzed under a Zeiss Axioscope A1 microscope equipped with 4 fluorescence channels with widefield IF. Microphotographs were collected using a Zeiss Axiocam 503 color digital camera with Zen 2.0 software (Zeiss). Quantitative analyses of immunofluorescence staining were performed by calculating the average percentage of positive cells in 5 nonoverlapping fields at medium-power magnification (×200) using HALO image analysis software (v3.2.1851.229, Indica Labs). To gain a better understanding of macrophage polarization in our cohort, we applied multiplex immunofluorescence (mIF) to FFPE BL cases with granulomatous reactions. Recent studies have indicated that C-Maf is a specific marker for M2 macrophages10 (a detailed description of mIF is provided in the supplemental Data).
SOPHiA DDM Lymphoma Solution
DNA library preparation was performed using the SOPHiA DNA Library Prep Kit II (SOPHiA GENETICS, Lausanne, Switzerland), covering 73 lymphoma-related genes (a detailed description of Next Generation Sequencing [NGS] is provided in the supplemental Data).
All procedures performed in this study involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. This was a noninterventional study of archived tissue samples.
Results
Patient information
The case cohort comprised 10 females and 13 males, ranging in age from 5 to 70 years. Anatomical localization included the gastrointestinal tract, oral mucosa, bone marrow, lymph nodes, and abdominal mass. We analyzed 3 groups of BLs composed of 8 EBV– BL with starry sky pattern, 8 EBV+ BL with starry sky pattern, and 7 BL with granulomatous reaction. Among the latter, 5 of 7 cases were characterized by a diffuse granulomatous reaction that partially obscured neoplastic proliferation and 2 of 7 cases were characterized by a partial granulomatous reaction. The cases with granulomatous reactions were stage I/II and 3 of the 7 cases showed spontaneous regression. Two patients were in complete remission after 5 years of treatment 19, whereas 1 case was lost to follow-up after 3 years. In contrast, cases with typical starry sky, EBV+ or EBV–, were mainly stage III or IV, with bulky disease, significantly different from cases with granulomatous reaction (P < .001; Table 1). The diagnosis of BL was issued by expert hematopathologists. All the cases had typical morphology and immunophenotype features of BL (CD20+, CD10+, BCL6+, LMO2–, BCL2–, Myc >80%, and Ki67 >95%) and were harbored in 21 of 23 cases of IGH::MYC translocation, and 2 of the 20 IGL::MYC translocation demonstrated using fusion fluorescence in situ hybridization. BCL2 and BCL6 rearrangement and 11q aberration were not detected.
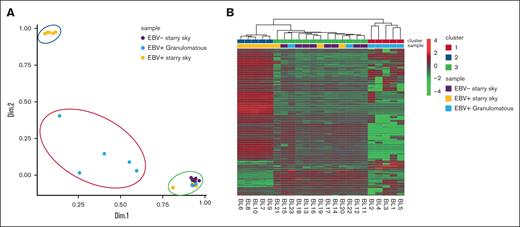
PCA and heat map graph identify 3 groups of BL
The NanoString panel analysis included all 23 BL FFPE samples that satisfied the RNA quality criteria. PCA and heat map graph revealed the presence of 3 distinct clusters (Figure 1A-B). Specifically, cluster 1 consisted of 5 BL with a diffuse granulomatous reaction, including 3 of 5 cases with spontaneous regression. Cluster 2 had 5 EBV+ BL with typical starry sky pattern and cluster 3 was enriched in EBV– BL with a typical starry sky, but also included 3 cases of EBV+ BL with a typical starry sky and 2 cases of BL with a partial granulomatous reaction (Table 1). GEP of BL samples showed significant heterogeneity in the immune response functional categories (a detailed description of GEP is reported in supplemental Table 1). Interestingly, clusters 2 and 3 showed similarities and differences in GEP. Indeed, some features were exclusively linked to the presence of EBV, as shown by the differentially enriched genes in the NF-κB, JAK-STAT, and B-cell receptor signaling pathways (supplemental Figure 1A-C, respectively).
PCA and heat map showed 3 clusters of BL. PCA (A) and heat map (B) revealed the presence of 3 distinct clusters. Cluster 1 consisted of the 5 BL with a diffuse granulomatous reaction (blue), cluster 2 had 5 EBV+ BL with typical starry sky pattern (orange), and cluster 3 includes 8 EBV– BL with typical starry sky (violet), 2 cases of BL with a partial granulomatous reaction, and 3 cases of EBV+ BL with typical starry sky. Heat map visualizing the expression levels of immune-related genes (rows) in each BL sample (column). Upregulated or downregulated genes were selected for subsequent analysis if their expression values were found to exceed an adjusted P value cutoff of <.05, after multiple testing correction using a moderated t statistic.
PCA and heat map showed 3 clusters of BL. PCA (A) and heat map (B) revealed the presence of 3 distinct clusters. Cluster 1 consisted of the 5 BL with a diffuse granulomatous reaction (blue), cluster 2 had 5 EBV+ BL with typical starry sky pattern (orange), and cluster 3 includes 8 EBV– BL with typical starry sky (violet), 2 cases of BL with a partial granulomatous reaction, and 3 cases of EBV+ BL with typical starry sky. Heat map visualizing the expression levels of immune-related genes (rows) in each BL sample (column). Upregulated or downregulated genes were selected for subsequent analysis if their expression values were found to exceed an adjusted P value cutoff of <.05, after multiple testing correction using a moderated t statistic.
GEP and GSEA of immune-related genes among BL cases show differences in terms of cytokines, chemokines, macrophage polarization, and immune checkpoint molecules
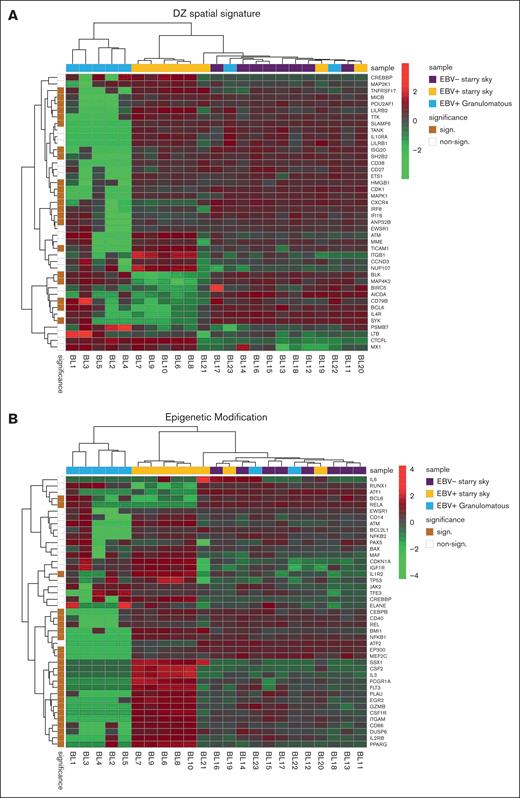
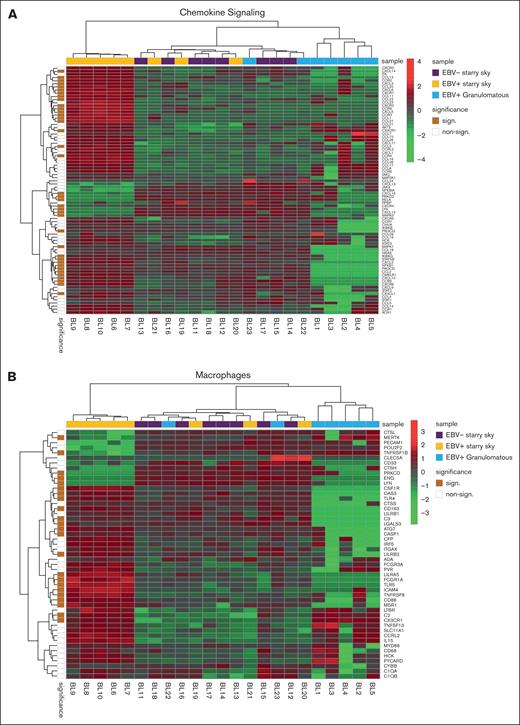
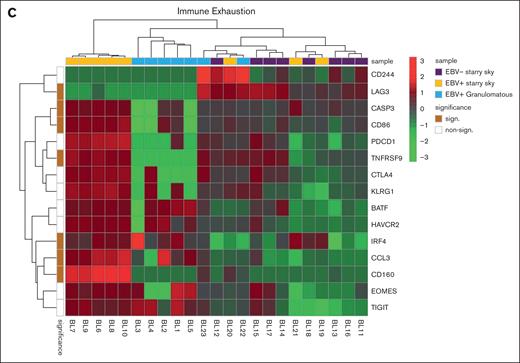
We performed GSEA to assess the degree of association between gene signatures derived from the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Reactome databases and BL cases. It is worth noting that the genes included in significant pathways were found mostly enriched in patients with BL EBV+ and EBV– with starry sky pattern (supplemental Tables 1 and 2). Specifically, genes within the interleukin-4 (IL-4)/IL-13 pathway were enriched in these 2 groups, showing upregulation of CD36 and IL13RA231,32 (supplemental Table 1). In addition, they were characterized by overexpression of CCL17, CCL22, CCL2, CCL18, CCR1, and CCR4, which are M2-related chemokines33-35 (Figure 2A). Indeed, BL with starry sky also showed upregulation of M2 immune response genes (CD163, LILRB1, LGALS3, and CSF1R) in comparison with cluster 1 (Figure 2B).36-39 Furthermore, our results demonstrated the enrichment of the immune exhaustion signature in BL with starry sky, both EBV+ and EBV–, as compared with BL with a granulomatous reaction. Specifically, our results showed the upregulation of several immune checkpoint molecules (IRF4, PDCD1, HAVCR2/TIM3, and CTLA4) (Figure 2C).40-42 In contrast, BL with granulomatous reaction showed downregulation of all M2-related genes (CD163, LILRB1, LGALS3, CCL17, CCL22, CCL2, CCR4, CD36, and IL13RA2). Interestingly, IRF3 and IFNG1, which contribute to the induction of M1 polarization, were upregulated (supplemental Table 1).43 Because the cell of origin of BL is characterized by a dark zone (DZ) signature,44 we considered a group of 169 DZ-UP genes differentially expressed between the DZ and LZ regions of reactive germinal centers profiled using the GeoMx Digital Spatial Profiler.45 This new gene set identified again 3 distinct clusters. In particular, BL with starry sky pattern in both EBV+ and EBV– was characterized by overexpression of DZ-expressed genes, whereas BL with granulomatous reaction showed downregulation of DZ genes. The differentially expressed genes belonged to immune evasion (LILRB1, IRF8, ITGB1, CTCFL, MICB, and CD38), M2-polarization (LILRB2, SLAMF6, HMGB1, ATM, AICDA, and POU2AF1) but also epigenetic modification (CREBBP, ETS1, HMGB1 CDK1, EWSR, ATM, and SH2B2) (Figure 3A-B).46-61 As the epigenetic cross talk between tumor cells and TME component activates signal cascades that result in immune evasion and T-cell exhaustion, we also considered the Epigenetic Modification NanoString pathway, which reveals different expression in epigenetic modulation genes between BL with starry sky, both EBV+ and EBV–, and BL with granulomatous reaction. Indeed, genes involved in epigenetic modulation (CREBBP, EP300, EWSR1, and ATM)56,59,60,62 were upregulated in BL with starry sky compared with BL with a granulomatous reaction. Finally, we performed target sequencing analysis in 3 cases of BL with granulomatous reaction for which the material was sufficient. The findings presented in supplemental Figure 2 are compared with the mutational landscape of classic BL, as reported in previous studies by us and others.63-65 BL with granulomatous reactions exhibits a low number of mutations, and notably, lacks mutations in genes related to epigenetic mechanisms, such as SMARC4A, ARID1A, and DDX3X, which are typically found in classical BL.64
BL with starry sky was characterized by DZ signatures and epigenetic modification signatures. Analysis of additional signatures showed that BL with starry sky was enriched in DZ signature (A) genes and epigenetic modulation signature (B).
BL with starry sky was characterized by DZ signatures and epigenetic modification signatures. Analysis of additional signatures showed that BL with starry sky was enriched in DZ signature (A) genes and epigenetic modulation signature (B).
CIBERSORTx platform: BL with granulomatous reactions were enriched in proinflammatory cells
Focusing on the deconvolution of the gene expression data, we applied the CIBERSORTx platform to better identify the immune cell proportions. Our results revealed a high percentage of CD8 and CD4 T cells, T follicular helper cells, and M1-macrophages in BL with granulomatous as compared with BL with starry sky, which showed a high prevalence of M2-macrophages (supplemental Table 3).
Validation of GEP analysis by multiplex immunostaining
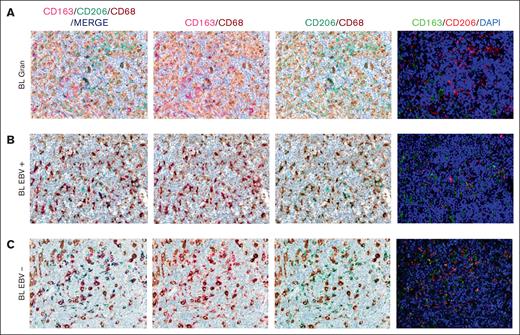
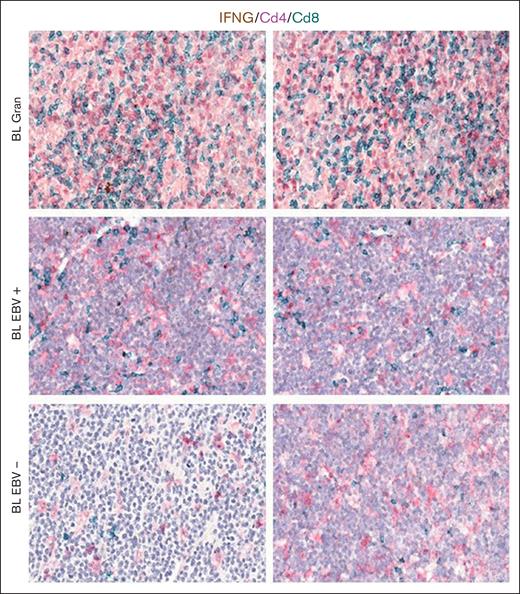
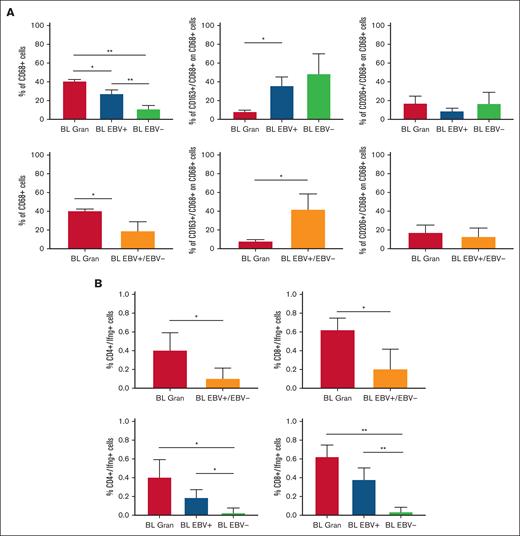
Based on the CIBERSORTx results, we decided to validate the M1 and M2 polarization and CD8+ and CD4+ T cells on the tissue section by applying multiplex immunostaining. In situ multiplexing immunostaining for CD68 with the M2 markers CD163 and CD206 was performed in BL with granulomatous reaction and BL with starry sky. In BL with granulomatous cases, the fraction of total macrophages was significantly higher, but with a lower density of M2/TAM polarized macrophages and starry sky histiocytes (Figure 4). These results were consistent with the increased infiltration of CD4 and CD8 T cells. Indeed, to further characterize the expression of IFNG in EBV+ BL with granulomatous reaction, we performed in situ mRNA hybridization of IFNG mRNA combined with immunohistochemistry for CD4 and CD8. Upon analyzing the results, we found that IFNG (Figure 5) was actively produced by tumor-infiltrating lymphocytes and was primarily associated with CD8+ cytotoxic lymphocytes, mainly at the edges of the neoplastic cells. These results are supported by the quantitative analysis of the signals performed with HALO image analysis software and statistical analysis using PRISM (Figure 6A-B). Additionally, these cases were triple-stained for CD68, CD163, and C-Maf. Overall, in all BL with granulomatous reaction, mIF showed a prevalence of M1 macrophage defined as CD68+/CD163–/C-Maf– cells, accounting for 80% to 95% of the total macrophages (supplemental Figure 5).
Multiplex immunostaining to validate M1 and M2 polarization. In BL granulomatous cases, the fraction of macrophages was significantly high, with a lower density of M2/TAM polarized macrophages (A). Conversely, BL with starry sky pattern, both EBV+ (B) and EBV– (C) showed an higher density of M2 macrophages. Original magnification, ×40.
Multiplex immunostaining to validate M1 and M2 polarization. In BL granulomatous cases, the fraction of macrophages was significantly high, with a lower density of M2/TAM polarized macrophages (A). Conversely, BL with starry sky pattern, both EBV+ (B) and EBV– (C) showed an higher density of M2 macrophages. Original magnification, ×40.
mRNA in situ hybridization for IFNG and immunohistochemistry (IHC) of CD4 and CD8. Representative microphotographs and quantitative analyses of mRNA in situ hybridization for IFNG and IHC for CD4 and CD8 showing the increase in the prevalence of CD8+ cytotoxic lymphocytes expressing IFNG in BL case. Original magnification, ×200 and ×400.
mRNA in situ hybridization for IFNG and immunohistochemistry (IHC) of CD4 and CD8. Representative microphotographs and quantitative analyses of mRNA in situ hybridization for IFNG and IHC for CD4 and CD8 showing the increase in the prevalence of CD8+ cytotoxic lymphocytes expressing IFNG in BL case. Original magnification, ×200 and ×400.
Quantitative analysis of immunostaining. HALO image analysis software and statistical analysis using PRISM supported the results of immunostaining (A) and combined RNAscope and IHC.
Quantitative analysis of immunostaining. HALO image analysis software and statistical analysis using PRISM supported the results of immunostaining (A) and combined RNAscope and IHC.
Discussion
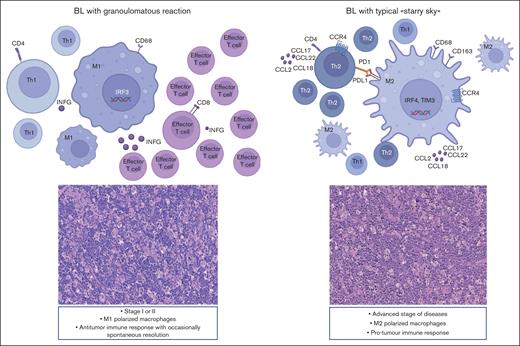
The lymphoma microenvironment is a complex and dynamic network that includes immune cells, stromal cells, cytokines, blood vessels, and extracellular matrix components. The arrangement of this network is guided by neoplastic cells and can influence tumor initiation, progression, resistance to cell death, evasion from growth suppressors, and drug resistance.66,67 Although there have been numerous studies investigating TME expression in B-cell lymphomas, there has been a limited amount of research conducted in BL.19 Recent data using multiplex immunohistochemistry show that the TME of BL with granulomatous reaction is different, being characterized by a prevalence of M1 macrophages and proinflammatory response, which may possibly explain at times the spontaneous regression.10 In this study, we further portrayed the immune landscape of BL using GEP with NanoString technologies, focusing on stromal cells, soluble molecules, and immune gene categories using a 730 immune-related genes panel. Our analysis led to the separation of BL into 3 clusters by GEP analysis based on immune gene categories. Cluster 1 consisted of 5 BL cases with diffuse granulomatous reaction, whereas clusters 2 and 3 were enriched of cases with BL with typical starry sky pattern, including EBV+ or EBV–, respectively. Some features could be linked exclusively to the presence of EBV by comparing the TME of EBV+ BL and EBV– BL. Indeed, our results confirm previous findings of an upregulation of the NF-κB and JAK-STAT pathways in EBV+ BL, whereas a “tonic” activation of B-cell receptor signaling was more prevalent in EBV– BL.68-74 In addition to our previous studies, here we demonstrated an increased expression of M1 genes such as IRF3 and INFG, in BL with a granulomatous reaction. IRF3 is constitutively expressed in various cell types and binds to conserved sequences known as IFN–stimulated response elements, which induce transcription of type I IFN genes (IFNG1).74,75 By applying in situ mRNA hybridization and immunolocalization analyses, we were able to show that a fraction of effector T cells was characterized by active IFNG production within the inflammatory milieu of such peculiar BL cases. In particular, INFG was found to be mainly expressed in CD8+ cells, suggesting its role as an effector cell in the immune response against neoplastic cells. In contrast, we demonstrated an upregulation of the IL-4/IL-13 pathway (CD36 and IL13RA2),31,32 M2-secreted chemokines (CCL17, CCL22, CCL2, CCR4, CCL18, and CCR1),33-35 and M2-immune response genes (CD163, LILRB1, LGALS3, and CSF1R)36-39 in cases of BL with the typical starry sky. These results were also validated by multiplex immunostaining, which showed the prevalence of M2-macrophages in BL with starry sky associated with a low content of T cells (cold lymphomas), as compared with BL with granulomatous reaction characterized by a high proportion of M1-macrophages and reactive T lymphocytes (hot lymphomas). Intriguingly, a recent paper reported that targeting the chemokine CCL22 induces an intense granulomatous reaction and limits EBV infection from spreading in an experimental model.40 Furthermore, here we demonstrated that BL with starry sky shows overexpression of immune checkpoint genes (IRF4, PDCD1, HAVCR2/TIM3, and CTLA4), which favors the immune escape of tumor.41-43 These results were further confirmed by the analysis of additional signatures. In particular DZ signature genes (CD38, CDK1, SLAMF6, CD27, TNFRSF17, MAPK1, SH2B2, CREBBP, LILRB1, EWSR1, ATM, AICDA, ETS1, HMGB1, POU2AF1, TTK, TANK, and IFI16) and the epigenetic modulation signature (CREBBP, EP300, EWSR1, and ATM)47-63,76 were upregulated in BL with starry sky both EBV+ and EBV–. Although these differences may be related to diverse tumor cell components, we cannot completely exclude a different cell of origin during the B-cell differentiation steps, as differences in TME-related genes, not typical of the DZ, in addition to tumor-related genes (epigenetic mechanism and metabolism) were identified.20,77 The upregulation of the epigenetic modulation genes may activate a signal cascade that results in immune evasion and T-cell exhaustion in these cases.78 The genetic landscape also revealed a low number of mutations and a lack of mutations in genes related to epigenetic mechanisms and immunescape in BL with granulomatous reaction. Although our data do not provide clear evidence of intrinsic differences in the tumor cells, our results revealed differences related to genes involved in both genetic and epigenetic mechanisms that are at work favoring the immunescape in BL with a starry sky pattern, both EBV+/EBV–.64-66 Finally, our series showed that BL with a diffuse granulomatous reaction is typically diagnosed at an early disease stage, namely, stage I or II. Interestingly, 3 of 5 cases showed spontaneous regression, and 2 patients were in complete remission after 5 years of follow-up. In contrast, BLs with a starry sky pattern, both EBV+ and EBV–, were significantly associated with advanced stages of disease (stage III or IV and bulky disease). Thus, based on our results, we may envision biological-clinical settings characterized by an immune response able to control the neoplastic growth in cases with a diffuse granulomatous reaction and a protumor immune response in BL with starry sky pattern (Figure 7). The dynamic process of macrophage plasticity may be responsible for this scenario. Novel therapeutic strategies, able to induce TAM repolarization and target epigenetic regulators, have been introduced and may have the potential to be a game-changer in the fight against BL lymphoma and improve patient outcomes, opening up alternative therapeutic avenues for patients refractory to therapy.78,79
BL can show 2 different biological and clinical settings. M1 polarized macrophages prevail, and the immune response may be able to control neoplastic growth. In contrast, in cases characterized by the starry sky pattern, M2 macrophages dominate and may be responsible for a protumor immune response, resulting in disease rapid progression and dissemination.
BL can show 2 different biological and clinical settings. M1 polarized macrophages prevail, and the immune response may be able to control neoplastic growth. In contrast, in cases characterized by the starry sky pattern, M2 macrophages dominate and may be responsible for a protumor immune response, resulting in disease rapid progression and dissemination.
Acknowledgment
This study was supported by the Associazione Italiana per la Ricerca sul Cancro 5 × 1000 grant (number 21198 [S.P.]).
Authorship
Contribution: M.C.S. designed, performed, and interpreted experiments bioinformatic analyses, and wrote the manuscript; G.B. performed and interpreted the statistical and bioinformatic analyses; M.D.C. and M.R.S. performed bioinformatic analyses; G.M. performed multiplex immunostaining and combined RNAscope and immunohistochemistry; S.T., G.B., C.I.L., E.F., B.B., F.A., R.G., and R.B. performed experiments; M.G., M.V., V.M., N.O., J.N., R.S., G.D.S., D.F., C.B., T.M., G.O., R.B., L.Q.F., F.F., P.M., C.T., S.P., S.L., and L.L. provided and analyzed the molecular and clinical data of patients with Burkitt lymphoma; S.L. and L.L. designed and supervised the experiments, analyzed the data, and wrote the manuscript; and all authors discussed the results and commented on the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stefano Lazzi, Department of Medical Biotechnology, Anatomical Pathology Section, University of Siena, via delle Scotte, Siena 53100, Italy; email: lazzi2@unisi.it.
References
Author notes
M.C.S. and G.B. contributed equally to this study.
Data are available on request from coauthor, Maria Chiara Siciliano (siciliano10@student.unisi.it).
The full-text version of this article contains a data supplement.