Key Points
Platelet Ido1 is upregulated in response to IFN-γ including in Plasmodium infection.
Platelets modulate plasma tryptophan metabolites altering kynurenine-to-tryptophan ratios.
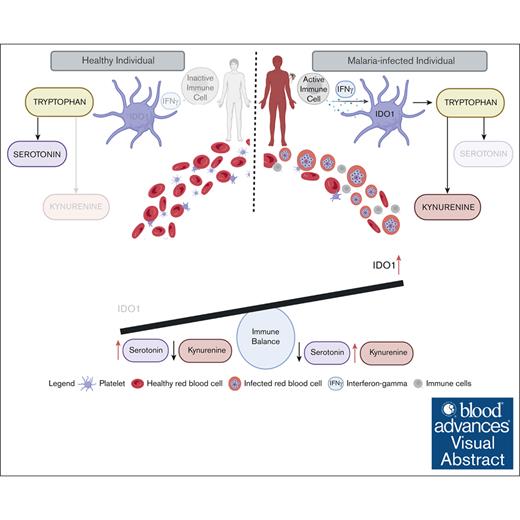
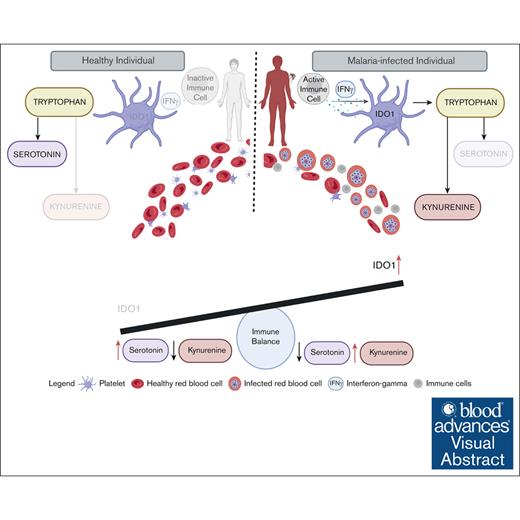
Visual Abstract
Platelets are immune responsive in many diseases as noted by changes in platelet messenger RNA in conditions such as sepsis, atherosclerosis, COVID-19, and many other inflammatory and infectious etiologies. The malaria causing Plasmodium parasite is a persistent public health threat and significant evidence shows that platelets participate in host responses to infection. Using a mouse model of nonlethal/uncomplicated malaria, non-lethal Plasmodium yoelii strain XNL (PyNL)-infected but not control mouse platelets expressed Ido1, a rate limiting enzyme in tryptophan metabolism that increases kynurenine at the expense of serotonin. Interferon-γ (IFN-γ) is a potent inducer of Ido1 and mice treated with recombinant IFN-γ had increased platelet Ido1 and IDO1 activity. PyNL-infected mice treated with anti-IFN-γ antibody had similar platelet Ido1 and metabolic profiles to that of uninfected controls. PyNL-infected mice become thrombocytopenic by day 7 after infection and transfusion of platelets from IFN-γ–treated wild-type mice but not Ido1−/− mice increased the plasma kynurenine-to-tryptophan ratio, indicating that platelets are a source of postinfection IDO1 activity. We generated platelet-specific Ido1 knockout mice to assess the contribution of platelet Ido1 during PyNL infection. Platelet-specific Ido1−/− mice had increased death and evidence of lung thrombi, which were not present in infected wild-type mice. Platelet Ido1 may be a significant contributor to plasma kynurenine in IFN-γ-driven immune processes and the loss of platelets may limit total Ido1, leading to immune and vascular dysfunction.
Introduction
Platelets are small anucleate cells released from megakaryocytes,1 most studied for their role in hemostasis and thrombosis, but platelets also participate in wound healing,2 immune sensing,3-7 and immune regulation.8,9 Platelets have central roles in innate and adaptive immune responses during infections,10 including malaria,11 contributing to both infection resolution12 and disease pathology.13 Similar to how “typical” immune cells adapt their responses to immune stimuli, immune molecule expression and immune functions of platelets may also be influenced by inflammatory or vascular diseases.6,14
Metabolism is rapidly becoming a better-understood regulator of immune cell adaptation to shifts in environmental demands.15-17 Glycolysis is a fuel for platelet activation,18 but platelet metabolism beyond glycolysis has received little attention. Indoleamine 2,3-dioxygenase (IDO1) is an interferon-gamma (IFN-γ)-induced immunomodulatory rate-limiting enzyme that catalyzes the first step in tryptophan (TRP) catabolism toward the kynurenine (KYN) pathway and away from the serotonin (5-hydroxytryptamine) pathway.19 IDO1 is expressed by few cells under basal conditions but is increased by inflammation.20,21 KYN is a metabolic product of IDO1, most associated with limiting immune cell activity.22 Increased IDO1 and its enzymatic activity lead to alterations in the KYN-to-TRP ratio (KTR) that has been noted in human patients with Plasmodium vivax infection.23 Mouse malaria models have also indicated that Ido1 expression is increased by infection, leading to changes in TRP metabolism, and inhibiting Ido1 provided mice with protection against cerebral malaria.24
Despite significant research investment, malaria remains prevalent in many regions of the world,25 with >240 million infections and 600 000 deaths annually.26 There is a need for a deeper understanding of malaria pathogenesis to develop adjunctive therapies. The role of platelets in malaria is complex and the literature presents seemingly contradicting evidence for platelet roles in mouse models of malaria infection.27 Some studies have shown that platelet activation drives the cerebral vascular pathology associated with cerebral or severe malaria,28,29 and other studies have indicated that platelets may limit parasitemia in uncomplicated malaria models.12,30,31 Thrombocytopenia is a frequent complication of all forms of malaria32-34 and is associated with a higher mortality rate, further complicating an understanding of platelets in malaria infections.34-36 The causes of malaria-associated thrombocytopenia are poorly understood but may be due to, in part, decreased platelet production, increased platelet clearance, a shortened platelet life span, increased apoptosis, and/or platelet sequestration.37-41 Alterations in platelet count and/or responses affect the host’s immune responses to infection.42
A gap in understanding malaria immune responses is the immunometabolic potential of platelets. Emerging evidence shows that P vivax, a malaria strain previously thought to be more benign than P falciparum, has been increasing in severity and mortality.43 Considering the increasing severity of P vivax infection, and particularly its significant association with thrombocytopenia, there is a need to investigate the immunometabolic contributions of platelets and their impact on infection outcomes. We have now found that infection-associated IFN-γ increased platelet Ido1, which contributes to higher plasma KYN levels. Furthermore, our findings show that mice lacking platelet Ido1 had increased lung thrombi and P yoelii–associated death compared with control infected mice.
Materials and methods
Detailed materials and methods can be found in the supplemental Materials.
Ethics statement and study approval
All animal experiments performed were approved by the University of Rochester Medical Center institutional animal care and use committee under the protocol number 101984/2009-022E.
Mice studies
All mice used were on a C57BL/6J background. Control C57BL/6J mice (stock number: 000664) and Ido1−/− mice (stock number: 005867) were from The Jackson Laboratory. Ido1flox/flox mice from Matthew Ciorba (Washington University School of Medicine, St Louis, MO) were crossed with platelet Factor 4 cre-recominase (PF4-Cre) mice (The Jackson Laboratory, stock number: 008535) to generate platelet-specific Ido1−/− (Plt-Ido1−/−) mice. Studies used both female and male mice aged 8 to 12 weeks with no significant sex-based differences. All mouse studies, including infections, treatments, and sample collections were performed in the mornings between 8 and 10 AM.
P yoelii infection
The non-lethal Plasmodium yoelii strain XNL (PyNL) was obtained from BEI Resources, National Institute of Allegery and Infectious Diseases, National Institutes of Health: Plasmodium yoelii subspecies Yoelii, strain 17XNL (1.1), MRA-593. A total of 5 × 105 infected red blood cells (RBCs) were intraperitoneally (IP) injected into C57BL/6J surrogate mice to establish a blood-stage malaria infection. Blood was collected from surrogate mice 3 to 7 days after infection and used to infect experimental mice. Parasitemia was assessed using Wright-Giesma–stained thin blood or by flow cytometry. SYBR (fluorescein isothiocyanate) Green I Nucleic Acid Gel Stain (Invitrogen, S7567) was diluted (1:10 000) into phosphate-buffered saline (PBS) and 1 μL of Ter119-APC antibody (Biolegend, 116212) was added (1:100) and adjusted to 20 μL final volume with 3 μL of blood to identify RBCs. After 15 minutes of room-temperature incubation in the dark, the cells were washed with 1 mL of PBS, and analysis was carried out using Accuri C6. Parasitized RBCs were identified as SYBR+/Ter119+ cells. The analysis was performed with the FlowJo software version 10.0.7.
Platelet transfusions
Wild-type (WT) platelet donor mice received IP injections of either 100 μL of PBS or 1 μg/100 μL of IFN-γ (Fisher Scientific, 485MI100CF). For IFN-γ treatment, mice received IP injections of 1 μg/100 μL IFN-γ for 3 consecutive days. On transfusion days, platelets were collected from donor mice via retro-orbital bleed into heparinized Tyrodes, the platelet pellet was resuspended in PBS, and platelet counts were adjusted to be equal among transfusion groups, with 700 × 103 to 800 × 103 platelets per μL from 5 donor mice. Platelet transfusions were carried out using 100 μL of platelets injected retro-orbitally. On day 14, mice were lethally bled to collect samples for subsequent analyses. See supplemental Materials for a detailed timeline and experimental setup (supplemental Table 1).
In the anti-IFN-γ treatment experiments, mice were treated with either 200 μg of anti-IFN-γ (BioXCell, BE0055) or immunoglobulin G (IgG; BioXCell, BE0088) at days 0, 3, and 6. All reagents were diluted in sterile PBS.
RNA-seq
RNA sequencing (RNA-seq) was performed on platelets isolated from healthy controls or patients infected with P vivax as well as from P yoelii–infected mice, as detailed in supplemental Methods.
ELISAs
Enzyme-linked immunosorbent assays (ELISAs) were performed following the manufacturers’ instructions to analyze mouse plasma for serotonin (Fitzgerald Industries, 55R-1865).
Plasma sample preparation for LC-MS/MS analysis
Plasma supernatants were thawed at room temperature, diluted 1:5 with a mixture of high-performance liquid chromatography (HPLC)-grade methanol (MeOH) and water (80:20; Omnisolv MeOH number MX0488-1 and Omnisolv water number WX0004-1), centrifuged at 21 380g for 10 minutes to remove precipitates, and the clear supernatants were transferred to new tubes. Samples were then divided for underivatized and derivatized method runs. Approximately 100 μL of underivatized samples were transferred to HPLC microsampling vials (Thermo Fisher, 05-704-225) and sealed with cap closures (Thermo Fisher, 14-823-380) and used for KYN and 5-hydroxyindoleacetic acid (5-HIAA) analysis. For amino acid analysis, 90 μL of diluted plasma with 5 μL of trimethylamine (Sigma number 471283) and 1 μL of benzylchloroformate (Acros number 152940050) were incubated for 5 minutes at room temperature, centrifuged at 21 380g for 10 minutes to remove precipitates, and the supernatants were transferred to HPLC microsampling vials and sealed with cap closures. HPLC-grade MeOH and water mixture was used as blank control.
In some samples (Figure 5), additional cleanup steps were incorporated to eliminate lipids and debris from plasma to simplify machine care.44 These changes were compared and validated alongside our original method. For solid-phase extraction cleanup, 1% ammonium hydroxide (NH4OH; Fisher Scientific, AC390070010) in HPLC-grade water (volume per volume [v/v])–to-MeOH (50:50, v/v) and 1% formic acid (Fisher Scientific, A117-50) in H2O (v/v) were prepared and stored at 4°C. Plasma samples (200 μL) were processed with each cartridge within a Visiprep SPE vacuum manifold (Millipore Sigma, 57030-U) with disposable liners (Millipore Sigma, 57059). Waters hydrophilic-lipophilic balance cartridges (3 cc) were used (Mildford, MA, part no. WAT094226), and borosilicate glass tubes were placed in the manifold to collect the elute (VWR, 47729-568). The cartridge was conditioned without drying, starting with 500 μL of MeOH followed by 500 μL of 1% formic acid. A 200 μL plasma sample was loaded onto the cartridge, followed by 250 μL of 1% formic acid. The loaded cartridge was washed with 250 μL of 1% formic acid, and then dried for 10 minutes using a slight vacuum. After drying, the absorbed analytes were eluted with 1 mL of 1% NH4OH in MeOH: water, and the eluent was transferred to a 15 mL conical tube and evaporated under a gentle stream of nitrogen at room temperature. The dried residue was reconstituted with 100 μL of 1% NH4OH in MeOH-water mixture and transferred to an HPLC autosampler vial for injection into the HPLC–tandem mass spectrometry (LC-MS/MS) system for underivatized analysis.
Analysis of metabolites
For the data generated in Figures 2, 3, and 4, LC-MS/MS analysis used selected-reaction monitoring (supplemental Table 2) using a Shimadzu high-performance LC system coupled (with reversed phase chromatography with an ion-pairing reagent) to a Thermo Quantum triple-quadrupole mass spectrometer, which is run in negative mode with selected-reaction monitoring specific scans as previously described.45-47 Detailed LC-MS/MS and metabolic analysis methods can be found in supplemental Methods.
Immunohistochemistry
WT and psIdo1-/- mouse lungs were collected and placed in 60% MeOH, 10% acetic acid, 30% distilled water, paraffin embedded, and sectioned. For immunostaining, slides were deparaffinized and rehydrated and placed into 3% hydrogen peroxide for 15 minutes, then washed 3 times with PBS. Slides were incubated in Dako target retrieval solution (catalog S1699) for 15 minutes in a pressure cooker, washed 3 times in PBS, and incubated in Animal-Free Blocker and diluent (catalog SP-5035) for 30 minutes. Fibrinogen α-chain recombinant rabbit monoclonal antibody (Invitrogen, catalog no. MA5-37943) was diluted 1:200 into Dako antibody diluent (catalog S0809) and incubated overnight at 4°C. CD42c (Sapphire North America, catalog DF9759) was diluted 1:250 into Dako antibody diluent and incubated overnight at 4°C. After incubation, slides were rinsed in PBS and covered with ImmPRESS horseradish peroxidase anti-rabbit IgG reagent (Vector Laboratories, MP-7451) and were incubated for 30 minutes at room temperature. Slides were rinsed 3 times with PBS for 3 minutes each. 3,3' diaminobenzidine peroxidase (horseradish peroxidase) substrate (Vector Laboratories, SK-4100) were added to the slides for 5 minutes. Slides were washed in distilled H2O for 5 minutes, counterstained, and coverslips were added.
Using a BX51 microscope, images were taken under ×40 or ×20 magnification. Images were acquired with Olympus DP74 with the cellSens standard software. For analysis with ImageJ, each image was converted to 8-bit; threshold was adjusted to differentiate between positive cells (dark brown) and negative (blue) and measured by pixel intensity. For CD42c immunostains, a scale was used to measure thrombi of <10 μm or >10 μm. Data analysis was performed with GraphPad Prism.
Statistics
GraphPad Prism version 9 was used for statistical analysis. Data are presented as mean ± standard error of the mean. The data were assessed using an unpaired 2-tailed Student t test, and significance was defined as a P value < .05. The significance levels are represented by asterisks, such as ∗ for P < .05, ∗∗ for P < .01, ∗∗∗ for P < .001, and ∗∗∗∗ for P < .0001.
Results
IDO1 messenger RNA (mRNA) is expressed in platelets from humans infected with P vivax and mice infected with P yoelii
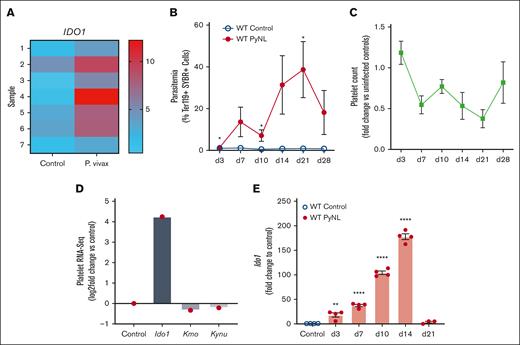
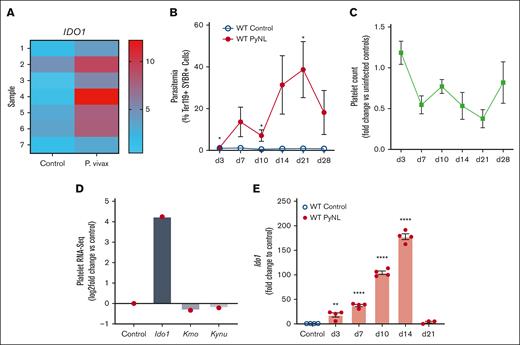
Platelets have important roles in immune responses and their mRNA expression is changed during infections and inflammatory conditions. RNA-seq analysis of human platelets isolated from healthy controls or individuals infected by P vivax revealed an upregulation of IDO1 in platelets from those who were infected (Figure 1A, platelet donor demographics in supplemental Table 4). P yoelii is a nonlethal (PyNL) mouse model of uncomplicated malaria that has an infectious time course of ∼28 days (Figure 1B) with mice becoming thrombocytopenic before clearing the infection (Figure 1C). Platelets were isolated from infected and uninfected control mice on day 7 after infection to isolate mRNA for RNA-seq. RNA-seq revealed increased Ido1 in infected mouse platelets and changes in enzymes involved with the downstream KYN pathway (Figure 1D). IDO1 is an inducible enzyme with increased expression in response to IFN-γ, a potent cytokine released during Plasmodium infection.21,48-50 To determine platelet Ido1 expression during PyNL infection, platelets were isolated from control or infected mice at multiple time points after infection to isolate platelet mRNA for quantitative reverse transcription polymerase chain reaction. Platelet Ido1 was greatly increased over the first 14 days after infection but was not expressed in control mouse platelets (Figure 1E). These data indicated that Plasmodium infection increased platelet Ido1.
Plasmodium infection increased platelet Ido1. (A) Heat map of RNA-seq for IDO1 expression in platelets isolated from patients infected with P vivax and healthy controls. Individual bars represent samples derived from 7 controls and 7 patients with P vivax infection. (B) Percentage of SYBR+ parasitized RBCs measured at multiple time points using flow cytometry. (n = 5, ∗P < .05). (C) Platelet counts were obtained on multiple days after infection and expressed as fold change vs uninfected controls (n = 5). (D) RNA-seq of mouse platelets also revealed changes in Ido1 in a mouse infection model. (E) Platelet RNA was isolated from PyNL-infected mice and polymerase chain reaction performed for Ido1 expression at multiple time points. (∗P < 0,5, ∗∗P < .01, and ∗∗∗∗P < .0001).
Plasmodium infection increased platelet Ido1. (A) Heat map of RNA-seq for IDO1 expression in platelets isolated from patients infected with P vivax and healthy controls. Individual bars represent samples derived from 7 controls and 7 patients with P vivax infection. (B) Percentage of SYBR+ parasitized RBCs measured at multiple time points using flow cytometry. (n = 5, ∗P < .05). (C) Platelet counts were obtained on multiple days after infection and expressed as fold change vs uninfected controls (n = 5). (D) RNA-seq of mouse platelets also revealed changes in Ido1 in a mouse infection model. (E) Platelet RNA was isolated from PyNL-infected mice and polymerase chain reaction performed for Ido1 expression at multiple time points. (∗P < 0,5, ∗∗P < .01, and ∗∗∗∗P < .0001).
Metabolic profiling of TRP and KYN pathways reveals changes associated with Ido1 expression during PyNL infection
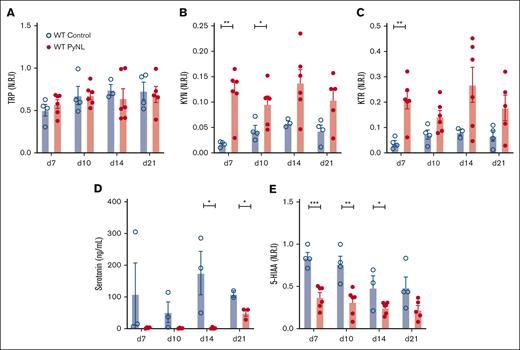
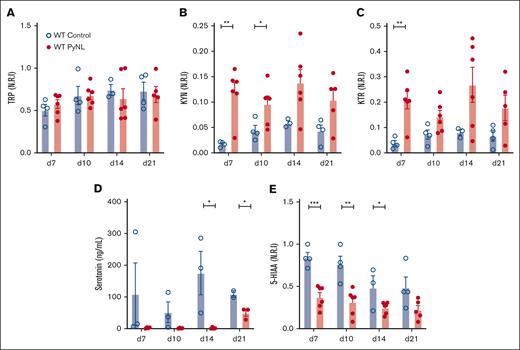
IDO1 metabolizes TRP to KYN and away from serotonin. Plasma was isolated from control and PyNL-infected mice at multiple time points and processed for LC-MS/MS. Intensity level measurements were normalized to the maximum metabolite found and reported as normalized relative intensity. TRP levels remained consistent throughout infection compared with controls, which indicated that TRP’s bioavailability itself is not a major limiting factor in KYN metabolism (Figure 2A). Plasma KYN was increased during infection (Figure 2B). KTR is a measurement of Ido1 activity and was increased in infected mice (Figure 2C). Serotonin and 5-HIAA (the metabolic breakdown product of serotonin)51 were less in infected plasma samples than in controls (Figure 2D-E). These data indicate that TRP is shunted away from the serotonin pathway and to the KYN pathway during PyNL infection.
Metabolic alterations within TRP/KYN pathway during P yoelii infection. Plasma was obtained from female and male control and infected mice at multiple time points. Normalized relative intensity measurements of metabolites were obtained from LC-MS/MS. (A) TRP (B) KYN (C) KTR as a measurement of Ido1 activity. (D) Serotonin levels measured in an ELISA and (E) normalized relative intensity of serotonin metabolite, 5-HIAA (∗P < .05, ∗∗P < .01, ∗∗∗P < .001).
Metabolic alterations within TRP/KYN pathway during P yoelii infection. Plasma was obtained from female and male control and infected mice at multiple time points. Normalized relative intensity measurements of metabolites were obtained from LC-MS/MS. (A) TRP (B) KYN (C) KTR as a measurement of Ido1 activity. (D) Serotonin levels measured in an ELISA and (E) normalized relative intensity of serotonin metabolite, 5-HIAA (∗P < .05, ∗∗P < .01, ∗∗∗P < .001).
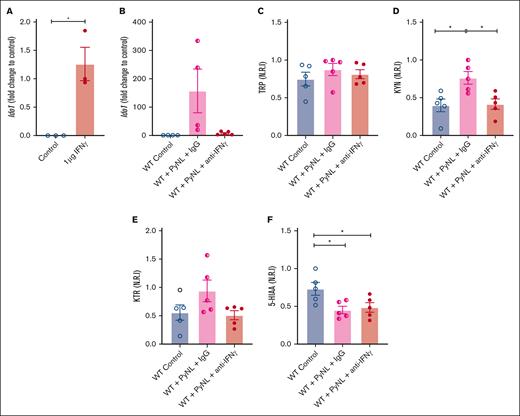
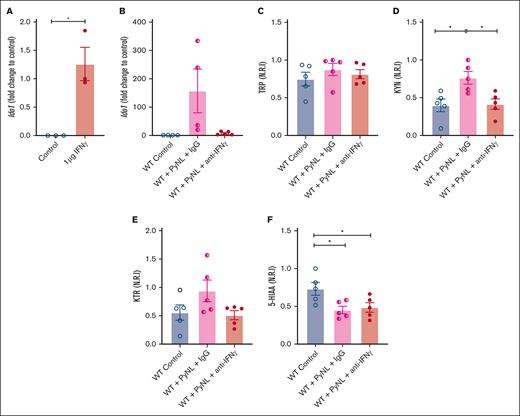
IFN-γ is a potent inducer of Ido1 and is increased in malaria infection.52-54 We determined the role of IFN-γ in platelet Ido1 expression by treating WT mice with control buffer or 1 μg of IFN-γ daily for 3 days. On day 4, platelet mRNA was isolated to quantify Ido1 by quantitative reverse transcription polymerase chain reaction. Platelets from control mice had no Ido1 whereas Ido1 was increased in IFN-γ-treated mice (Figure 3A). Next, we determined whether IFN-γ increased platelet Ido1 in PyNL infection. Infected mice received control IgG or anti-IFN-γ antibody at days 0, 3, and 6 after infection. On day 7, platelets and plasma were isolated. Isolated platelets from infected mice treated with IgG had increased Ido1, but mice that received anti-IFN-γ antibody did not (Figure 3B), whereas plasma TRP did not change (Figure 3C). Control uninfected mice and PyNL-infected mice treated with anti-IFN-γ antibody had similar levels of KYN, whereas PyNL-infected mice that received IgG had increased plasma KYN (Figure 3D). The KTR in the uninfected control group was similar to that in PyNL-infected mice that received anti-IFN-γ but not the control IgG treatment group (Figure 3E). 5-HIAA was reduced in both infected groups regardless of the treatment conditions compared with the control group (Figure 3F). Taken together, the data indicate that IFN-γ increased platelet Ido1 in PyNL infection and is a major regulator of plasma KYN concentrations.
IFN-γ increases platelet Ido1 expression. (A) IFN-γ treatment increased platelet Ido1 expression (n = 3). (B) Anti-IFN-γ treatment during PyNL infection inhibited platelet Ido1 expression at day 7 after infection. Normalized relative intensity levels on day 7 of (C) TRP, (D) KYN, and (E) KTR as an indicator of IDO1 activity. (F) Normalized relative intensity levels of 5-HIAA at day 7 after infection (∗P < .05).
IFN-γ increases platelet Ido1 expression. (A) IFN-γ treatment increased platelet Ido1 expression (n = 3). (B) Anti-IFN-γ treatment during PyNL infection inhibited platelet Ido1 expression at day 7 after infection. Normalized relative intensity levels on day 7 of (C) TRP, (D) KYN, and (E) KTR as an indicator of IDO1 activity. (F) Normalized relative intensity levels of 5-HIAA at day 7 after infection (∗P < .05).
Assessment of platelet contribution to TRP metabolism
Given that platelet Ido1 was increased in Plasmodium infection as well as plasma KYN, we sought to determine whether platelets contributed to plasma KYN changes in PyNL infection. PyNL infection results in thrombocytopenia, meaning that infection-associated loss of platelets may affect TRP metabolism. To begin to explore the role of platelets in TRP metabolism, we used platelet transfusions with the expectation that platelets from IFN-γ-treated mice would express Ido1 and increase KTR (experimental setup shown in Figure 4A). Mice were given transfusions of control WT platelets, platelets from IFN-γ-treated WT mice to induce IDO1, or platelets from Ido1−/− mice treated with IFN-γ, on days 7 and 10 after infection, and plasma collected on day 14 (note: carboxyfluorescein diacetate succinimidyl ester–labeled transfused platelets remained in circulation of infected mice for up to 3 days after transfusion; supplemental Data; Figure 1). PyNL-infected mice given platelets from IFN-γ-treated WT mice had reduced plasma TRP, perhaps due to increased IDO1 activity (Figure 4B). Plasma KYN was increased in PyNL-infected mice that received either WT- or IFN-γ-treated WT mouse platelet transfusions (Figure 4C). As expected, IDO1 activity was greatest in the WT IFN-γ-treated mouse platelet transfusion group (Figure 4D). There was no significant difference in platelet transfusions from Ido1−/− that received IFN-γ. We also assessed plasma serotonin levels by ELISA (Figure 4E) and plasma 5-HIAA by LC-MS/MS (Figure 4F). Serotonin and 5-HIAA were less in all infected groups than in control mice, but serotonin in particular was increased in the Ido1−/− + IFN-γ platelet transfusion group (Figure 4E). Taken together, these data show that platelets modulate TRP metabolic phenotype during infection and that infection-induced thrombocytopenia may contribute to immunometabolic alterations.
Platelet transfusions alter plasma TRP metabolites in P yoelii infection. (A) Schematic diagram of platelet transfusion experimental set up (created with BioRender.com). (B) TRP and (C) KYN levels obtained from LC-MS/MS. (D) KTR as a measurement of IDO1 activity. (E) Serotonin quantification measured by ELISA, and (F) 5-HIAA levels obtained from LC-MS/MS (∗P < .05, ∗∗P < .01, ∗∗∗P < .001).
Platelet transfusions alter plasma TRP metabolites in P yoelii infection. (A) Schematic diagram of platelet transfusion experimental set up (created with BioRender.com). (B) TRP and (C) KYN levels obtained from LC-MS/MS. (D) KTR as a measurement of IDO1 activity. (E) Serotonin quantification measured by ELISA, and (F) 5-HIAA levels obtained from LC-MS/MS (∗P < .05, ∗∗P < .01, ∗∗∗P < .001).
Platelet IDO1 may contribute to infection outcomes and subsequent metabolic changes
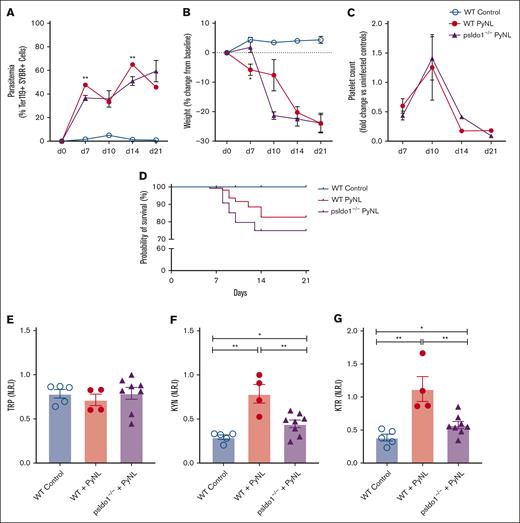
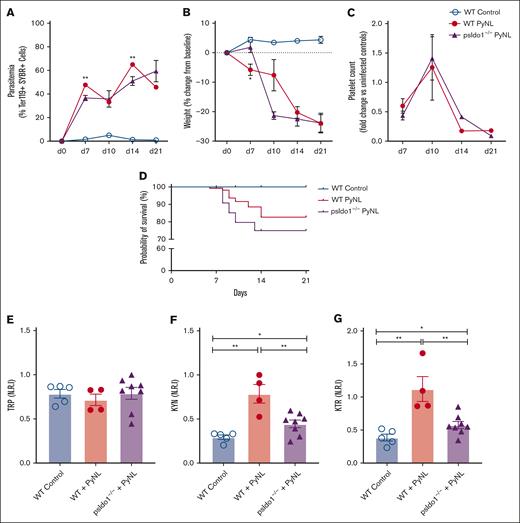
To directly assess the role of platelet IDO1 in plasma TRP metabolism during Plasmodium infection, we generated platelet Ido1−/− mice (psIdo1−/−) by crossing PF4cre and Ido1flox/flox mice, and WT and psIdo1−/− mice were infected with PyNL. Parasitemia was greater in psIdo1−/− infected mice (Figure 5A) and psIdo1−/− mice lost more weight early after infection than WT infected mice (Figure 5B). Platelet counts were tracked for all groups and there were no significant differences in platelet counts between WT and psIdo1−/− infected mice, with the exception of WT mice having fewer platelets at day 14 than psIdo1−/−mice (Figure 5C). WT and psIdo1−/− infected mice were tracked for 21 days to assess survival. Infected psIdo1−/− mice were ∼0% less likely to survive than WT infected mice, indicating that platelet Ido1 may contribute to infection survival (Figure 5D). The plasma metabolic phenotypes were similar between WT controls and psIdo1−/− infected mice on all days except day 10. Day 10 is a time point at which there is a consistent increase in infected mouse platelet counts compared with days 7 or 14, meaning platelets are more likely to contribute to plasma metabolites at the time point. On day 10, both groups of mice had similar platelet counts and parasite burden and all mice had similar plasma TRP (Figure 5E). However, there was a greater plasma KYN in the WT infected group (Figure 5F) than infected psIdo1−/− mice. The KTR was less in psIdo1−/− infected mice than the WT infected mice (Figure 5G). Because PF4 can be expressed by cells other than platelets in some inflammatory states55 and liver Kupffer cells (macrophage cell type) are important in platelet clearance and immune responses, we used PF4cre-iDTR mice,42 and treated the mice with diphtheria toxin to eliminate PF4-expressing cells. There were no changes in liver Kupffer cell numbers (supplemental Figure 2) in these mice, indicating that the clearance of RBCs and platelets in infected psIdo1−/− mice is unlikely to be a contributing factor in the observed phenotype.
Platelet-specific Ido1−/− mice have altered plasma metabolites and decreased P yoelii infection survival. (A) Parasitemia was assessed by SYBR+ RBCs quantified by flow cytometry. No significant differences were found comparing both groups during PyNL infection. (B) Weight loss was tracked across time points for all groups and was similar except at day 10. (C) Platelet counts, shown as fold change to control, were similar between WT and psIdo1−/− mice during infection. (D) Kaplan Meier survival curve. PsIdo1−/− mice had decreased survival compared with WT control infected mice. (E-G) Normalized relative intensity on day 10 after infection for plasma (E) TRP, (F) KYN, and (G) KTR as a measurement of Ido1 activity. Data presented as mean ± standard error of the mean (SEM; ∗P < .05, ∗∗P < .01, ∗∗∗P < .001). Data were analyzed using unpaired 2-tailed Student t test.
Platelet-specific Ido1−/− mice have altered plasma metabolites and decreased P yoelii infection survival. (A) Parasitemia was assessed by SYBR+ RBCs quantified by flow cytometry. No significant differences were found comparing both groups during PyNL infection. (B) Weight loss was tracked across time points for all groups and was similar except at day 10. (C) Platelet counts, shown as fold change to control, were similar between WT and psIdo1−/− mice during infection. (D) Kaplan Meier survival curve. PsIdo1−/− mice had decreased survival compared with WT control infected mice. (E-G) Normalized relative intensity on day 10 after infection for plasma (E) TRP, (F) KYN, and (G) KTR as a measurement of Ido1 activity. Data presented as mean ± standard error of the mean (SEM; ∗P < .05, ∗∗P < .01, ∗∗∗P < .001). Data were analyzed using unpaired 2-tailed Student t test.
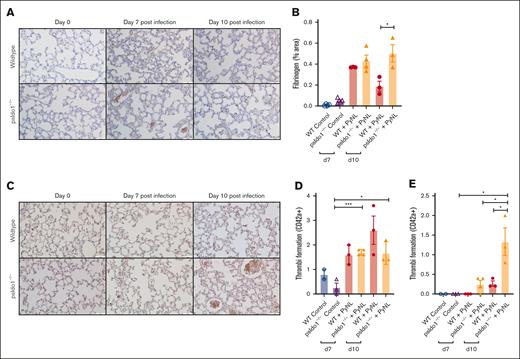
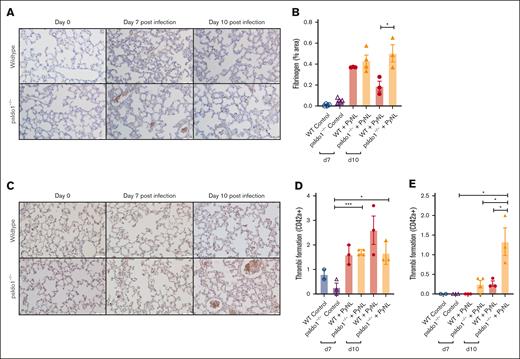
Because infected psIdo1−/− mice had increased death, we assessed lung fibrinogen as a marker of thrombosis. More fibrinogen was noted in the lungs of the psIdo1−/− infected mice than in WT infected mice (Figure 6A-B). This indicated that platelet Ido1 may be important in modulating thrombo-inflammatory responses. By immunostaining for platelets (anti-CD42c) we detected similar numbers of thrombi of >10 μm (Figure 6C-D) but increased thrombi of >10 μm in the lungs of psIdo1−/− compared with WT infected mice (Figure 6E). These data indicate that platelet IDO1 has a role in shifting TRP metabolism toward the KYN pathway during infection. The loss of platelet IDO1 may lead to increased thrombi and decreased survival during PyNL infection.
Platelet-specific Ido1−/− mice had increased thrombi in their lungs after infection compared with WT infected mice. (A) Fibrinogen was stained and quantified on day 7 and 10 lung sections to detect thrombi. Images were taken at ×40 original magnification, scale bar represents 50 μm, and (B) immunohistochemistry quantification with ImageJ. (C) Lung sections were immunostained for CD42c on day 7 and 10 after infection to detect thrombi. Images were taken at ×40 original magnification, scale bar represents 25 μm. (D-E) Quantification of (D) thrombi <10 μm or (E) >10 μm averaged over 5 fields per mouse. Data presented as mean ± SEM (∗P < .05, ∗∗P < .01, ∗∗∗P < .001). Data were analyzed using unpaired 2-tailed Student t test.
Platelet-specific Ido1−/− mice had increased thrombi in their lungs after infection compared with WT infected mice. (A) Fibrinogen was stained and quantified on day 7 and 10 lung sections to detect thrombi. Images were taken at ×40 original magnification, scale bar represents 50 μm, and (B) immunohistochemistry quantification with ImageJ. (C) Lung sections were immunostained for CD42c on day 7 and 10 after infection to detect thrombi. Images were taken at ×40 original magnification, scale bar represents 25 μm. (D-E) Quantification of (D) thrombi <10 μm or (E) >10 μm averaged over 5 fields per mouse. Data presented as mean ± SEM (∗P < .05, ∗∗P < .01, ∗∗∗P < .001). Data were analyzed using unpaired 2-tailed Student t test.
Discussion
Thrombocytopenia is common in patients with malaria. However, studies on how thrombocytopenia affects malaria immune responses, particularly immunometabolic alterations, remain limited. This study is, to our knowledge, the first to show that in infection, platelets can express Ido1 and that platelet Ido1 is part of the immunometabolic response. We also show a relationship between IFN-γ and platelet Ido1 during PyNL infection that may be part of other disease processes including COVID-19 and influenza. Given that IFN-γ is a major cytokine mediator of viral responses, our study has broad implications beyond the malaria model. IFN-γ, IDO1, and the KYN pathway’s involvement have also been reported in several autoimmune disease contexts, such as systematic lupus erythematosus,56 multiple sclerosis,57 and several autoimmune endocrinopathies,58 extending the potential impact of these data. In addition, a shift from serotonin to KYN may also have yet explored impacts on platelet activation and vascular tone.
This study marks an initial exploration into the immunometabolic capabilities of platelets and potential roles for platelet metabolic processes beyond thrombosis. Given that platelets are the most numerous metabolically active cells in the circulation59 and possess metabolic flexibility,18,60,61 platelets may regulate metabolic pathways that impact both thrombotic and immune responses. Platelets from patients with COVID-19 were shown to have metabolic depression and impaired metabolic flexibility.62 A decrease in platelet count may lead to systemic metabolic alterations, subsequently modifying immune responses. With significant advances in understanding immunometabolism and nonthrombotic roles for platelets, there is a critical need to improve our understanding of how platelet functions affect metabolism in both healthy and pathological contexts. Because platelets express Ido1 in an IFN-γ-dependent manner and constitute the most abundant immune cell in the bloodstream, actively participating in both innate and adaptive immune response systems, they might also contribute to downstream immune responses through metabolic pathway modulations. It is important to note that every mouse model has caveats in translating to a human disease context, including the use of Cre drivers, because few, if any, are completely cell specific. It is possible that a small portion of the leukocyte population may be affected by using PF4cre mice.63 Despite this limitation, PF4cre mice remains the best available option for our experiments in order to delineate on the role of platelet IDO1. In addition, given the nature of this Cre model system, we cannot truly delineate direct effects from megakaryocytes vs circulating platelets in TRP metabolism.
The number of platelets transfused to mice in our studies represented ∼25% of the total platelet count. Although increased numbers of transfused platelets may lead to more robust changes in the plasma metabolites, more platelets may not be a positive contribution. Activated platelets also drive inflammation that can have negative outcomes depending on the disease context. Therefore, finding a fine line between the beneficial and deleterious effects of platelet transfusions in malaria and other diseases is a focus of future research.
Although this project is focused on malaria, it has direct implications on understanding the immunometabolic role of platelets in general that is likely to affect our understanding of many inflammatory disease pathologies, because Ido1 is expressed in many disease pathologies, emphasizing a need to improve our understanding of the metabolic capabilities of platelets. Platelets are capable of regulating amino acid metabolic pathways that could affect both thrombotic and immune responses. Further understanding of this interplay between platelets and biochemical pathways may provide an understanding of the impact of thrombocytopenia in diseases beyond malaria and provide a means to improve malaria infection responses, as well as improved platelet-derived therapeutics in many hematological, metabolic, and immune diseases.
Acknowledgments
The authors are grateful to Matthew Ciobra from Washington University, St. Louis for kindly gifting the Ido1 flox mice.
This work was supported by National Institutes of Health (NIH), Heart Blood and Lung Institute grant 1F31HL159933-01, American Heart Association predoctoral fellowship award GR835752 (S.K.B.-N.), and project support by NIH grants R01 HL160610, R01 HL141106, and R01 HL142152 (C.N.M.).
Authorship
Contribution: C.N.M., M.T.R., and S.K.B.-N. designed this study; C.N.M. and S.K.B.-N. contributed to the drafting of the manuscript; and S.K.B.-N., C.N.M., S.K.T., X.L.S., J.C.M., A.C.L., C.L., and P.M. performed experiments and analyzed data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Craig N. Morrell, Aab Cardiovascular Research Institute, University of Rochester School of Medicine, 601 Elmwood Ave, Box CVRI, Rochester, NY 14642; email: craig_morrell@urmc.rochester.edu.
References
Author notes
For original data, please contact the corresponding author, Craig Morrell (craig_morrell@urmc.rochester.edu); the RNA-sequencing data will be deposited as National Institutes of Health Gene Expression Omnibus files. The mouse data accession number: Submission ID: SUB14649913 BioProject ID: PRJNA1146398. Project information will be accessible with the following link: http://www.ncbi.nlm.nih.gov/bioproject/1146398; for the human RNA-sequencing data: transcriptomic data from patients with malaria are available upon reasonable request to Matthew Rondina at matthew.rondina@hsc.utah.edu&rdquo.
The full-text version of this article contains a data supplement.