Key Points
TLX3 transactivates LINC00478, the host gene of oncogenic miR-125b-2 in T-ALL.
TLX3 and miR-125b contribute to the differentiation arrest and the expansion of transformed T cells.
Abstract
The oncogenic mechanisms driven by aberrantly expressed transcription factors in T-cell acute leukemia (T-ALL) are still elusive. MicroRNAs (miRNAs) play an important role in normal development and pathologies. Here, we examined the expression of 738 miRNA species in 41 newly diagnosed pediatric T-ALLs and in human thymus-derived cells. We found that expression of 2 clustered miRNAs, miR-125b/99a, peaks in primitive T cells and is upregulated in the T leukemia homeobox 3 (TLX3)–positive subtype of T-ALL. Using loss- and gain-of-function approaches, we established functional relationships between TLX3 and miR-125b. Both TLX3 and miR-125b support in vitro cell growth and in vivo invasiveness of T-ALL. Besides, ectopic expression of TLX3 or miR-125b in human hematopoietic progenitor cells enhances production of T-cell progenitors and favors their accumulation at immature stages of T-cell development resembling the differentiation arrest observed in TLX3 T-ALL. Ectopic miR-125b also remarkably accelerated leukemia in a xenograft model, suggesting that miR125b is an important mediator of the TLX3-mediated transformation program that takes place in immature T-cell progenitors. Mechanistically, TLX3-mediated activation of miR-125b may impact T-cell differentiation in part via repression of Ets1 and CBFβ genes, 2 regulators of T-lineage. Finally, we established that TLX3 directly regulates miR-125b production through binding and transactivation of LINC00478, a long noncoding RNA gene, which is the host of miR-99a/Let-7c/miR-125b. Altogether, our results reveal an original functional link between TLX3 and oncogenic miR-125b in T-ALL development.
Introduction
T-cell acute lymphoblastic leukemias (T-ALLs) are aggressive hematologic tumors resulting from the malignant transformation of T-cell progenitors. T-ALLs are characterized by multiple genetic lesions including constitutive activation of the NOTCH1 signaling pathway,1 inactivation of tumor suppressor genes (PTEN, NF1, and PHF6), gene amplification (NUP214-ABL1, MYB), and inappropriate expression of transcription factors (TAL1, LMO1, LMO2, LYL1, TLX1, and TLX3).2 Several of these features are unique and mutually exclusive, thereby endowing T-ALL subgroups with distinct biological properties. T leukemia homeobox 3 (TLX3)-rearranged T-ALL cells phenotypically resemble thymocytes arrested at the early cortical stage.3-5 TLX3 is overexpressed in up to 25% of pediatric T-ALL and is rare in adult T-ALL. The orphan homeobox TLX3 gene is involved in the regulation of embryonic brain development.6 Aberrant expression of TLX3 in T-ALL arises commonly from the t(5; 14)(q35; q32) translocation which places this gene under the influence of enhancer regions downstream of the BCL11B gene. Alternative rearrangements of the 5q35 locus affecting expression of TLX3 have also been reported.7-9 How TLX3 contributes to leukemia development is still elusive. Recent studies suggest that TLX3 as well as its homolog TLX1 may be recruited by the ETS1 transcription factor to the core of the T-cell receptor α (TCRα) enhancer (Eα) and repress its activity. This repression incites the epigenetic silencing of the TCRα locus and maturation arrest.10 TLX1/TLX3-rearranged T-ALLs are highly related in terms of global gene expression and tend to cluster together, away from other T-ALL subtypes.4,5,11 Moreover, TLX1 and TLX3 have been shown to be master regulators of an oncogenic transcriptional circuit centered around RUNX1, a main target of their inhibitory action.12
T-cell development is dynamically regulated by microRNAs (miRNAs).13,14 Combinatorial action of miRNAs optimizes the expression patterns of protein-coding genes and so provides a mechanism for fine-tuning signaling pathways and main cell functions.15 Genetic lesions in human leukemia often perturb miRNA expression, unleashing their tumor suppressor or oncogenic activity.16-19 In T-ALL, a small subset of 5 oncogenic miRNAs was found to cooperatively downregulate tumor suppressor genes like PTEN and FBXW7. These miRNAs are highly expressed across different cytogenetic groups of T-ALL, and their ectopic expression accelerates development of leukemia in a NOTCH-induced T-ALL mouse model.20 The alternative group of tumor suppressor miRNAs is downregulated in T-ALL upon MYC activation. Their loss relieves the high expression of B-MYB and HPB1 oncogenic proteins and thus promotes the leukemia onset in mice.21
We hypothesized that miRNA deregulation could be triggered by abnormal expression of transcription factors frequently activated in pediatric leukemia and so contribute to oncogenic transformation. Comparative expression profiling of 738 miRNAs in a panel of 41 newly diagnosed pediatric T-ALLs and in 7 human thymus-derived cell subsets revealed the increased expression of 2 clustered miRNAs: miR-125b and miR-99a in T-ALL with deregulated expression of TLX3 in accordance with previous work.20 We found that TLX3 directly transactivates the expression of long intergenic nonprotein coding RNA 478 (LINC00478), the host gene of the miR-99a/let-7c/125b-2 cluster, an original result in comparison with earlier reported miRNA profiling. Furthermore, we provide convincing evidence that TLX3-mediated overexpression of oncogenic miR-125b in early T-cell progenitors impairs normal T-cell development inducing proliferation and differentiation arrest of immature T cells.
Methods
T-ALL samples
T-ALL cells were collected at diagnosis upon informed consent of patient’s relatives in accordance with the Declaration of Helsinki and national ethics guidelines. The INSERM ethical committee (IRB000003888) approved the project (evaluation number 13-105-1). Primary T-ALL cells were cultured in contact with mouse MS5/DL1 stromal cells as described22 (see details in supplemental Methods).
Normal human T-cell subsets
Pieces of postnatal thymi were obtained from 5-day- to 4-year-old children undergoing corrective cardiac surgery (Service de Chirurgie Cardiaque Pédiatrique, Hôpital Necker, Paris, France) with the parental informed consent and according to the French Ethical Committee’s regulations. Lymphocyte subsets were purified by sorting on an INFLUX cell sorter (BD Biosciences) with combinations of surface markers (supplemental Methods).
miRNA profiling
Total RNA was isolated using mirVana or mirAqueous RNA isolation kits (Ambion and Life Technologies) following the standard protocols for specific miRNA retention. RNA integrity was estimated by capillary electrophoresis using the Agilent 2100 Bioanalyzer (Agilent Technologies) and samples with RNA integrity number >8 were used in TaqMan low-density array (TLDA) analysis. miRNA profiling was performed according to the manufacturers’ protocol (Applied Biosystems and Life Technologies).23 miRNA data processing and statistics are detailed in supplemental Methods. Individual miRNAs were validated by the single miRCURY LNA microRNA PCR system (Exiqon).
Human T-cell differentiation
Cord blood (CB) samples were collected from healthy newborns with the informed written consents of the mothers. All experiments were done in accordance with the ethics evaluation committee of INSERM (IRB000003888, FWA00005831) and with collaborative protocols signed with the Hôpital Antoine Beclère (Clamart, France) and Clinique des Noriets (Vitry-sur-Seine, France). CD34+/CD38−/low/CD45RA−/CD90− hematopoietic progenitor cells (HPCs) were sorted, transduced with lentiviral vectors, and cocultured with MS5/DL1 cells as described.24 Cell counting and immunophenotyping of culture-derived human T cells were performed every week starting from day 21 using FACSCalibur and LSR-II flow cytometers (BD) (supplemental Methods).
Human CD34+/CD38−/low HPCs were also transduced with control (CTRL) (70% transduction efficiency), TLX3/OE (42%), and 125b/OE (42%) vectors and injected into NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice recipients (5 × 104 cells per mouse). Mouse bone marrow (BM), spleen, and thymus were recovered 90 days after transplantation; cells were numbered and immunophenotyped using anti-human specific antibodies (supplemental Methods).
Lentiviral vector transduction
Constructs and generation of lentiviral supernatants are detailed in supplemental Methods and in Amsellem et al25 and Gerby et al26 ; T-ALL samples and T-ALL cell lines were transduced with G/IL-7SUx envelope-pseudotyped vectors. Human CB-derived HPCs were transduced with VSV/G-pseudotyped vectors.27
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) experiments on DND41, HPB, and Jurkat cell lines or T-ALL samples were performed according to the Agilent protocol (version 10.0), using anti-TLX3 (H-55 X; Santa Cruz Biotechnology), anti-H3ac (06-599; Millipore), normal rabbit and mouse immunoglobulin Gs (IgGs) (sc-2027 and sc-2025; Santa Cruz Biotechnology). ChIP sequencing (ChIP-seq) of a TLX3+ T-ALL sample was performed using a customized antibody10 ; sequencing and data proceedings are detailed in supplemental Methods.
Results
miRNA profiling shows increased levels of miR-125b in pediatric TLX3 T-ALL
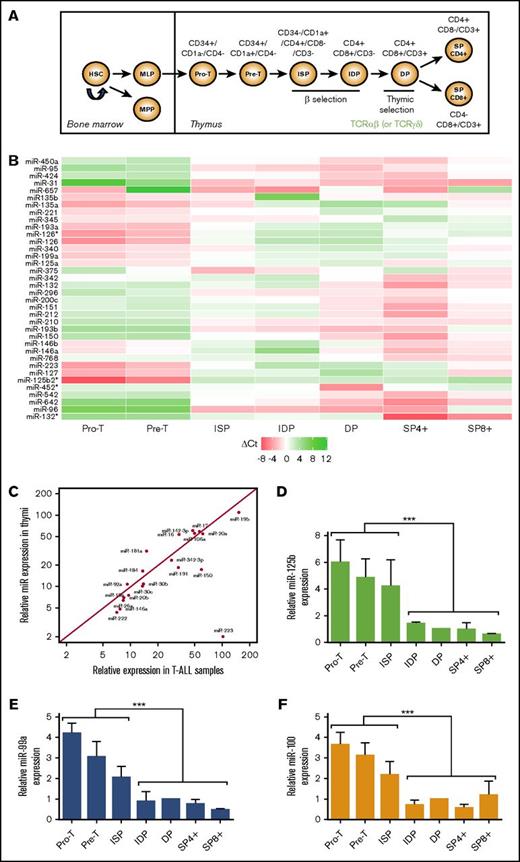
We first assessed the expression of 738 miRNAs in a panel of 41 newly diagnosed pediatric T-ALLs by high-throughput real-time quantitative polymerase chain reaction (R-Q-PCR) analysis (TLDA). Blast cell specimens were divided into 3 molecular groups: (1) a TAL1-expressing group (“TAL1”; n = 19) which included samples with SIL-TAL1 deletions (n = 10), TAL1 rearrangements with either TCRA/D or TCRB loci (n = 3), and TAL1 overexpression in the absence of the above chromosomal alterations (n = 6); (2) a “TLX3”-expressing group (n = 8); and (3) an “other” group (n = 14) which was characterized by the lack of both TAL1 and TLX3 expression and the absence of CALM-AF10, inv(7), TLX1, LMO1/2 rearrangements or overexpression (supplemental Tables 1-2). The number of expressed miRNAs per sample ranged between 407 and 541, which corresponded to 51% and 70% of the miRNA species available on the TLDA cards. Overall, expression of 532 miRNAs was detected in >75% of samples (supplemental Table 3) and considered in further analysis. Some of them were highly expressed in all T-ALL samples (Figure 1A). No statistical difference in the expression of the “top 20” most abundant miRNAs was found among the 3 TAL1/TLX3/other oncogenic groups. When analyzing all miRNAs, analysis of variance (ANOVA) and pairwise analyses revealed a small subset of miRNAs significantly upregulated in the TLX3 T-ALL. This miRNA subset comprised clustered miR-125b, miR-99a, miR-100, and nonclustered miR-153, miR-135a, and miR-506 (Figure 1B). Relative expression of the TLX3-associated miR-125b, miR-99a, and miR-100 was substantially higher compared with miR-135a, miR-153, and miR-506 (supplemental Table 3). Overexpression of miR-125b and miR-99a, but not miR-100 in TLX3 T-ALL was further confirmed by R-Q-PCR analysis including an independent series of primary T-ALLs (Figure 1C-E).
miRNA expression in pediatric T-ALL. (A) Twenty most abundantly expressed miRNAs in different cytogenetic subgroups of 41 T-ALL clinical specimens as assessed by TLDA and normalized to the RNU48 expression in a given sample (2−∆Ct). miRNAs are ranked by expression levels (given as percentage of RNU48 expression). (B) Heatmap of miRNAs differentially expressed in 3 cytogenetic subgroups of T-ALL. Normalized expressions (ΔCt z-values) were color-coded by a gradient from green (underexpressed) to red (overexpressed) relative to the mean (ANOVA adjusted P values: .005 [miR-135a], .005 [miR-125b], .013 [miR-100], .03 [miR-506], .03 [miR-99a], .04 [miR-153]). (C-E) Comparison of miR-125b, miR-99a, and miR-100 levels in TLX3 vs non-TLX3 T-ALL samples by R-Q-PCR (Exiqon). miRNA expression is normalized to RNU44; levels in DP T cells are used as calibrator (2−∆∆Ct): (C) miR-125b (n = 25 vs n = 33), (D) miR-99a (n = 16 vs n = 16), and (E) miR-100 (n = 16 vs n = 16). ***P < .001, Mann-Whitney U test. Ct, cycle threshold; NS, not significant.
miRNA expression in pediatric T-ALL. (A) Twenty most abundantly expressed miRNAs in different cytogenetic subgroups of 41 T-ALL clinical specimens as assessed by TLDA and normalized to the RNU48 expression in a given sample (2−∆Ct). miRNAs are ranked by expression levels (given as percentage of RNU48 expression). (B) Heatmap of miRNAs differentially expressed in 3 cytogenetic subgroups of T-ALL. Normalized expressions (ΔCt z-values) were color-coded by a gradient from green (underexpressed) to red (overexpressed) relative to the mean (ANOVA adjusted P values: .005 [miR-135a], .005 [miR-125b], .013 [miR-100], .03 [miR-506], .03 [miR-99a], .04 [miR-153]). (C-E) Comparison of miR-125b, miR-99a, and miR-100 levels in TLX3 vs non-TLX3 T-ALL samples by R-Q-PCR (Exiqon). miRNA expression is normalized to RNU44; levels in DP T cells are used as calibrator (2−∆∆Ct): (C) miR-125b (n = 25 vs n = 33), (D) miR-99a (n = 16 vs n = 16), and (E) miR-100 (n = 16 vs n = 16). ***P < .001, Mann-Whitney U test. Ct, cycle threshold; NS, not significant.
miRNA expression in sorted T-cell progenitors
We next performed miRNA profiling of 7 thymus-derived cell subsets corresponding to different stages of human T-cell development: from immature double-negative (DN) including pro-T and pre-T, immature single-positive (ISP), immature double-positive (IDP), mature double-positive (DP), and mature single-positive (SP4) and (SP8) thymocytes28 (Figure 2A). Comparison of miRNA profiles of these fractions permitted identification of 36 miRNAs whose expression changed significantly (false discovery rate < 0.05, ANOVA test; LIMMA package) throughout T-cell maturation (Figure 2B; supplemental Table 4). Among these most variable miRNAs, only 4 overlapped with a set of highly expressed miRNAs found in T-ALL analysis, namely miR-223, miR-150, miR-146a, and miR-342. Relative expression of miR-223 peaked in early thymic precursors and decreased in cortical thymocytes, whereas expression of miR-150, miR-146a, and miR-342 increased along the differentiation. On the other hand, 17 of the 20 most abundant miRNAs in T-ALL were expressed through all stages of normal T-cell development at levels similar to those observed in T-ALL except miR-223, showing 50-fold higher expression in T-ALL (Figure 2C).
miRNA expression during human T-cell development. miR-125b is highly expressed at early immature stages. (A) Schematic representation of human T-cell development. Human thymocytes obtained from healthy donors were sorted according to established cell surface markers: immature DN including pro-T (CD34+/CD7+CD1a−/CD4−) and pre-T (CD34+/CD7+/CD1a+/CD4−), ISP (CD34−/CD1a+/CD4+/CD8−/CD3−), IDP (CD4+/CD8+/CD3−), mature DP (CD4+/CD8+/CD3+), and mature SP (SP4: CD4+/CD8−/CD3+ and SP8: CD4−/CD8+/CD3+) thymocytes. (B) Heatmap of the 36 most differentially expressed miRNAs at 7 sequential stages of T-cell development (n = 3). Shown are miRNA expression levels relative to the average of the row. The white color corresponds to the mean expression across all stages (ΔCt = 0). Red and green colors correspond, respectively, to overexpression (ΔCt < 0) and underexpression (ΔCt > 0). (C) Comparative expression of the 20 most abundant miRNAs in T-ALLs and in thymi. miRNA expression is given as percentage of RNU48 control. (D-F) Relative miRNA expression in T-cell subsets (2−∆∆Ct). Levels in DP cells were used as calibrator (n = 3) (D) miR-125b; (E) miR-99a; (F) miR-100. ***P < .001, Mann-Whitney U test. HSC, hematopoietic stem cell; MLP, multilymphoid progenitor; MPP, multipotent progenitor.
miRNA expression during human T-cell development. miR-125b is highly expressed at early immature stages. (A) Schematic representation of human T-cell development. Human thymocytes obtained from healthy donors were sorted according to established cell surface markers: immature DN including pro-T (CD34+/CD7+CD1a−/CD4−) and pre-T (CD34+/CD7+/CD1a+/CD4−), ISP (CD34−/CD1a+/CD4+/CD8−/CD3−), IDP (CD4+/CD8+/CD3−), mature DP (CD4+/CD8+/CD3+), and mature SP (SP4: CD4+/CD8−/CD3+ and SP8: CD4−/CD8+/CD3+) thymocytes. (B) Heatmap of the 36 most differentially expressed miRNAs at 7 sequential stages of T-cell development (n = 3). Shown are miRNA expression levels relative to the average of the row. The white color corresponds to the mean expression across all stages (ΔCt = 0). Red and green colors correspond, respectively, to overexpression (ΔCt < 0) and underexpression (ΔCt > 0). (C) Comparative expression of the 20 most abundant miRNAs in T-ALLs and in thymi. miRNA expression is given as percentage of RNU48 control. (D-F) Relative miRNA expression in T-cell subsets (2−∆∆Ct). Levels in DP cells were used as calibrator (n = 3) (D) miR-125b; (E) miR-99a; (F) miR-100. ***P < .001, Mann-Whitney U test. HSC, hematopoietic stem cell; MLP, multilymphoid progenitor; MPP, multipotent progenitor.
Interestingly, expression levels of miR-125b, miR-99a, and miR-100 peaked in pro-T, pre-T, and ISP cells and decreased throughout further T-cell maturation, reaching the lowest values in surface CD3+ cells (Figure 2D-F). We hypothesized that the high expression of miR-125b and miR-99a observed in TLX3 T-ALL may interfere with normal T-cell differentiation and contribute to T-ALL development. Recently described oncogenic features of miR-125b in ALL29,30 and the major impact of miR-125b and not miR-99a on hematopoietic stem cell growth31 prompted us to focus our study on the functional relationship between miR125b and TLX3 in T-ALL.
TLX3 knockdown reduces miR-125b levels and impairs T-ALL growth
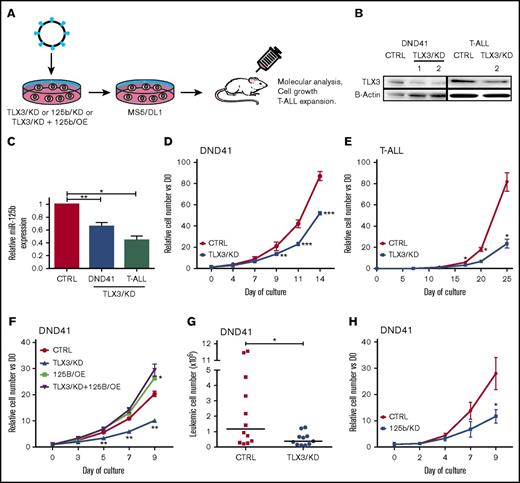
We first tested the effect of TLX3 knockdown (TLX3/KD) on miR-125b expression in the DND41 cell line and in 3 primary TLX3 T-ALLs (Figure 3A). Two short hairpin RNA (shRNA) vectors efficiently decreased TLX3 protein levels in transduced cells by 60% to 70% (Figure 3B) in correlation with a nearly twofold decrease in miR-125b levels in leukemic cells (Figure 3C). Functionally, TLX3/KD slowed down the in vitro growth of DND41 or T-ALL cells (Figure 3D-E), a defect that was rescued by ectopic expression of miR-125b (125b/OE) (Figure 3F). Impaired tumorigenic behavior of TLX3/KD DND41 cells was also observed in vivo. Six weeks after transplant, 10 times fewer leukemic cells were found in spleens of TLX3/KD DND41-transplanted mice compared with CTRL (Figure 3G) in parallel to lower spleen weight (supplemental Figure 1A). Leukemic BM infiltration was not modified (supplemental Figure 1B), suggesting a delay of extramedullary leukemia propagation or that our mouse analyses were done too late in the leukemic progression. Finally, knocking down miR-125b (125b/KD) (supplemental Figure 2) in DND41 T-ALL cells phenocopied the in vitro growth inhibitory effect observed with TLX3/KD (Figure 3H). Altogether, these results show a correlation between TLX3 and miR125b expression in T-ALL, both factors being implicated in the control of leukemic cell growth.
TLX3 knockdown correlates with reduced expression of miR-125b and slows proliferation of T-ALL. (A) Flowchart of TLX3/KD and 125/KD experiments. A lentivirus-based system was used to downregulate TLX3 or miR-125b in T-ALL cells. CTRL eGFP and mCherry vectors express, respectively, irrelevant shRNA and “sponge” RNA. Sorted transduced T-ALL cells were cocultured with MS5-DL1 stromal cells. The DND41 cell line was grown in stroma-free standard culture conditions for functional and molecular analyses. (B) Western blot analysis of TLX3 silencing. Two TLX3/KD vectors efficiently silenced TLX3 protein in DND41 cells (left panel), of which one (right panel) repressed TLX3 in primary T-ALL. (C) miR-125b expression in TLX3/KD vs CTRL in DND41 cells and in primary T-ALLs. Results of R-Q-PCR analyses performed in (n = 7) independent experiments for DND41 cells and in (n = 3) independent T-ALL samples. (D-E) In vitro cell growth of TLX3/KD cells: (D) DND41 cell line (n = 3) and (E) primary T-ALL (n = 1) made with 4 technical replicates. Data represent the mean ± standard error of the mean (SEM) of 3 experiments for DND41 and of 4 technical replicates for T-ALL. (F) Ectopic expression of miR-125b rescues growth inhibitory effect of TLX3 knockdown. DND41 cells were doubly transduced in the following conditions: (CTRL): CTRL/eGFP and CTRL/mCherry that do not express any miRNA; (TLX3/KD): CTRL/mCherry and TLX3/KD(eGFP); (125b/OE): CTRL/eGFP and 125b/OE(mCherry); (TLX3/KD+125b/OE): TLX3/KD(eGFP) and 125b/OE(mCherry). Transduced cells were sorted and cultured in standard conditions. GFP/mCherry DP cells were enumerated by flow cytometer; (n = 3) independent transduction experiments. (G) Reduced leukemic propagation of TLX3/KD DND41 cells in mice. Shown are engraftment levels in spleens 6 weeks after transplantation. Bars represent median values (n = 2). (H) In vitro growth of 125b/KD DND41 cells (n = 3). Data represent the mean value ± SEM. *P < .05, **P < .01, ***P < .001, Mann-Whitney U test.
TLX3 knockdown correlates with reduced expression of miR-125b and slows proliferation of T-ALL. (A) Flowchart of TLX3/KD and 125/KD experiments. A lentivirus-based system was used to downregulate TLX3 or miR-125b in T-ALL cells. CTRL eGFP and mCherry vectors express, respectively, irrelevant shRNA and “sponge” RNA. Sorted transduced T-ALL cells were cocultured with MS5-DL1 stromal cells. The DND41 cell line was grown in stroma-free standard culture conditions for functional and molecular analyses. (B) Western blot analysis of TLX3 silencing. Two TLX3/KD vectors efficiently silenced TLX3 protein in DND41 cells (left panel), of which one (right panel) repressed TLX3 in primary T-ALL. (C) miR-125b expression in TLX3/KD vs CTRL in DND41 cells and in primary T-ALLs. Results of R-Q-PCR analyses performed in (n = 7) independent experiments for DND41 cells and in (n = 3) independent T-ALL samples. (D-E) In vitro cell growth of TLX3/KD cells: (D) DND41 cell line (n = 3) and (E) primary T-ALL (n = 1) made with 4 technical replicates. Data represent the mean ± standard error of the mean (SEM) of 3 experiments for DND41 and of 4 technical replicates for T-ALL. (F) Ectopic expression of miR-125b rescues growth inhibitory effect of TLX3 knockdown. DND41 cells were doubly transduced in the following conditions: (CTRL): CTRL/eGFP and CTRL/mCherry that do not express any miRNA; (TLX3/KD): CTRL/mCherry and TLX3/KD(eGFP); (125b/OE): CTRL/eGFP and 125b/OE(mCherry); (TLX3/KD+125b/OE): TLX3/KD(eGFP) and 125b/OE(mCherry). Transduced cells were sorted and cultured in standard conditions. GFP/mCherry DP cells were enumerated by flow cytometer; (n = 3) independent transduction experiments. (G) Reduced leukemic propagation of TLX3/KD DND41 cells in mice. Shown are engraftment levels in spleens 6 weeks after transplantation. Bars represent median values (n = 2). (H) In vitro growth of 125b/KD DND41 cells (n = 3). Data represent the mean value ± SEM. *P < .05, **P < .01, ***P < .001, Mann-Whitney U test.
Overexpression of TLX3 or miR125b induces expansion of normal T cells and blocks differentiation at immature stages in vitro and in vivo
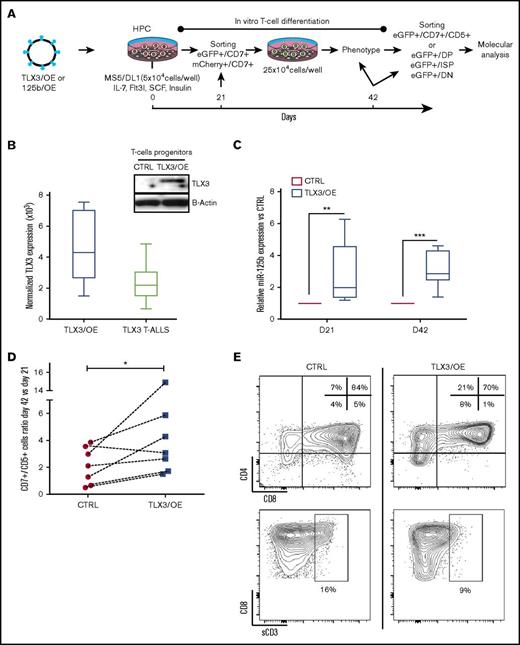
We next ectopically expressed TLX3 (TLX3/OE) during in vitro differentiation of normal human T cells and tested the possible interplay between TLX3 and miR-125b expression. Gene-modified human CD34+/CD38−/low/CD45RA−/CD90− HPCs were induced to differentiate in vitro24 and monitored during 42 days (Figure 4A). Molecular analyses of total CD7+/CD5+/enhanced green fluorescent protein–positive (eGFP+) T cells and DN, ISP, and DP T-cell subsets consistently demonstrated an association between ectopic TLX3 expression and concomitant upregulation of miR-125b and miR-99a, and, to a lesser extent, low abundant miR-100, in newly generated T cells (Figure 4B-C and not shown).
Overexpression of TLX3 induces miR-125b expression and correlates with expansion and differentiation arrest of T-cell progenitors. (A) Design of gain-of-function experiments. Human HPCs isolated from UCB were transduced with lentiviral expression vectors encoding, respectively, TLX3 (TLX3/OE) or eGFP alone (CTRL), miR-125b (125b/OE), or mCherry alone (CTRL) and cocultured with the murine MS5 stromal cells constitutively expressing human NOTCH ligand Δ-like 1 (MS5/DL1) in the presence of a cytokine cocktail. (CD7+/eGFP+) or (CD7+/mCherry+) T/NK progenitors were sorted and replated at day 21 of coculture with MS5/DL1 stromal cells. Thereafter, differentiation was weekly monitored and HPC-derived T cells were fractionated at day 42. (B) Left, Comparison of TLX3 expression in transduced T cells and in primary TLX3 T-ALL by R-Q-PCR (n = 8). Shown are absolute copy numbers of TLX3 normalized to housekeeping gene ABL quantified by using external DNA dilution standards. Top right, Western blot analysis illustrating TLX3 protein expression in sorted CD7+/eGFP+ T cells at day 21. (C) miR-125b expression levels in TLX3/OE vs CTRL CD7+/CD5+ T cells measured at day 21 (N = 5) and 42 (N = 7) of differentiation. Bar represents medians. ***P < .001, **P < .01, Mann-Whitney U test. (D) TLX3 induces proliferation of T-cell progenitors. Ratios of T-cell numbers at day 42 vs day 21 obtained in 7 independent experiments. *P < .05, Wilcoxon test. (E) Fluorescence-activated cell sorting (FACS) data from a representative experiment at day 42 of culture are shown. Proportions of T-cell fractions are given relative to gated eGFP+/CD7+/CD5+ T cells. IL, interleukin; sCD3, soluble CD3; SCF, stem cell factor.
Overexpression of TLX3 induces miR-125b expression and correlates with expansion and differentiation arrest of T-cell progenitors. (A) Design of gain-of-function experiments. Human HPCs isolated from UCB were transduced with lentiviral expression vectors encoding, respectively, TLX3 (TLX3/OE) or eGFP alone (CTRL), miR-125b (125b/OE), or mCherry alone (CTRL) and cocultured with the murine MS5 stromal cells constitutively expressing human NOTCH ligand Δ-like 1 (MS5/DL1) in the presence of a cytokine cocktail. (CD7+/eGFP+) or (CD7+/mCherry+) T/NK progenitors were sorted and replated at day 21 of coculture with MS5/DL1 stromal cells. Thereafter, differentiation was weekly monitored and HPC-derived T cells were fractionated at day 42. (B) Left, Comparison of TLX3 expression in transduced T cells and in primary TLX3 T-ALL by R-Q-PCR (n = 8). Shown are absolute copy numbers of TLX3 normalized to housekeeping gene ABL quantified by using external DNA dilution standards. Top right, Western blot analysis illustrating TLX3 protein expression in sorted CD7+/eGFP+ T cells at day 21. (C) miR-125b expression levels in TLX3/OE vs CTRL CD7+/CD5+ T cells measured at day 21 (N = 5) and 42 (N = 7) of differentiation. Bar represents medians. ***P < .001, **P < .01, Mann-Whitney U test. (D) TLX3 induces proliferation of T-cell progenitors. Ratios of T-cell numbers at day 42 vs day 21 obtained in 7 independent experiments. *P < .05, Wilcoxon test. (E) Fluorescence-activated cell sorting (FACS) data from a representative experiment at day 42 of culture are shown. Proportions of T-cell fractions are given relative to gated eGFP+/CD7+/CD5+ T cells. IL, interleukin; sCD3, soluble CD3; SCF, stem cell factor.
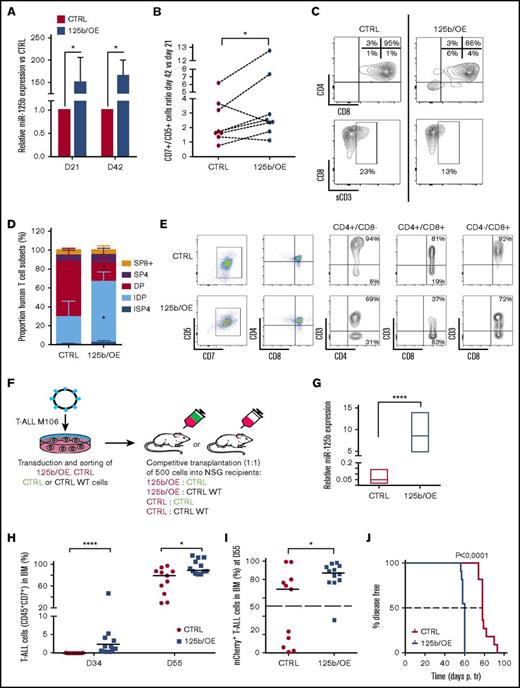
In vitro human T-cell development proceeds through a first wave of T-cell production (21-28 days) followed by a regular decrease in T-cell numbers (28-42 days). In TLX3/OE conditions, the late drop in T-cell numbers was not recurrently observed (supplemental Figure 3A). TLX3/OE in HPCs allowed twofold to fourfold more CD7+/CD5+ T cells to be kept between 21 and 42 days of culture compared with CTRL cells (Figure 4D). Phenotype analysis of newly produced T cells revealed a higher percentage of CD4+/CD8− ISP and a significant decrease of mature CD4+/CD8+/CD3+ DP subsets in TLX3/OE conditions (Figure 4E; supplemental Figure 3B), indicating that TLX3 not only maintains T-cell production during cell culturing but also impacts their differentiation. Similar expansion of early T cells was observed when ectopic miR-125b (125b/OE) was delivered into HPCs instead of TLX3 (Figure 5A-B). We found low but significant accumulation of immature DN progenitors as well as a significant decrease of cell surface CD3+CD4+CD8+ DP cells (Figure 5C; supplemental Figure 4A). The blocking stage of 125b/OE T cells did not strictly coincide with TLX3/OE conditions in which T-cell differentiation was stopped at the early cortical stage close to β-selection but both severely impacted CD3 surface expression. It should be noted that the phenotypes of TLX3/OE and 125b/OE T cells resembled the different TLX3 T-ALL blasts stuck at the immature cortical stages, extending from pro-T to IDP.5
Ectopic expression of miR-125b in HPCs mimics the effects of TLX3 on T-cell differentiation. Sorted human HPCs were transduced with either miR-125b/OE or CTRL mCherry-traceable vectors. T-cell differentiation was followed as described in Figure 4A. (A) Relative miR-125b expression at days 21 (N = 3) and 42 (N = 5) in miR-125b/OE cells compared with CTRL cells. *P < .05, Mann-Whitney U test. (B) miR-125b/OE enhances T-cell growth. Cumulative T-cell numbers at day 42 compared with day 21 of culture for miR-125b/OE and CTRL cells (n = 8). *P < .05, Wilcoxon test. (C) miR-125b/OE induces accumulation of immature DN cells and blocks sCD3 acquisition by DP cells. Shown are the data from the representative experiment at day 42. Proportions of T-cell fractions are relative to gated mCherry+/CD7+/CD5+ T cells. (D-E) Human T-cell development in mouse xenograft model. Sorted human HPCs were transduced with either miR-125b/OE or CTRL mCherry-traceable vectors and IV injected into NSG recipients. Mice were sacrificed at day 90 posttransplantation. (D) Proportion of ISP4, IDP, DP, SP4, and SP8 populations recovered from total thymi was estimated within the mCherry+/CD7+/CD5+ compartment (n = 3). *P < .05, multiple Student t test. (E) Shown are the representative plots; 1 mouse per group. Proportions are given relative to gated CD4+/CD8−; CD4+/CD8+ and CD4+/CD8+ fractions. (F) Flowchart of leukemia competitive transplantation experiments. Sorted T-ALL (M106) infected cells (mCherry+) expressing or not miR-125b were mixed at a 1:1 ratio with sorted control (eGFP+) or sorted wild-type (WT) cell populations and 500 cells were transplanted into NSG recipients. The hosts were monitored for T-ALL development by BM analysis. (G) Relative miR-125 expression (R-Q-PCR), normalized to RNU44 (2−∆Ct), in M106 T-ALL blasts yielded from BM of recipient mice at sacrifice (n = 4 for CTRL; n = 3 for 125b/OE). (H) T-ALL cell expansion in periphery assessed by CD45+/CD7+ cells in BM at 2 time points. The percentage of CD45+/CD7+ cells was measured gating on live cells. (I) Percentage of (mCherry+) CD45+/CD7+ T-ALL cells in BM was measured at day 55. The dashed line indicates the percentage of mCherry+ cells (50%) at the day of injection. (H-I) CTRL (n = 11); 125b/OE (n = 12). (J) Kaplan-Meier survival plot depicting accelerated leukemia onset in the mouse recipients with ectopic expression of miR-125b. Death points of the recipients were recorded when found dead or moribund. The dashed line indicates median survival time. Data: (G-I) ****P < .0001; *P < .05, Mann-Whitney U test; (J) Mantel-Cox test.
Ectopic expression of miR-125b in HPCs mimics the effects of TLX3 on T-cell differentiation. Sorted human HPCs were transduced with either miR-125b/OE or CTRL mCherry-traceable vectors. T-cell differentiation was followed as described in Figure 4A. (A) Relative miR-125b expression at days 21 (N = 3) and 42 (N = 5) in miR-125b/OE cells compared with CTRL cells. *P < .05, Mann-Whitney U test. (B) miR-125b/OE enhances T-cell growth. Cumulative T-cell numbers at day 42 compared with day 21 of culture for miR-125b/OE and CTRL cells (n = 8). *P < .05, Wilcoxon test. (C) miR-125b/OE induces accumulation of immature DN cells and blocks sCD3 acquisition by DP cells. Shown are the data from the representative experiment at day 42. Proportions of T-cell fractions are relative to gated mCherry+/CD7+/CD5+ T cells. (D-E) Human T-cell development in mouse xenograft model. Sorted human HPCs were transduced with either miR-125b/OE or CTRL mCherry-traceable vectors and IV injected into NSG recipients. Mice were sacrificed at day 90 posttransplantation. (D) Proportion of ISP4, IDP, DP, SP4, and SP8 populations recovered from total thymi was estimated within the mCherry+/CD7+/CD5+ compartment (n = 3). *P < .05, multiple Student t test. (E) Shown are the representative plots; 1 mouse per group. Proportions are given relative to gated CD4+/CD8−; CD4+/CD8+ and CD4+/CD8+ fractions. (F) Flowchart of leukemia competitive transplantation experiments. Sorted T-ALL (M106) infected cells (mCherry+) expressing or not miR-125b were mixed at a 1:1 ratio with sorted control (eGFP+) or sorted wild-type (WT) cell populations and 500 cells were transplanted into NSG recipients. The hosts were monitored for T-ALL development by BM analysis. (G) Relative miR-125 expression (R-Q-PCR), normalized to RNU44 (2−∆Ct), in M106 T-ALL blasts yielded from BM of recipient mice at sacrifice (n = 4 for CTRL; n = 3 for 125b/OE). (H) T-ALL cell expansion in periphery assessed by CD45+/CD7+ cells in BM at 2 time points. The percentage of CD45+/CD7+ cells was measured gating on live cells. (I) Percentage of (mCherry+) CD45+/CD7+ T-ALL cells in BM was measured at day 55. The dashed line indicates the percentage of mCherry+ cells (50%) at the day of injection. (H-I) CTRL (n = 11); 125b/OE (n = 12). (J) Kaplan-Meier survival plot depicting accelerated leukemia onset in the mouse recipients with ectopic expression of miR-125b. Death points of the recipients were recorded when found dead or moribund. The dashed line indicates median survival time. Data: (G-I) ****P < .0001; *P < .05, Mann-Whitney U test; (J) Mantel-Cox test.
The effects of TLX3/OE and miR125b/OE on T-cell differentiation were also tested in vivo using transplantation of transduced human CD34+/CD38− progenitor/stem cells into NSG mice. The results confirm that high miR125b increases cell proliferation (supplemental Figure 4B) and modifies the ratio of newly generated T cells. IDP cells especially were significantly enhanced at the expense of the DP cells (Figure 5D-E). Importantly, surface CD3 expression was decreased by enhanced miR125b expression as observed in vitro (Figure 5C). Intriguingly, although the control and miR125b-expressing cells did engraft and develop into human T cells in NSG mice, TLX3/OE CD34+/CD38−/low cells did not engraft the mouse BM, spleen, and thymus. These results show that TLX3, which is not expressed in normal immature progenitors, somehow interferes with the human hematopoiesis process in those conditions.
Altogether, these results support our hypothesis that oncogenic protein TLX3 and miR-125b both contribute to the development of a pre-leukemic T-cell state by sustaining T-cell production and blocking normal differentiation.
miR-125b enhances leukemia development in T-ALL xenograft
Forced expression of TLX3 in 2 negative cell lines (PEER and Jurkat), and in mature cortical SIL-TAL1 rearranged primary T-ALL (hereafter M106), did not result in miR-125b induction (supplemental Figure 5). Assuming that TLX3 may operate at an earlier T-cell development stage in immature cortical TLX3 T-ALL compared with mature TAL1 T-ALL, we bypassed TLX3 and enforced miR-125b expression in M106 T-ALL. Leukemia propagation in vivo was studied using (1:1) competitive transplantation of mCherry+ 125b/OE T-ALL cells with nontransduced wild-type or eGFP+ CTRL cells (Figure 5F-G). We observed that leukemia development was enhanced and skewed toward 125b/OE cells that overgrew wild-type or CTRL/eGFP+ cells. Indeed, mice transplanted with 125b/OE leukemia cells contained nonrandomly distributed transduced blasts compared with control mice (Figure 5H-I). Importantly, the median survival time was shorter for 125b/OE T-ALL mouse recipients compared with CTRLs (Figure 5J). This strong oncogenic potential of miR-125b was further confirmed in additional experiments using 2 other 125b/OE TAL1 T-ALLs (supplemental Figure 6). Although these results do not provide information in the context of TLX3, they further outline the strong oncogenic potential of miR125b in human T-ALL.
ETS1 and CBFb expression inversely correlates with TLX3 and miR125b
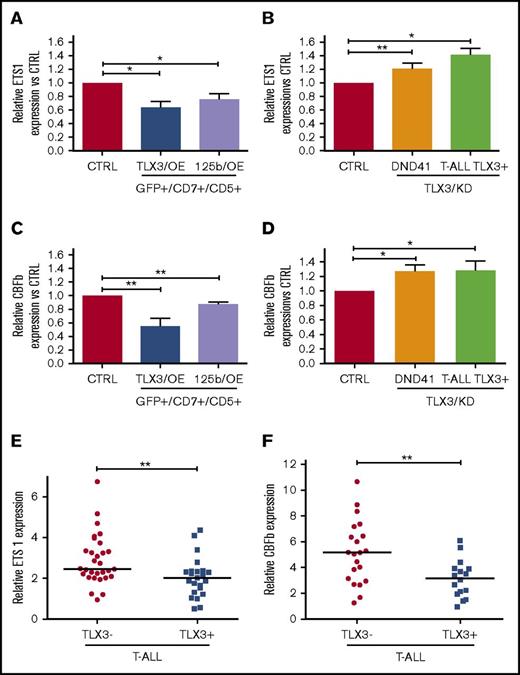
Given the fact that both 125b/OE and TLX3/OE block human T-cell differentiation and that TLX3 T-ALLs exhibit a predominantly immature phenotype, we looked for candidate target proteins involved in T-cell differentiation. Transcription factor ETS1 is a known miR-125b target in human invasive breast cancer and it is an essential regulator of the activity of the TCRα enhancer and the rearrangement of the TCRα locus.10 The core-binding factor β (CBFβ) is another miR125b target in hematopoietic cells.32 It takes part in RUNX1-CBFβ complexes regulating TCRβ rearrangement and β-selection of developing T cells. We observed that ETS1 as well as CBFβ were downregulated in TLX3/OE and in 125b/OE HPC-derived T cells and upregulated in TLX3/KD DND41 cells and primary T-ALLs (Figure 6A-D). We also found significantly lower expression levels of ETS1 and CBFβ in TLX3 T-ALL specimens compared with non-TLX3 T-ALLs (Figure 6E-F). The inverse correlation between TLX3 and ETS1 or CBFβ levels, 2 miR-125b target genes, supports our hypothesis that miR-125b takes part in T-cell differentiation arrest caused by TLX3.
miR-125b regulates ETS1 and CBFβ genes critical for T-cell differentiation. (A-D) ETS1 and CBFβ expression negatively correlates with TLX3 and miR-125b expression in T-ALLs and T-cell progenitors. (A) Relative ETS1 messenger RNA (mRNA) levels in TLX3/OE (n = 6) and miR-125b/OE (n = 7) HPC-derived T cells at day 42 of culture. (B) Relative ETS1 mRNA levels in TLX3/KD DND41 cell line (n = 19) and primary T-ALL TLX3+ (n = 3). (C) Relative CBFβ mRNA levels in TLX3/OE (n = 3) and miR-125b/OE (n = 7) HPC-derived T cells at day 42 of culture. (D) Relative CBFβ mRNA levels in TLX3/KD DND41 cell line (n = 4) and primary T-ALL TLX3+ (n = 5). (E-F) ETS1 (E) and CBFβ (F) levels in TLX3− compared with TLX3+ T-ALLs (respectively, n = 30 vs n = 20 for ETS1 and n = 21 vs n = 16 for CBFβ). Shown are relative mRNA expressions normalized to ABL housekeeping gene (2−∆Ct). (A-F) Data are presented as mean ± SEM of values. *P < .05; **P < .01, Mann-Whitney U test.
miR-125b regulates ETS1 and CBFβ genes critical for T-cell differentiation. (A-D) ETS1 and CBFβ expression negatively correlates with TLX3 and miR-125b expression in T-ALLs and T-cell progenitors. (A) Relative ETS1 messenger RNA (mRNA) levels in TLX3/OE (n = 6) and miR-125b/OE (n = 7) HPC-derived T cells at day 42 of culture. (B) Relative ETS1 mRNA levels in TLX3/KD DND41 cell line (n = 19) and primary T-ALL TLX3+ (n = 3). (C) Relative CBFβ mRNA levels in TLX3/OE (n = 3) and miR-125b/OE (n = 7) HPC-derived T cells at day 42 of culture. (D) Relative CBFβ mRNA levels in TLX3/KD DND41 cell line (n = 4) and primary T-ALL TLX3+ (n = 5). (E-F) ETS1 (E) and CBFβ (F) levels in TLX3− compared with TLX3+ T-ALLs (respectively, n = 30 vs n = 20 for ETS1 and n = 21 vs n = 16 for CBFβ). Shown are relative mRNA expressions normalized to ABL housekeeping gene (2−∆Ct). (A-F) Data are presented as mean ± SEM of values. *P < .05; **P < .01, Mann-Whitney U test.
TLX3 binds and transactivates LINC00478, the host gene of miR-125b-2
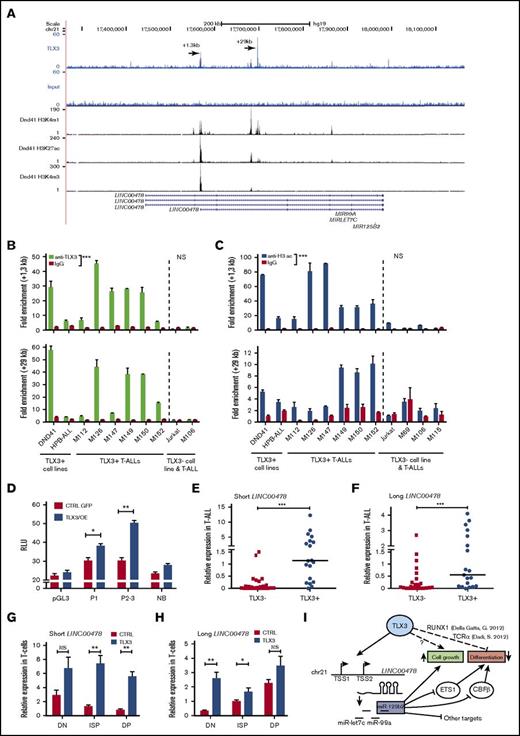
Coordinated expression of TLX3 and miR125b suggests that TLX3 may be directly involved in miR-125b regulation. As mature miR-125b-1 and miR-125b-2 have identical sequences, we tested TLX3 binding to regulatory regions near genomic loci encoding 2 paralogous miR clusters: MIR100/LET-7A/MIR125B-1 and MIR99A/LET-7C/MIR125B-2 on the corresponding 11 and 21 chromosomes. ChIP analysis of primary T-ALL (hereafter M126) coupled to deep sequencing (ChIP-seq) showed the presence of 2 peaks of TLX3 binding in the locus encoding LINC00478, a predicted host gene of miR-99a/let-7c/mir-125b-2 (Figure 7A). Of note, TLX3 binding was not detected in the vicinity of the genetic locus harboring MIR100/LET7a/MIR125B1 and overlapping the MIR100HG gene (supplemental Figure 7). Interestingly, 1 of the TLX3-binding regions on chromosome 21 maps inside of the LINC00478 gene, near the transcription start site (TSS) of its shorter transcript (+1.3 kb) and the second one (+29 kb) is located in the second intron of the same transcript. Both TLX3-binding sites were validated by ChIP-Q-PCR analysis of blasts obtained from 6 diagnosed TLX3 T-ALL patients as well as from TLX3-expressing DND41 and HPB cell lines. These binding sites were not detected in the non-TLX3 Jurkat cell line and T-ALL M106 (Figure 7B). The enrichments of 2 TLX3-binding regions in T-ALL were of similar order of magnitude as the enrichment of a well-characterized TCRα enhancer-associated sequence,10 used as a positive control in ChIP experiments (supplemental Figure 8). Interestingly, in DND41 cells, both genomic regions were also enriched with histone H3K4me3 and H3K27ac marks, indicative of an active chromatin state and promoter and enhancer elements (Figure 7A and ENCODE data). These observations were confirmed upon ChIP analysis of 6 TLX3 T-ALLs and 2 cell lines, whereas no active chromatin state of the LINC00478 locus was found in non-TLX3 T-ALLs and Jurkat cells (Figure 7C). Two fragments of the +1.3-kb binding region were cloned in the pGL3 promoter (SV40) vector (supplemental Figure 9A). Both fragments increased the activity of the heterologous promoter in the presence of ectopic TLX3 in 293T cells (Figure 7D). MiR-99a, let-7, and miR-125b-2 sequences overlap with the last introns of annotated LINC00478 transcripts having different TSSs and alternative splicing isoforms (supplemental Figure 9B). In agreement with our hypothesis, 2 LINC00478 transcripts were expressed at significantly higher levels in TLX3 T-ALLs compared with the non-TLX3 T-ALLs (Figure 7E-F) and in newly produced TLX3/OE T cells compared with CTRL T cells (Figure 7G-H). As mentioned in “Overexpression of TLX3 or miR125b induces expansion of normal T cells and blocks differentiation at immature stages in vitro and in vivo,” mature miR-125b and miR-99a were induced in TLX3/OE HPC-derived T cells while expression of miR-100 mapping to the chromosome 11 was considerably weaker. Accordingly, analysis of publicly available RNA-seq data obtained from 2 independent cohorts of T-ALL patients33,34 showed significantly higher levels of the LINC00478 transcript in TLX3+ than in TLX3− T-ALL (supplemental Figure 10), whereas MIR100HG expression is restricted to normal thymus cells. Taken together, our results indicate that TLX3 transcriptionally regulates the production of miRNAs, including miR125b, encoded in the intron of LINC00478 in T-ALL.
TLX3 directly regulates miR-125b-2 expression in T-ALL cells. (A) TLX3 ChIP-seq analysis performed for 1 newly diagnosed primary T-ALL. Gene tracks represent binding at the MIR-125B-2 locus on chromosome 21. Y-axis values show the number of sequence reads. Positions of 2 major binding peaks are indicated relative to the TSS of the shorter transcript of LINC00478, which overlaps with the MIR-99a/LET7C/MIR125B-2 locus. Two TLX3-binding regions are also enriched in H3K4me1 and H3K4me3, marking promoter/transcriptional initiation sites and H3K27ac marking active regulatory elements in the DND41 cell line (ENCODE). (B) TLX3 ChIP-Q-PCR analysis of DND41 and HPB-ALL TLX3–expressing cell lines and of 6 primary TLX3 T-ALLs. Shown are fold enrichment values at +1.3-kb and +29-kb binding regions of LINC00478 locus relative to the IGX1A ORF-free intergenic region containing no TSS. All ChIP experiments were performed at least twice. (C) Histone H3 acetylation at the +1.3-kb and +29-kb chromatin regions in TLX3+ and TLX3− cell lines and T-ALLs assessed by ChIP-Q-PCR analysis. Fold enrichment values relative to the IGX1 locus is shown. (D) Luciferase reporter assay in 293T cells. The empty pGL3 promoter plasmid and nonbinding 980-bp fragment immediately downstream of TLX3 binding “+1.3-kb” region served as negative controls. Relative luminescence units (RLU) were determined by normalizing firefly to Renilla luciferase activity (n = 4). (E-H) Relative expression of 2 LINC00478 transcripts normalized to ABL1 (2−ΔCt × 100): (E-F) in TLX3− (N = 26) and TLX3+ (N = 19) T-ALLs and (G-H) in TLX3/OE and CTRL HPC-derived T cells at day 42 of differentiation. (B-H) Data are presented as mean ± SEM of values: ***P < .001; **P < .01; *P < .05: Mann-Whitney U test. (I) A simplified view of TLX3-miR-125b relations in T-ALL and involvement of miR-125b target genes in the blockage of T-cell development.
TLX3 directly regulates miR-125b-2 expression in T-ALL cells. (A) TLX3 ChIP-seq analysis performed for 1 newly diagnosed primary T-ALL. Gene tracks represent binding at the MIR-125B-2 locus on chromosome 21. Y-axis values show the number of sequence reads. Positions of 2 major binding peaks are indicated relative to the TSS of the shorter transcript of LINC00478, which overlaps with the MIR-99a/LET7C/MIR125B-2 locus. Two TLX3-binding regions are also enriched in H3K4me1 and H3K4me3, marking promoter/transcriptional initiation sites and H3K27ac marking active regulatory elements in the DND41 cell line (ENCODE). (B) TLX3 ChIP-Q-PCR analysis of DND41 and HPB-ALL TLX3–expressing cell lines and of 6 primary TLX3 T-ALLs. Shown are fold enrichment values at +1.3-kb and +29-kb binding regions of LINC00478 locus relative to the IGX1A ORF-free intergenic region containing no TSS. All ChIP experiments were performed at least twice. (C) Histone H3 acetylation at the +1.3-kb and +29-kb chromatin regions in TLX3+ and TLX3− cell lines and T-ALLs assessed by ChIP-Q-PCR analysis. Fold enrichment values relative to the IGX1 locus is shown. (D) Luciferase reporter assay in 293T cells. The empty pGL3 promoter plasmid and nonbinding 980-bp fragment immediately downstream of TLX3 binding “+1.3-kb” region served as negative controls. Relative luminescence units (RLU) were determined by normalizing firefly to Renilla luciferase activity (n = 4). (E-H) Relative expression of 2 LINC00478 transcripts normalized to ABL1 (2−ΔCt × 100): (E-F) in TLX3− (N = 26) and TLX3+ (N = 19) T-ALLs and (G-H) in TLX3/OE and CTRL HPC-derived T cells at day 42 of differentiation. (B-H) Data are presented as mean ± SEM of values: ***P < .001; **P < .01; *P < .05: Mann-Whitney U test. (I) A simplified view of TLX3-miR-125b relations in T-ALL and involvement of miR-125b target genes in the blockage of T-cell development.
Discussion
Here, we characterized miRNA expression patterns in pediatric T-ALLs and compared them with the most variable miRNAs detected throughout human T-cell maturation. Nine of the 20 most abundant miRNAs we detected were previously reported as highly expressed in T-ALLs and 5 of them are known regulators of tumor suppressor genes involved in the pathogenesis of T-ALL. In particular, high expression of miR-223 is found in all T-ALL specimens in comparison with normal T-cell precursors. Downregulation of miR-223 target protein E3-ligase FBXW7 and stabilization of c-MYC, MYB, and NOTCH1 oncoproteins have been previously proposed as a part of the oncogenic network in T-ALL.20 Our results thus consolidate established links between miRNAs and T-ALL development while extending miRnome analysis to pediatric T-ALLs. We also provide highly accurate miRNA profiling of early T-cell precursors during 7 specific maturation stages, including uncharacterized pro-T, pre-T, and ISP cell subsets and uncover the 36 most variable miRNAs that distinguish the different T-cell maturation stages.
In pairwise comparison of the miRNA signatures of the 3 cytogenetic T-ALL groups, miR-125b and miR-99a segregated with TLX3 expression. Taking into account that elevated expression of miR-125b was previously observed but not explored in TLX3/TLX1 T-ALL,35 and that enforced constitutive overexpression of miR125b in mice induces 3 types of neoplasms including T-ALL,29,30,36 we investigated the functional relationships between miR-125b and TLX3. We found that miR-125b expression levels in TLX3 T-ALL are as high as in early cortical thymocytes from the pro-T to ISP stages of differentiation where TCRβ gene rearrangement, pre-TCR surface expression, and extensive cell expansion take place. miR-125b expression drops substantially in normal IDP, DP, SP4, and SP8 thymocytes, suggesting that miR-125b levels of TLX3 T-ALL mirror the differentiation arrest at the immature cortical stage reported for this pediatric T-ALL subgroup.10 Relative miR-125b levels in TLX3 T-ALL blasts are up to 10 times higher than in normal DP progenitors and the growth rate of TLX3 T-ALL depends on the miR-125b expression.
miR-125b is transcribed from 2 chromosomal loci, 11q23 (hsa-miR-125b-1) and 21q21 (hsa-miR-125b-2). Genetic rearrangements of both loci are involved in the development of leukemia. MIR125b-1 is targeted by t(11;14)(q24; q32) and t(2;11)(p21; q23-q24) translocations in B-lymphoid and myeloid malignancies, respectively.17,18,37 Upregulation of miR-125b-2 linked to structural aberrations of chromosome 21 was observed in megakaryocytic leukemia associated with Down syndrome38 and in childhood ETV/RUNX1 ALL.39,40 In the case of the t(11;14) B-ALL, inappropriate MIR125b-1 expression is controlled by immunoglobulin heavy chain (IGH) regulatory elements.18,37 In myelodysplasia and acute myeloid leukemia (AML), mechanisms of MIR125B1 activation are not understood.17 Published genetic analyses of TLX3 T-ALL did not describe rearrangements of chromosomes 11 and 21 with breakpoints close to MIR125B1 and MIR125B2 loci.41 Comparative genomic hybridization analysis of 12 TLX3 T-ALL from our cohort did not detect copy-number variations of the loci (data not shown). These data argued for direct transcriptional TLX3-mediated miR-125b regulation, further supported by the fact that upregulation and downregulation of TLX3 caused coordinated changes in miR-125b expression. Our data demonstrate that intronic miR-99a/let-7c/miR-125b-2 are processed from the LINC00478 transcript. Three common TSSs for the miR-99a, let-7c, and miR-125b-2 tricistronic region were previously predicted42 and recently confirmed in megakaryoblastic cell lines.43 Interestingly, the homeobox transcription factor HOXA10 and its cofactor PBX1 involved in the regulation of the miR-99a, let-7c, and miR-125b-2 tricistron in hematopoietic stem cells occupy binding sites located at −18 to +866 bp from TSS2 of LINC00478.43 We show that TLX3 binds to 2 transcriptionally active genomic regions mapping to the LINC00478 gene. However, the in-fine molecular mechanism of activation awaits further investigations as no consensus TLX3-binding sites and few regulatory partners were reported. TLX1 and TLX3 transcription factors have nearly identical homeodomains, although blast alignment shows that overall identity positions between these proteins is 54%. Genome-wide binding profiles of TLX1, in the context of T-ALL, showed the enrichment of RUNX- and ETS-binding sites in the promoters of TLX1-repressed genes.12,44 Computing analysis of the +1.3-kb TLX3-binding region of LINC00478 indicated close RUNX (WAACCAC) and ETS (AGGAARY) motifs and other 3 separate ETS motifs. However, our ChIP-seq experiments with ETS1 in the same patient did not reveal ETS1 binding at these locations, nor at the +29-kb region (data not shown). At least for this particular case, ETS1 does not seem to be directly involved in the regulation of LINC00478.
ChIP-seq analysis of primary T-ALL did not find any TLX3-binding sites near the MIR125B1 locus despite very similar genomic organization of MIR125B1 and MIR125B2 clusters. Independent regulation of miRNAs residing in these clusters is described in myeloid malignancies and in nonhematopoietic context via regulatory binding sites close to the pre-miR-125b-1 sequence.45 The differences in regulation of miRNAs from these 2 clusters may rely on accelerated evolution of regulatory elements compared with the coding sequences.
Let-7c expression did not correlate with miR-125b-2 and miR-99a. Uncoordinated expression profiles of several clustered miRNAs related to the posttranscriptional modulation of the miRNA maturation process have been reported.46,47
Our hypothesis that miR-125b is implicated in the genesis of T-ALL relies on the sustained cell growth and impaired differentiation observed upon enforced expression of TLX3 or miR-125b in T-cell progenitors. 125b/OE in mature T-ALL enhances leukemia propagation, outlining the oncogenic potential of miR-125b in T-ALL. Both TLX3/OE- and 125b/OE-transduced human T-cell progenitors have similar differentiation blocks at immature DN/ISP stages and proliferative advantage. In normal T-cell development, miR-125b expression drops in early progenitors and this process may help the maturation process. Transcription factor ETS-1 promotes early thymocyte differentiation mediated by the pre-TCR complex.48 In Ets1-deficient mice, the numbers of thymocytes and lymph node resident T cells are markedly decreased and T cells undergo apoptosis.49 TLX1 and TLX3 proteins physically interact with ETS1 and thus repress TCRα enhanceosome activity leading to the blockage of TCR-Jα rearrangement and T-cell development.10 On the other hand, bioinformatic tools (PICTAR-VERT, TARGETSCAN-VERT) predict 2 recently validated conserved miR-125b–binding sites in the 3′ untranslated region of ETS1 mRNA.50 Our results show inverse correlation of ETS1 and miR-125 expression in the model of T-cell differentiation perturbed by ectopic TLX3 and in TLX3 T-ALLs. ETS1 functionally interacts with the RUNX-CBFβ CBF complex where the CBFβ subunit stabilizes the binding of runt-related transcription factors RUNX1 or RUNX3 to target DNA sequences, including TCR enhancers.51 Of note, CBFβ is also a validated target of miR-125b in hematopoietic cells.52 Development of early T-cell progenitors is very sensitive to an incremental reduction of CBFβ levels in the mouse model carrying hypomorphic alleles of the gene53 with differentiation blockage occurring in CD4/CD8− DN1, DN2, and DN3 stage and affecting TCRβ expression.53 These observations may explain the drastic effect of ectopic miR-125b in our in vitro differentiation model as human T-cell progenitors show the differentiation block at the DN stage and the levels of lentiviral miR-125b expressions are higher (up to 30-fold) than those induced by TLX3.
In conclusion, our results demonstrate that TLX3 and TLX3-induced LINC00478/miR125b cooperate in oncogenic transformation of human T cells. miR-125b contributes to maturation arrest in T-ALL via downregulation of ETS1 and CBFβ genes (Figure 7I) that might be a part of a mechanism leading to disease progression. Our study uncovers a novel regulatory pathway, which offers new opportunities for the development of original therapeutic approaches.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors acknowledge all patients who allowed them to use their cells for research, B. Canque for sharing thymic specimens, and the medical staff who collected samples at the following French hospitals: A. Trousseau and R. Debré in Paris, Institut d'Hématologie et d'Oncologie Pédiatrique in Lyon, Noriets in Vitry-sur-Seine, and Antoine-Béclère in Clamart. Cell sorting was performed at the cytometry platform of the Institut de Radiobiologie Cellulaire et Moléculaire with N. Dechamps. P. Fouchet helped isolating T-cell subsets. The authors are grateful to M.-L. Arcangeli for expertise and kind help in cytometry analysis, and P. H. Romeo and A. Gortchakov for very helpful critical reading of the manuscript.
This work was supported by Commissariat à l’Energie Atomique et aux Energies Alternatives, INSERM, Université Paris Diderot, and Université Paris Sud, Association pour la Recherche contre le Cancer (ARC; 07/1/4959), Association Laurette Fugain (08/02), and Ligue contre le Cancer (LNCC; Équipe Labellisée).
L.R. was a fellow of the French Ministry of Education and ARC.
Authorship
Contribution: L.R., C.D., J.C., B.U., A. Benyoucef, I.N., F.P., and A.C. performed experiments; I.N., F.P., L.R., and P.B. designed the experiments, analyzed the results, and wrote the manuscript; P.Y.-B. provided statistical analysis; S.S., M.B., and V.A. provided ChIP-seq and RNA-seq bioinformatic analyses; P.Y.-B., S.S., and V.A. critically read the manuscript; and A. Baruchel, J.L.-P., and P.B. provided patient samples and shared crucial information.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Irina Naguibneva, Laboratoire des Cellules Souches Hématopoïétiques et Leucémiques, Equipe Labellisée Ligue contre le Cancer, IRCM, CEA, 18 route du Panorama, 92260 Fontenay-aux-Roses, France; e-mail: irina.naguibneva@cea.fr; or Paola Ballerini, Service d’Hématologie Pédiatrique, Hôpital A. Trousseau, Assistance Publique–Hôpitaux de Paris, Paris, France; e-mail: paola.ballerini@trs.aphp.fr.
References
Author notes
F.P., P.B., and I.N. are joint supervisors.

![Figure 1. miRNA expression in pediatric T-ALL. (A) Twenty most abundantly expressed miRNAs in different cytogenetic subgroups of 41 T-ALL clinical specimens as assessed by TLDA and normalized to the RNU48 expression in a given sample (2−∆Ct). miRNAs are ranked by expression levels (given as percentage of RNU48 expression). (B) Heatmap of miRNAs differentially expressed in 3 cytogenetic subgroups of T-ALL. Normalized expressions (ΔCt z-values) were color-coded by a gradient from green (underexpressed) to red (overexpressed) relative to the mean (ANOVA adjusted P values: .005 [miR-135a], .005 [miR-125b], .013 [miR-100], .03 [miR-506], .03 [miR-99a], .04 [miR-153]). (C-E) Comparison of miR-125b, miR-99a, and miR-100 levels in TLX3 vs non-TLX3 T-ALL samples by R-Q-PCR (Exiqon). miRNA expression is normalized to RNU44; levels in DP T cells are used as calibrator (2−∆∆Ct): (C) miR-125b (n = 25 vs n = 33), (D) miR-99a (n = 16 vs n = 16), and (E) miR-100 (n = 16 vs n = 16). ***P < .001, Mann-Whitney U test. Ct, cycle threshold; NS, not significant.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/1/12/10.1182_bloodadvances.2017005538/3/m_advances005538f1.jpeg?Expires=1765918628&Signature=rPbHau4kJKRpPGbqZC0zrm1CBb-2Mn8pxg-BWmli5L2Y9JPhdfsis6s~XKjx8p1LW1hpkfOFDoD7TzUqaUwHY8ImRQRtWFG9OlESUoBBf~c~HaTyRdnCwL-w-hatHsSumwimcpsIZU-YkXRZfrrBB2WfNabt1zk2Rcu71X5k98bYr9EPrqj~clveTbNtHpNM2n2LY1jR7ZR94I--4qw0HDdBeeAATQ~JOp-U0MC0e1zAVkyHddT2~UMEiN~cOVg9DQKw4UvBOQak3dxnIQpEr0c6C7yuEqG56wphaoOI38jiSuC8w5d7M5EAkcIGVD1cuvSgnOa1HIcYGqOP~FOo~w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)