Key Points
We present the first in-depth analysis of platelet PtdIns(3,4,5)P3-binding proteins, providing a valuable resource for future studies.
The PtdIns(3,4,5)P3-binding protein, DAPP1, negatively regulates glycoprotein VI–driven platelet activation and thrombus formation.
Abstract
The class I phosphoinositide 3-kinase (PI3K) isoforms play important roles in platelet priming, activation, and stable thrombus formation. Class I PI3Ks predominantly regulate cell function through their catalytic product, the signaling phospholipid phosphatidylinositol 3,4,5-trisphosphate [PtdIns(3,4,5)P3], which coordinates the localization and/or activity of a diverse range of binding proteins. Notably, the complete repertoire of these class I PI3K effectors in platelets remains unknown, limiting mechanistic understanding of class I PI3K–mediated control of platelet function. We measured robust agonist-driven PtdIns(3,4,5)P3 generation in human platelets by lipidomic mass spectrometry (MS), and then used affinity-capture coupled to high-resolution proteomic MS to identify the targets of PtdIns(3,4,5)P3 in these cells. We reveal for the first time a diverse platelet PtdIns(3,4,5)P3 interactome, including kinases, signaling adaptors, and regulators of small GTPases, many of which are previously uncharacterized in this cell type. Of these, we show dual adaptor for phosphotyrosine and 3-phosphoinositides (DAPP1) to be regulated by Src-family kinases and PI3K, while platelets from DAPP1-deficient mice display enhanced thrombus formation on collagen in vitro. This was associated with enhanced platelet α/δ granule secretion and αIIbβ3 integrin activation downstream of the collagen receptor glycoprotein VI. Thus, we present the first comprehensive analysis of the PtdIns(3,4,5)P3 signalosome of human platelets and identify DAPP1 as a novel negative regulator of platelet function. This work provides important new insights into how class I PI3Ks shape platelet function.
Introduction
Platelets are small, anucleate cells that play an essential role in hemostasis, but can contribute critically to the pathogenesis of cardiovascular disease.1 Their function is coordinated by an array of cell-surface receptors coupled to diverse intracellular signaling effectors, including class I phosphoinositide 3-kinases (PI3Ks).2 The use of gene-targeted mice and small molecule inhibitors has revealed important roles for the 4 class I PI3K isoforms (PI3Kα, β, δ, and γ) in platelet priming, activation, and thrombus formation.3-7 PI3Kβ appears to be the predominant class I isoform in platelets, being important for glycoprotein VI (GPVI), protease-activated receptor (PAR), and P2Y12 signaling in addition to bidirectional αIIbβ3 integrin function.6,8-10 This translates to a broad and important role for this isoform in platelet activation and subsequent stable thrombus formation, which has attracted PI3Kβ considerable attention as a potential antithrombotic target.8,11,12 This is supported by the observation that genetic loss or pharmacological inhibition of PI3Kβ provides protection from occlusive arterial thrombus formation in animal models.8,9 Furthermore, AZD6482, a selective PI3Kβ inhibitor, has demonstrated promising antiplatelet effects and tolerance in humans.11,12 Thus, PI3Kβ inhibition appears to afford protection from occlusive arterial thrombosis while demonstrating limited bleeding risk,6,8,9,12 although the potential for embolization with this strategy needs additional investigation.13,14
Despite extensive confirmation of the importance of the class I PI3Ks to platelet function, detailed mechanistic understanding of the events downstream of PI3K activation remains limited. Although class I PI3Ks may have protein kinase activity15 and scaffolding roles,16 they predominantly regulate cell function through the product of their lipid kinase activity, phosphatidylinositol 3,4,5-trisphosphate [PtdIns(3,4,5)P3].17 PtdIns(3,4,5)P3 is generated by the class I PI3K–catalyzed phosphorylation of phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2] and serves to coordinate the localization and/or activity of a range of binding proteins.17-19 Known PtdIns(3,4,5)P3-binding proteins often possess a conserved pleckstrin homology (PH) domain and span a range of protein functional classes.17,20,21 Much of the focus with platelets has been on the serine/threonine kinase, AKT (protein kinase B [PKB]), the archetypal class I PI3K effector, which undergoes membrane recruitment on binding of its PH domain to PtdIns(3,4,5)P3 and has important roles in platelet function.6,22 Although a limited number of other PtdIns(3,4,5)P3-binding proteins have received attention in platelets,23-25 the current understanding of class I PI3K effectors in this cell type is poor, in large part because the full repertoire of PtdIns(3,4,5)P3-binding proteins in platelets remains unknown.
Mass spectrometry (MS) has allowed unprecedented global insights into platelet biology in recent years26-28 and is a powerful approach for the characterization of platelet subproteomes and specific signaling networks. In this article, we have used MS to conduct a detailed analysis of the PtdIns(3,4,5)P3 signalosome of human platelets. Using lipidomic MS, we observed robust PtdIns(3,4,5)P3 generation in response to PAR and GPVI receptor activation. We then conducted a global, unbiased screen for PtdIns(3,4,5)P3-binding proteins in human platelets using affinity capture coupled to high resolution proteomic MS. Our approach identified an extensive PtdIns(3,4,5)P3 interactome, including many proteins previously uncharacterized in this cell type. Of these, we define dual adaptor for phosphotyrosine and 3-phosphoinositides (DAPP1/Bam32/PHISH), shown previously to be an important regulator of leukocyte function,29-33 as a Src family kinase (SFK)- and PI3K-regulated protein that serves to restrain GPVI-mediated platelet activation.
Materials and methods
Human platelet preparation
Venous blood anticoagulated with 4% trisodium citrate (1:10, volume-to-volume) was obtained from healthy volunteers after obtaining informed consent, with the approval of the local research ethics committee at the University of Bristol. Platelets were isolated as previously described34 with the following modifications to minimize plasma, erythrocyte, and leukocyte contamination (Figure 2A); (1) only the upper two-thirds of platelet-rich plasma were collected; (2) this platelet-rich plasma was diluted with prewarmed N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES)-Tyrode’s medium supplemented with 0.1% (weight-to-volume) d-glucose, 10 µM indomethacin, and 0.02 U/mL apyrase (HT+++) before centrifugation; (3) platelets were washed twice in HT+++ plus acid citrate dextrose; (4) the final platelet suspension was passed through leukocyte removal filters. Automated hematology analysis was conducted to confirm the absence of detectable levels of contaminating erythrocytes and leukocytes. The platelet suspension was allowed to rest for 30 minutes at 30°C before use.
Identification of platelet PtdIns(3,4,5)P3-binding proteins
Purified platelets were centrifuged (520g, 10 min, room temperature), providing an additional wash step, and the pellet lysed in ice-cold lysis buffer (20 mM HEPES [pH 7.4], 120 mM NaCl, 0.5% NP40, 5 mM EGTA, 5 mM EDTA, 5 mM β-glycerophosphate, 10 mM NaF, 1 mM sodium orthovanadate, cOmplete mini protease inhibitor tablet [Roche]). Phosphatase inhibitors were included to preserve the identity of PtdIns(3,4,5)P3 on the beads.19 Lysates were freeze-thawed, vortexed, and centrifuged (12 000g, 10 min, 4°C) to provide final clarified samples. A total of 8 × 108 platelets were used per sample for proteomics experiments. Affinity capture was performed by incubating lysates with control or PtdIns(3,4,5)P3-coupled beads for 90 minutes at 4°C. Additional control lysates were preincubated with 40 µM free PtdIns(3,4,5)P3 for 30 minutes prior to PtdIns(3,4,5)P3 bead incubation. Beads were washed 3 times with lysis buffer, and proteins were eluted in NuPAGE LDS sample buffer (plus 50 mM dithiothreitol). Eluates were subjected to western blotting, or the proteins were fractionated by gel walking, trypsin digested, and the resulting peptides fractionated using an Ultimate 3000 nano–high-performance liquid chromatography (HPLC) system in line with an Orbitrap Fusion Tribrid mass spectrometer. The MS data have been deposited to the ProteomeXchange Consortium via the PRIDE35 partner repository with the data set identifier PXD003777.
Mice
Animal studies were approved by the local research ethics committee at the University of Bristol, and mice were bred and maintained under a UK Home Office project license (PPL30/2908). Generation of DAPP1−/− (knockout [KO]) mice has been previously described.36 Experiments were performed on C57BL/6 DAPP1−/− mice from heterozygote breeders, with wild-type littermate controls sex-matched where possible. Blood was obtained by cardiac puncture of sacrificed mice, and washed platelets were prepared as previously described.37
In vitro thrombus formation
In vitro thrombus formation assays were performed under noncoagulating conditions, as previously described.38 Mouse blood was drawn by cardiac puncture into a syringe containing 4% trisodium citrate (1:10, volume-to-volume), 2 U/mL heparin, and 40 μM PPACK. Samples were imaged by using a 40× oil immersion objective on a Leica DM IRE2 inverted epifluorescent microscope attached to a Leica TCS-SP2-AOBS confocal laser scanning microscope. Quantification was performed by using Volocity 6.1.1 Quantitation software.
Aggregometry
Platelet aggregation assays were performed as previously described.4 Briefly, washed platelets at 2 × 108/mL were stimulated with agonist while monitoring for aggregation by using a Chronolog 490-4D aggregometer at 37°C with continuous stirring at 1200 rpm.
Flow cytometry analysis
Flow cytometry analysis of platelets was performed as previously described.5 Samples were analyzed on a BD FACSCanto II by using FACSDiva software (10 000 platelet events per sample). Subsequent analysis was performed by using Flowing Software 2.5.
Additional details are provided in supplemental Methods.
Results
Characterizing the PtdIns(3,4,5)P3 signalosome of human platelets
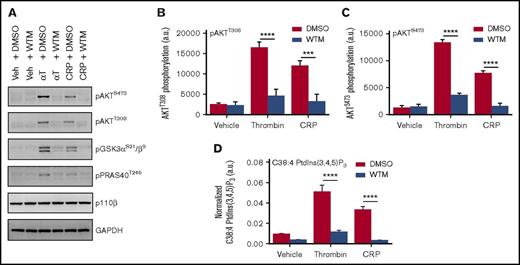
Class I PI3K activation in platelets is most commonly inferred from the phosphorylation status of the downstream effector, AKT,39 which in turn propagates signal transduction via the regulation of its substrates, including GSK3 and PRAS4022,40 (Figure 1A-C). Recently, Clark et al41,42 developed a new method for the direct and sensitive measurement of cellular phosphoinositides by MS, including quantification of the specific molecular (fatty acyl) species of the class I PI3K catalytic product, PtdIns(3,4,5)P3. We applied this lipidomic approach to human platelets and observed robust generation of stearoyl/arachidonoyl (C38:4 or C18:0/C20:4) PtdIns(3,4,5)P3 in response to PAR or GPVI activation with thrombin or collagen-related peptide (CRP), respectively (Figure 1D). Notably, we were also able to quantify the less abundant C38:3 PtdIns(3,4,5)P3 in human platelets for the first time, the behavior of which mirrored the C38:4 form, in addition to multiple species of PtdInsP2 (supplemental Figure 1A-B).
Human platelets show robust PtdIns(3,4,5)P3generation and associated AKT pathway phosphorylation (p) in response to PAR and GPVI receptor activation. Washed human platelets were preincubated with dimethyl sulfoxide (DMSO) or 100 nM Wortmannin (WTM) for 10 minutes at 37°C before stimulation for 2 minutes with vehicle (Veh) (HEPES-Tyrode’s buffer), 0.2 U/mL thrombin (αT), or 5 μg/mL CRP. Each sample was divided in 2 for western blotting of class I PI3K pathway components (A-C) and parallel lipid extraction and measurement of C38:4 PtdIns(3,4,5)P3 by lipidomic MS (D). Quantified data represents the mean of 3 independent donors + standard error of the mean, with representative blotting presented for 1 of the 3 donors. PtdIns(3,4,5)P3 is normalized to C38:4 PtdIns, with each normalized to its own synthetic internal standard, as detailed in the supplemental Methods. Statistical analyses were performed by using 2-way analysis of variance with Bonferroni post-tests. ***P = .0001; ****P < .0001. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Human platelets show robust PtdIns(3,4,5)P3generation and associated AKT pathway phosphorylation (p) in response to PAR and GPVI receptor activation. Washed human platelets were preincubated with dimethyl sulfoxide (DMSO) or 100 nM Wortmannin (WTM) for 10 minutes at 37°C before stimulation for 2 minutes with vehicle (Veh) (HEPES-Tyrode’s buffer), 0.2 U/mL thrombin (αT), or 5 μg/mL CRP. Each sample was divided in 2 for western blotting of class I PI3K pathway components (A-C) and parallel lipid extraction and measurement of C38:4 PtdIns(3,4,5)P3 by lipidomic MS (D). Quantified data represents the mean of 3 independent donors + standard error of the mean, with representative blotting presented for 1 of the 3 donors. PtdIns(3,4,5)P3 is normalized to C38:4 PtdIns, with each normalized to its own synthetic internal standard, as detailed in the supplemental Methods. Statistical analyses were performed by using 2-way analysis of variance with Bonferroni post-tests. ***P = .0001; ****P < .0001. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Upon confirming robust PtdIns(3,4,5)P3 generation in human platelets, we sought to better understand how this phosphoinositide permits class I PI3K to regulate multiple, diverse aspects of platelet function. PtdIns(3,4,5)P3 is considered to regulate cell function predominantly through the recruitment and/or regulation of a range of binding proteins,17,19 the full platelet repertoire of which remains unknown. We were recently able to confirm the RAS/RAP-GAP, RASA3, as a platelet PtdIns(3,4,5)P3-binding protein by using PtdIns(3,4,5)P3 immobilized on agarose beads.43 We therefore set out to develop this affinity capture strategy to conduct the first high-resolution, global, unbiased proteomic analysis of the complete PtdIns(3,4,5)P3 interactome of human platelets. To do this, we developed a modified human platelet preparation protocol to minimize sample contamination with proteins derived from plasma or contaminating blood cells (Figure 2A), utilizing freshly isolated platelets to avoid proteome degradation.27 First, the eluates from affinity capture experiments with platelet lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and viewed by SYPRO Ruby gel staining. This revealed protein bands present specifically in PtdIns(3,4,5)P3 bead eluates (Figure 2B), suggesting our approach could successfully capture a number of human platelet PtdIns(3,4,5)P3-binding proteins. To obtain an in-depth PtdIns(3,4,5)P3 interactome, we reduced sample complexity and increased resolution by incorporating a gel walking step for protein fractionation and subjecting our samples to analysis on an Orbitrap Fusion Tribrid mass spectrometer. Because only a proportion of the proteins captured on the beads were likely to be genuine PtdIns(3,4,5)P3-regulated proteins, in addition to using blank beads, we incorporated additional control samples preincubated with competing PtdIns(3,4,5)P3 to confirm binding specificity and used label-free Top 3 Protein Quantification (T3PQ) of the MS data.44 This dual-controlled quantitative approach validated the specificity and reproducibility of our method across independent donors (Figure 2C) and enabled us to apply highly stringent filtering criteria to the proteomics data to define the human platelet PtdIns(3,4,5)P3 interactome.
Experimental workflow for proteomics experiments. (A) Pure platelet preparations were obtained from whole blood by using a multistep approach (see “Materials and methods”), and lysates were subjected to affinity capture of PtdIns(3,4,5)P3-binding proteins by using PtdIns(3,4,5)P3-coupled beads. Eluate sample complexity was reduced by sodium dodecyl sulfate-polyacrylamide gel electrophoresis separation of proteins, followed by trypsin digest, nano-HPLC, and MS analysis. Data were subject to stringent filtering and analysis using a range of bioinformatics tools. (B) Human platelet lysates were incubated with control (Ctl) or PtdIns(3,4,5)P3 [PIP3, or after preincubation with competing free PtdIns(3,4,5)P3 (PIP3+)]-coupled beads for 90 minutes at 4°C before washing, elution, and analysis by SYPRO Ruby gel staining. (C) Experiments conducted as described in panel B were subjected to liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis. Histograms demonstrate validation of the proteomics approach by quantitative analysis of known PtdIns(3,4,5)P3-binding proteins with the T3PQ method across the independent donors. Bars represent the mean of 3 independent donors + standard error of the mean. ACD, acid citrate dextrose; Ctl, control; HT, HEPES-Tyrode’s buffer; PRP, platelet-rich plasma.
Experimental workflow for proteomics experiments. (A) Pure platelet preparations were obtained from whole blood by using a multistep approach (see “Materials and methods”), and lysates were subjected to affinity capture of PtdIns(3,4,5)P3-binding proteins by using PtdIns(3,4,5)P3-coupled beads. Eluate sample complexity was reduced by sodium dodecyl sulfate-polyacrylamide gel electrophoresis separation of proteins, followed by trypsin digest, nano-HPLC, and MS analysis. Data were subject to stringent filtering and analysis using a range of bioinformatics tools. (B) Human platelet lysates were incubated with control (Ctl) or PtdIns(3,4,5)P3 [PIP3, or after preincubation with competing free PtdIns(3,4,5)P3 (PIP3+)]-coupled beads for 90 minutes at 4°C before washing, elution, and analysis by SYPRO Ruby gel staining. (C) Experiments conducted as described in panel B were subjected to liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis. Histograms demonstrate validation of the proteomics approach by quantitative analysis of known PtdIns(3,4,5)P3-binding proteins with the T3PQ method across the independent donors. Bars represent the mean of 3 independent donors + standard error of the mean. ACD, acid citrate dextrose; Ctl, control; HT, HEPES-Tyrode’s buffer; PRP, platelet-rich plasma.
Dissecting the platelet PtdIns(3,4,5)P3 interactome
Our analysis reveals an extensive platelet PtdIns(3,4,5)P3 interactome, including >40 proteins previously reported to show affinity for PtdIns(3,4,5)P3 in other cell types or in vitro assays (Table 1; supplemental Table 1). Indeed, our data set spans extensively established class I PI3K effectors, such as BTK, TEC, PDK1, and AKT,18 to additional proteins with previously reported PtdIns(3,4,5)P3 affinity, including RASA3, DAPP1, cytohesin 1-3, PHLDB1, DOCK and ELMO proteins, ADAP1, myosin 1G, SNX9, and TBC1D2B,20,45-53 the majority of which remain functionally uncharacterized in platelets. We also identified several potentially novel class I PI3K–regulated proteins, including FRMD4B, ARHGAP6, APPL2, and GRTP1. Furthermore, ARAP1,54 TNFAIP8 family proteins,55 SIN1,56 and P-REX1,57 all of which have reported PtdIns(3,4,5)P3 affinity, were also captured on our PtdIns(3,4,5)P3 beads, although falling below our stringent filtering criteria, further confirming the comprehensive nature of our approach.
Selected components of the human platelet PtdIns(3,4,5)P3 interactome
| Accession No. . | Gene name . | Protein description . | Mean area . | Mean score . | Mean coverage (%) . | Mean peptides . | Mean PSM . | Times identified, n/3 . | Characterized in platelets? . |
|---|---|---|---|---|---|---|---|---|---|
| Q06187 | BTK | Tyrosine-protein kinase BTK | 8.56e+9 | 3109.1 | 75.4 | 61 | 1228 | 3 | Yes |
| Q14644 | RASA3 | Ras GTPase-activating protein 3 | 4.58e+9 | 2083.6 | 77.3 | 70 | 953 | 3 | Yes |
| Q9UN19 | DAPP1 | Dual adapter for phosphotyrosine and 3-phosphoinositides/Bam32 | 2.24e+9 | 777.1 | 80.2 | 24 | 325 | 3 | No |
| P42680 | TEC | Tyrosine-protein kinase Tec | 1.02e+9 | 522.7 | 67.2 | 47 | 253 | 3 | Yes |
| Q99418 | CYTH2 | Cytohesin-2/ARNO | 9.58e+8 | 384.3 | 53.0 | 23 | 165 | 3 | Yes |
| O43739 | CYTH3 | Cytohesin-3/GRP1 | 8.02e+8 | 132.1 | 32.4 | 14 | 61 | 3 | No |
| Q15438 | CYTH1 | Cytohesin-1/PSCD1 | 7.12e+8 | 171.5 | 39.9 | 17 | 79 | 3 | No |
| Q15283 | RASA2 | Ras GTPase-activating protein 2 | 4.49e+8 | 131.0 | 34.7 | 27 | 76 | 3 | No |
| Q9Y2L6 | FRMD4B | FERM domain–containing protein 4B/GRSP1 | 3.26e+8 | 554.8 | 32.0 | 28 | 230 | 3 | No |
| O95782 | AP2A1 | AP-2 complex subunit α-1 | 2.96e+8 | 202.3 | 45.4 | 42 | 102 | 3 | No |
| Q92556 | ELMO1 | Engulfment and cell motility protein 1 | 1.11e+8 | 131.2 | 37.4 | 28 | 61 | 3 | No |
| Q15027 | ACAP1 | Arf-GAP with coiled-coil, ANK repeat, and PH domain–containing protein 1/CENTB1 | 8.56e+7 | 133.8 | 40.7 | 25 | 55 | 3 | No |
| P42566 | EPS15 | Epidermal growth factor receptor substrate 15 | 8.44e+7 | 148.4 | 39.0 | 31 | 68 | 3 | No |
| Q9H7D0 | DOCK5 | Dedicator of cytokinesis protein 5 | 6.62e+7 | 123.7 | 25.8 | 46 | 69 | 3 | No |
| Q86UU1 | PHLDB1 | PH-like domain family B member 1/LL5α | 5.95e+7 | 106.9 | 23.0 | 32 | 55 | 3 | No |
| Q8WWN9 | IPCEF1 | Interactor protein for cytohesin exchange factors 1 | 5.82e+7 | 69.7 | 31.4 | 13 | 29 | 3 | No |
| Q96JJ3 | ELMO2 | Engulfment and cell motility protein 2 | 5.72e+7 | 24.9 | 16.4 | 11 | 17 | 3 | No |
| Q14185 | DOCK1 | Dedicator of cytokinesis protein 1 | 4.96e+7 | 75.8 | 18.7 | 33 | 44 | 3 | No |
| A0FGR8 | ESYT2 | Extended synaptotagmin-2 | 4.86e+7 | 116.0 | 30.3 | 23 | 54 | 3 | No |
| O75689 | ADAP1 | Arf-GAP with dual PH domain–containing protein 1/CENTA1 | 3.85e+7 | 42.1 | 32.9 | 13 | 21 | 3 | No |
| O15530 | PDPK1 | 3-phosphoinositide–dependent protein kinase 1/PDK1 | 2.69e+7 | 46.3 | 35.9 | 17 | 25 | 3 | Yes |
| P07900 | HSP90AA1 | Heat shock protein HSP 90-α | 2.26e+7 | 50.6 | 21.9 | 16 | 27 | 3 | Yes |
| Q8WWW8 | GAB3 | GRB2-associated–binding protein 3 | 1.65e+7 | 37.1 | 20.6 | 11 | 18 | 3 | No |
| Q9ULP9 | TBC1D24 | TBC1 domain family member 24 | 1.53e+7 | 18.8 | 17.4 | 9 | 11 | 3 | No |
| Q5TC63 | GRTP1 | Growth hormone–regulated TBC protein 1/TBC1D6 | 1.07e+7 | 14.2 | 17.7 | 5 | 8 | 3 | No |
| O75563 | SKAP2 | Src kinase–associated phosphoprotein 2/SCAP2 | 1.00e+7 | 7.3 | 13.3 | 4 | 4 | 3 | Yes |
| Q92608 | DOCK2 | Dedicator of cytokinesis protein 2 | 9.28e+6 | 6.5 | 2.7 | 5 | 5 | 2 | No |
| B0I1T2 | MYO1G | Unconventional myosin-1G | 8.90e+6 | 24.8 | 14.5 | 13 | 16 | 3 | No |
| Q15057 | ACAP2 | Arf-GAP with coiled-coil, ANK repeat, and PH domain–containing protein 2/CENTB2 | 8.77e+6 | 40.3 | 19.5 | 12 | 18 | 3 | No |
| Q9Y5X1 | SNX9 | Sorting nexin-9 | 8.48e+6 | 14.1 | 13.5 | 6 | 8 | 3 | No |
| O00159 | MYO1C | Unconventional myosin-Ic | 7.22e+6 | 15.6 | 9.3 | 8 | 9 | 3 | No |
| Q9UQC2 | GAB2 | GRB2-associated–binding protein 2 | 7.19e+6 | 34.0 | 20.5 | 11 | 19 | 3 | Yes |
| O75791 | GRAP2 | GRB2-related adapter protein 2/GADS | 4.40e+6 | 9.9 | 18.4 | 5 | 5 | 3 | Yes |
| P98082 | DAB2 | Disabled homolog 2 | 3.37e+6 | 10.1 | 8.5 | 5 | 5 | 3 | Yes |
| P31751 | AKT2 | RAC-β serine/threonine-protein kinase/PKB β | 2.95e+6 | 10.2 | 12.1 | 5 | 6 | 3 | Yes |
| Q9UPU7 | TBC1D2B | TBC1 domain family member 2B | 2.16e+6 | 15.6 | 10.8 | 9 | 10 | 3 | No |
| Q14155 | ARHGEF7 | Rho guanine nucleotide exchange factor 7 | 2.12e+6 | 6.8 | 5.8 | 5 | 5 | 3 | No |
| Q0JRZ9 | FCHO2 | F-BAR domain only protein 2 | 1.80e+6 | 10.8 | 7.9 | 5 | 7 | 3 | No |
| P31749 | AKT1 | RAC-α serine/threonine-protein kinase/PKB α | 1.77e+6 | 1.8 | 3.3 | 1 | 1 | 3 | Yes |
| Q12965 | MYO1E | Unconventional myosin-Ie | 1.73e+6 | 10.0 | 5.2 | 5 | 5 | 3 | No |
| Q96HS1 | PGAM5 | Serine/threonine-protein phosphatase PGAM5 | 1.27e+6 | 3.4 | 9.7 | 3 | 4 | 3 | No |
| Q8NF50 | DOCK8 | Dedicator of cytokinesis protein 8 | 1.14e+6 | 8.0 | 2.4 | 4 | 4 | 3 | No |
| Q8NEU8 | APPL2 | DCC-interacting protein 13-β/DIP13b/APPL2 | 1.13e+6 | 7.8 | 7.1 | 4 | 4 | 3 | No |
| Q13480 | GAB1 | GRB2-associated–binding protein 1 | 8.18e+5 | 5.4 | 3.3 | 2 | 3 | 3 | Yes |
| Q6P1M0 | SLC27A4 | Long-chain fatty acid transport protein 4 | 7.11e+5 | 10.3 | 10.2 | 6 | 6 | 3 | No |
| O43182 | ARHGAP6 | Rho GTPase-activating protein 6 | 6.48e+5 | 7.7 | 7.5 | 5 | 5 | 2 | No |
| Q96N67 | DOCK7 | Dedicator of cytokinesis protein 7 | 6.27e+5 | 6.7 | 1.9 | 3 | 3 | 3 | No |
| Q9Y2X7 | GIT1 | ARF GTPase-activating protein GIT1 | 5.83e+5 | 4.6 | 2.5 | 2 | 2 | 3 | Yes |
| P52306 | RAP1GDS1 | Rap1 GTPase-GDP dissociation stimulator 1/GDS1 | 2.76e+5 | 2.4 | 2.0 | 1 | 1 | 3 | No |
| Q96P48 | ARAP1 | Arf-GAP with Rho-GAP domain, ANK repeat, and PH domain–containing protein 1/CENTD2 | 2.38e+6 | 22.3 | 8.6 | 11 | 11 | 2 | No |
| O00160 | MYO1F | Unconventional myosin-If | 1.84e+6 | 13.4 | 8.3 | 8 | 8 | 2 | No |
| O95379 | TNFAIP8 | Tumor necrosis factor α–induced protein 8 | 1.12e+6 | 0.9 | 4.0 | 1 | 1 | 2 | No |
| Q8WVP5 | TNFAIP8L1 | Tumor necrosis factor α–induced protein 8–like protein 1/TIPE1 | 3.18e+5 | 4.1 | 14.3 | 2 | 2 | 2 | No |
| Q9BPZ7 | MAPKAP1 | Target of rapamycin complex 2 subunit MAPKAP1/SIN1 | 2.93e+5 | 0.9 | 2.1 | 1 | 1 | 2 | No |
| Accession No. . | Gene name . | Protein description . | Mean area . | Mean score . | Mean coverage (%) . | Mean peptides . | Mean PSM . | Times identified, n/3 . | Characterized in platelets? . |
|---|---|---|---|---|---|---|---|---|---|
| Q06187 | BTK | Tyrosine-protein kinase BTK | 8.56e+9 | 3109.1 | 75.4 | 61 | 1228 | 3 | Yes |
| Q14644 | RASA3 | Ras GTPase-activating protein 3 | 4.58e+9 | 2083.6 | 77.3 | 70 | 953 | 3 | Yes |
| Q9UN19 | DAPP1 | Dual adapter for phosphotyrosine and 3-phosphoinositides/Bam32 | 2.24e+9 | 777.1 | 80.2 | 24 | 325 | 3 | No |
| P42680 | TEC | Tyrosine-protein kinase Tec | 1.02e+9 | 522.7 | 67.2 | 47 | 253 | 3 | Yes |
| Q99418 | CYTH2 | Cytohesin-2/ARNO | 9.58e+8 | 384.3 | 53.0 | 23 | 165 | 3 | Yes |
| O43739 | CYTH3 | Cytohesin-3/GRP1 | 8.02e+8 | 132.1 | 32.4 | 14 | 61 | 3 | No |
| Q15438 | CYTH1 | Cytohesin-1/PSCD1 | 7.12e+8 | 171.5 | 39.9 | 17 | 79 | 3 | No |
| Q15283 | RASA2 | Ras GTPase-activating protein 2 | 4.49e+8 | 131.0 | 34.7 | 27 | 76 | 3 | No |
| Q9Y2L6 | FRMD4B | FERM domain–containing protein 4B/GRSP1 | 3.26e+8 | 554.8 | 32.0 | 28 | 230 | 3 | No |
| O95782 | AP2A1 | AP-2 complex subunit α-1 | 2.96e+8 | 202.3 | 45.4 | 42 | 102 | 3 | No |
| Q92556 | ELMO1 | Engulfment and cell motility protein 1 | 1.11e+8 | 131.2 | 37.4 | 28 | 61 | 3 | No |
| Q15027 | ACAP1 | Arf-GAP with coiled-coil, ANK repeat, and PH domain–containing protein 1/CENTB1 | 8.56e+7 | 133.8 | 40.7 | 25 | 55 | 3 | No |
| P42566 | EPS15 | Epidermal growth factor receptor substrate 15 | 8.44e+7 | 148.4 | 39.0 | 31 | 68 | 3 | No |
| Q9H7D0 | DOCK5 | Dedicator of cytokinesis protein 5 | 6.62e+7 | 123.7 | 25.8 | 46 | 69 | 3 | No |
| Q86UU1 | PHLDB1 | PH-like domain family B member 1/LL5α | 5.95e+7 | 106.9 | 23.0 | 32 | 55 | 3 | No |
| Q8WWN9 | IPCEF1 | Interactor protein for cytohesin exchange factors 1 | 5.82e+7 | 69.7 | 31.4 | 13 | 29 | 3 | No |
| Q96JJ3 | ELMO2 | Engulfment and cell motility protein 2 | 5.72e+7 | 24.9 | 16.4 | 11 | 17 | 3 | No |
| Q14185 | DOCK1 | Dedicator of cytokinesis protein 1 | 4.96e+7 | 75.8 | 18.7 | 33 | 44 | 3 | No |
| A0FGR8 | ESYT2 | Extended synaptotagmin-2 | 4.86e+7 | 116.0 | 30.3 | 23 | 54 | 3 | No |
| O75689 | ADAP1 | Arf-GAP with dual PH domain–containing protein 1/CENTA1 | 3.85e+7 | 42.1 | 32.9 | 13 | 21 | 3 | No |
| O15530 | PDPK1 | 3-phosphoinositide–dependent protein kinase 1/PDK1 | 2.69e+7 | 46.3 | 35.9 | 17 | 25 | 3 | Yes |
| P07900 | HSP90AA1 | Heat shock protein HSP 90-α | 2.26e+7 | 50.6 | 21.9 | 16 | 27 | 3 | Yes |
| Q8WWW8 | GAB3 | GRB2-associated–binding protein 3 | 1.65e+7 | 37.1 | 20.6 | 11 | 18 | 3 | No |
| Q9ULP9 | TBC1D24 | TBC1 domain family member 24 | 1.53e+7 | 18.8 | 17.4 | 9 | 11 | 3 | No |
| Q5TC63 | GRTP1 | Growth hormone–regulated TBC protein 1/TBC1D6 | 1.07e+7 | 14.2 | 17.7 | 5 | 8 | 3 | No |
| O75563 | SKAP2 | Src kinase–associated phosphoprotein 2/SCAP2 | 1.00e+7 | 7.3 | 13.3 | 4 | 4 | 3 | Yes |
| Q92608 | DOCK2 | Dedicator of cytokinesis protein 2 | 9.28e+6 | 6.5 | 2.7 | 5 | 5 | 2 | No |
| B0I1T2 | MYO1G | Unconventional myosin-1G | 8.90e+6 | 24.8 | 14.5 | 13 | 16 | 3 | No |
| Q15057 | ACAP2 | Arf-GAP with coiled-coil, ANK repeat, and PH domain–containing protein 2/CENTB2 | 8.77e+6 | 40.3 | 19.5 | 12 | 18 | 3 | No |
| Q9Y5X1 | SNX9 | Sorting nexin-9 | 8.48e+6 | 14.1 | 13.5 | 6 | 8 | 3 | No |
| O00159 | MYO1C | Unconventional myosin-Ic | 7.22e+6 | 15.6 | 9.3 | 8 | 9 | 3 | No |
| Q9UQC2 | GAB2 | GRB2-associated–binding protein 2 | 7.19e+6 | 34.0 | 20.5 | 11 | 19 | 3 | Yes |
| O75791 | GRAP2 | GRB2-related adapter protein 2/GADS | 4.40e+6 | 9.9 | 18.4 | 5 | 5 | 3 | Yes |
| P98082 | DAB2 | Disabled homolog 2 | 3.37e+6 | 10.1 | 8.5 | 5 | 5 | 3 | Yes |
| P31751 | AKT2 | RAC-β serine/threonine-protein kinase/PKB β | 2.95e+6 | 10.2 | 12.1 | 5 | 6 | 3 | Yes |
| Q9UPU7 | TBC1D2B | TBC1 domain family member 2B | 2.16e+6 | 15.6 | 10.8 | 9 | 10 | 3 | No |
| Q14155 | ARHGEF7 | Rho guanine nucleotide exchange factor 7 | 2.12e+6 | 6.8 | 5.8 | 5 | 5 | 3 | No |
| Q0JRZ9 | FCHO2 | F-BAR domain only protein 2 | 1.80e+6 | 10.8 | 7.9 | 5 | 7 | 3 | No |
| P31749 | AKT1 | RAC-α serine/threonine-protein kinase/PKB α | 1.77e+6 | 1.8 | 3.3 | 1 | 1 | 3 | Yes |
| Q12965 | MYO1E | Unconventional myosin-Ie | 1.73e+6 | 10.0 | 5.2 | 5 | 5 | 3 | No |
| Q96HS1 | PGAM5 | Serine/threonine-protein phosphatase PGAM5 | 1.27e+6 | 3.4 | 9.7 | 3 | 4 | 3 | No |
| Q8NF50 | DOCK8 | Dedicator of cytokinesis protein 8 | 1.14e+6 | 8.0 | 2.4 | 4 | 4 | 3 | No |
| Q8NEU8 | APPL2 | DCC-interacting protein 13-β/DIP13b/APPL2 | 1.13e+6 | 7.8 | 7.1 | 4 | 4 | 3 | No |
| Q13480 | GAB1 | GRB2-associated–binding protein 1 | 8.18e+5 | 5.4 | 3.3 | 2 | 3 | 3 | Yes |
| Q6P1M0 | SLC27A4 | Long-chain fatty acid transport protein 4 | 7.11e+5 | 10.3 | 10.2 | 6 | 6 | 3 | No |
| O43182 | ARHGAP6 | Rho GTPase-activating protein 6 | 6.48e+5 | 7.7 | 7.5 | 5 | 5 | 2 | No |
| Q96N67 | DOCK7 | Dedicator of cytokinesis protein 7 | 6.27e+5 | 6.7 | 1.9 | 3 | 3 | 3 | No |
| Q9Y2X7 | GIT1 | ARF GTPase-activating protein GIT1 | 5.83e+5 | 4.6 | 2.5 | 2 | 2 | 3 | Yes |
| P52306 | RAP1GDS1 | Rap1 GTPase-GDP dissociation stimulator 1/GDS1 | 2.76e+5 | 2.4 | 2.0 | 1 | 1 | 3 | No |
| Q96P48 | ARAP1 | Arf-GAP with Rho-GAP domain, ANK repeat, and PH domain–containing protein 1/CENTD2 | 2.38e+6 | 22.3 | 8.6 | 11 | 11 | 2 | No |
| O00160 | MYO1F | Unconventional myosin-If | 1.84e+6 | 13.4 | 8.3 | 8 | 8 | 2 | No |
| O95379 | TNFAIP8 | Tumor necrosis factor α–induced protein 8 | 1.12e+6 | 0.9 | 4.0 | 1 | 1 | 2 | No |
| Q8WVP5 | TNFAIP8L1 | Tumor necrosis factor α–induced protein 8–like protein 1/TIPE1 | 3.18e+5 | 4.1 | 14.3 | 2 | 2 | 2 | No |
| Q9BPZ7 | MAPKAP1 | Target of rapamycin complex 2 subunit MAPKAP1/SIN1 | 2.93e+5 | 0.9 | 2.1 | 1 | 1 | 2 | No |
Presented are mean values from LC-MS/MS analysis of protein capture on PtdIns(3,4,5)P3-coupled beads across 3 independent donors. Proteins are ranked based on mean abundance. Shown are a spectrum of proteins identified in the screen, with the full data set presented in supplemental Table 1. Assessment of previous functional characterization in human or mouse platelets was carried out by literature searching. The final 5 proteins have previously reported PtdIns(3,4,5)P3 affinity and were captured on the PtdIns(3,4,5)P3 beads with lower stringency (see “Results”).
Area, T3PQ; score, the total score of the protein, which is the sum of all peptide XCorr values for that protein above the specified score threshold; coverage, the percentage of the protein sequence covered by the identified peptides; peptides, the number of unique peptide sequences identified for the protein; PSM, the total number of identified peptide sequences for the protein.
Ontology analysis58,59 revealed the enrichment of molecular functions and biological processes associated with class I PI3K,17,20,21 including regulation of small GTPase function (eg, cytohesin 1-3, ADAP1, and DOCK and ELMO proteins), intracellular transport (eg, adenosine 5′-diphosphate[ADP]-ribosylation factor [ARF]-guanine nucleotide exchange factors [GEFs]/GTPase-activating proteins [GAPs], MYO1G, and SNX9), signaling adaptors (eg, DAPP1, GAB1-3, and SKAP2), and kinases or phosphatases involved in phosphorylation events (eg, BTK, TEC, PDK1, and AKT) (supplemental Figure 2A). Enrichment analysis also confirmed the abundance of proteins bearing PH domains in our data set in addition to further protein domains associated with PtdIns(3,4,5)P3 binding and general cell signaling/adaptor function (supplemental Figure 2B). Analysis of our data set through literature searching and high-confidence network analysis using STRING60 suggested the majority of proteins were directly captured on the PtdIns(3,4,5)P3 beads, while also confirming a number to be present by virtue of protein-protein interactions as part of the wider PtdIns(3,4,5)P3 signalosome (supplemental Figure 3). These include AP-2 complex components/partners (eg, AP-2µ1, EPS15, and Stonin-2, potentially via AP-2α161 ), cytoskeletal components (eg, tubulin-β1 and tubulin-α4A), and protein chaperones (eg, HSP90α and TCP-1 complex components). Notably, FRMD4B, which lacks a signature PtdIns(3,4,5)P3-binding motif but has been reported to associate with cytohesin family proteins, was captured in abundance.62,63 Having previously identified a role for cytohesin-2 in platelet secretion,64 we hypothesized that FRMD4B may have been captured through its interaction with this ARF-GEF. In confirmation of this, we revealed an agonist-insensitive association of these proteins in human platelets (supplemental Figure 4), identifying a novel constitutive complex in this cell type.
DAPP1 is regulated by phosphoinositides and tyrosine phosphorylation in platelets
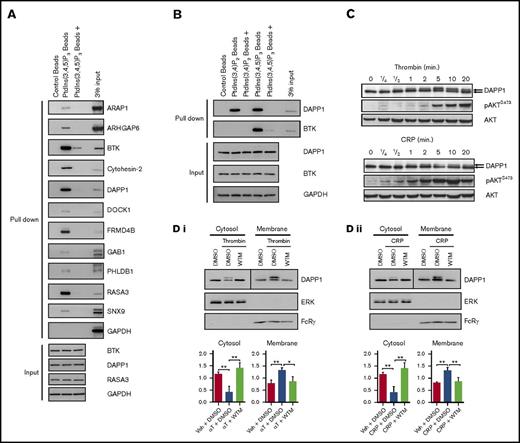
We verified the capture of a spectrum of proteins identified in our proteomics screen by western blotting (Figure 3A), including a number for which expression in human platelets has not previously been confirmed. By reference to the input material used for these experiments, this blotting also has the potential to provide more insight into the relative affinity of the proteins for PtdIns(3,4,5)P3 under our experimental conditions. In agreement with our proteomics data, proteins such as BTK, RASA3, and DAPP1 were captured in abundance, whereas others, such as ARAP1, were detectable in the bead eluates at lower levels, in line with previously reported affinity data.30,54,65,66 We also confirmed the capture of proteins utilizing liposomes comprising PtdIns(3,4,5)P3 in combination with the membrane glycerophospholipids, phosphatidylethanolamine and phosphatidylcholine, further validating our proteomics approach (supplemental Figure 5B).
Validation of the proteomics screen and characterization of DAPP1 as a PtdIns(3,4,5)P3- and PtdIns(3,4)P2-binding protein. (A) Human platelet lysates were incubated with control or PtdIns(3,4,5)P3-coupled beads for 90 minutes at 4°C, with (+) or without preincubation with competing free PtdIns(3,4,5)P3, before washing, elution, and western blotting analysis for a range of proteins identified in the proteomics screen. (B) Human platelet lysates were incubated with either PtdIns(3,4)P2- or PtdIns(3,4,5)P3-coupled beads as in panel A, and eluates were subjected to western blotting for DAPP1. (C) Human platelets were stimulated with 0.2 U/mL thrombin or 5µg/mL CRP for the indicated times, and lysates were blotted as indicated. The arrows indicate the molecular weight shift observed for DAPP1. (D) Human platelets stimulated with (i) thrombin (αT, 0.2 U/mL, 5 min) or (ii) CRP (5 μg/mL, 5 min) after 10 minutes of preincubation with dimethyl sulfoxide (DMSO) or 100 nM WTM were subjected to ultracentrifuge fractionation. Cytosol and membrane fractions were blotted as indicated. Histograms represent densitometry of blots from 3 independent experiments + standard error of the mean. Statistical analyses were performed by using 2-way analysis of variance with Bonferroni post-tests. *P < .05; **P < .001. Ctl, control; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Validation of the proteomics screen and characterization of DAPP1 as a PtdIns(3,4,5)P3- and PtdIns(3,4)P2-binding protein. (A) Human platelet lysates were incubated with control or PtdIns(3,4,5)P3-coupled beads for 90 minutes at 4°C, with (+) or without preincubation with competing free PtdIns(3,4,5)P3, before washing, elution, and western blotting analysis for a range of proteins identified in the proteomics screen. (B) Human platelet lysates were incubated with either PtdIns(3,4)P2- or PtdIns(3,4,5)P3-coupled beads as in panel A, and eluates were subjected to western blotting for DAPP1. (C) Human platelets were stimulated with 0.2 U/mL thrombin or 5µg/mL CRP for the indicated times, and lysates were blotted as indicated. The arrows indicate the molecular weight shift observed for DAPP1. (D) Human platelets stimulated with (i) thrombin (αT, 0.2 U/mL, 5 min) or (ii) CRP (5 μg/mL, 5 min) after 10 minutes of preincubation with dimethyl sulfoxide (DMSO) or 100 nM WTM were subjected to ultracentrifuge fractionation. Cytosol and membrane fractions were blotted as indicated. Histograms represent densitometry of blots from 3 independent experiments + standard error of the mean. Statistical analyses were performed by using 2-way analysis of variance with Bonferroni post-tests. *P < .05; **P < .001. Ctl, control; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Of the 3 most abundantly identified platelet PtdIns(3,4,5)P3-binding proteins in our proteomics screen, BTK has been shown previously to have a role in GPVI-mediated platelet activation,24 while we have recently revealed a role for RASA3 in integrin αIIbβ3 outside-in signaling.43 In contrast, the role of the PH and SH2 domain–containing DAPP1 in platelets remains unknown, despite important roles in multiple other cell types of hematopoietic origin.33,36,67-69 Some proteins are known to be regulated by multiple phosphoinositides, and PtdIns(3,4,5)P3 can be dephosphorylated by 5-phosphatases, such as SHIP1, to yield phosphatidylinositol 3,4-bisphosphate [PtdIns(3,4)P2],18,70 which can act in concert with PtdIns(3,4,5)P3 to regulate a subset of class I PI3K effectors, such as AKT.71 We confirmed that human platelet DAPP1 shows affinity for PtdIns(3,4)P2 in addition to PtdIns(3,4,5)P3 (Figure 3B), in agreement with the reported dual specificity of the DAPP1 PH domain.45 Additional proteins identified in our screen with the potential to be regulated by other phosphoinositides include PHLDB1,45 TAPP1/2,47 and SNX9.72,73
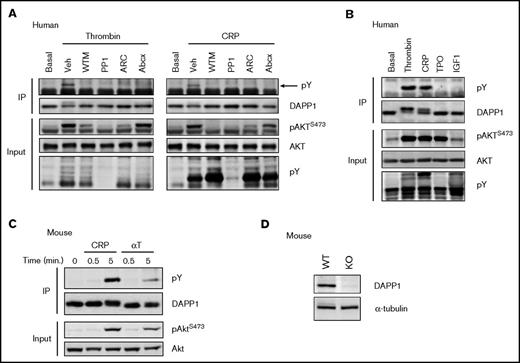
Upon stimulation of platelets with either thrombin or CRP, we observed a molecular weight shift in DAPP1 by western blotting (Figure 3C) and a PI3K-dependent increase in the proportion of DAPP1 present in the platelet membrane fraction (Figure 3D). The molecular weight shift is consistent with that observed for DAPP1 tyrosine phosphorylation in other cell types31,74,75 and was confirmed by western blotting of DAPP1 immunoprecipitates with the 4G10 antibody. This suggested that DAPP1 is recruited to membrane PtdIns(3,4,5)P3/PtdIns(3,4)P2 and tyrosine phosphorylated in activated platelets, and indeed the phosphorylation was dependent on both PI3K and SFK activity (Figure 4A). Furthermore, the P2Y12 inhibitor, AR-C66096, and the clinically used integrin αIIbβ3 antagonist, Abciximab, also inhibited DAPP1 tyrosine phosphorylation at this later time point (Figure 4A), revealing that ADP and integrin outside-in signaling contribute to DAPP1 phosphorylation in platelets, most likely through consolidation of PI3K activation.6 To investigate whether activation of PI3K alone is sufficient for DAPP1 tyrosine phosphorylation, we treated platelets with the primers, thrombopoietin and insulin-like growth factor-1, which signal to PI3K without triggering full platelet activation.3-5 Despite inducing a PI3K response, neither was able to induce DAPP1 tyrosine phosphorylation (Figure 4B), revealing differential integration of PI3K and SFK signaling downstream of platelet primers and full agonists.
DAPP1 is tyrosine phosphorylated in response to human and mouse platelet activation. (A) Western blotting of DAPP1 immunoprecipitates (IP) with the 4G10 antibody after thrombin (0.2 U/mL) or CRP (5 µg/mL) stimulation of human platelets for 5 minutes, after 10 minutes of preincubation with either Veh, WTM (100 nM), PP1 (10 µM), AR-C66096 (ARC, 1 µM) or Abciximab (Abcx, 1 μg/mL). The arrow indicates the position of tyrosine phosphorylated (pY) DAPP1. Corresponding whole-cell lysates were blotted for total AKT to confirm input loading and for AKT phosphorylation and global tyrosine phosphorylation to confirm the action of the agonists and inhibitors. (B) Western blotting of DAPP1 immunoprecipitates after treatment of human platelets for 5 minutes with the platelet primers, thrombopoietin (200 ng/mL), insulin-like growth factor-1 (200 nM), or the agonists described in panel A. (C) DAPP1 immunoprecipitates from mouse platelets stimulated for 5 minutes with CRP (10 μg/mL) or thrombin (αT, 0.5 U/mL) were blotted for 4G10 (pY) and DAPP1. (D) DAPP1 expression in wild-type (WT) and DAPP1−/− (KO) mouse platelets. Results are representative of at least 3 independent experiments.
DAPP1 is tyrosine phosphorylated in response to human and mouse platelet activation. (A) Western blotting of DAPP1 immunoprecipitates (IP) with the 4G10 antibody after thrombin (0.2 U/mL) or CRP (5 µg/mL) stimulation of human platelets for 5 minutes, after 10 minutes of preincubation with either Veh, WTM (100 nM), PP1 (10 µM), AR-C66096 (ARC, 1 µM) or Abciximab (Abcx, 1 μg/mL). The arrow indicates the position of tyrosine phosphorylated (pY) DAPP1. Corresponding whole-cell lysates were blotted for total AKT to confirm input loading and for AKT phosphorylation and global tyrosine phosphorylation to confirm the action of the agonists and inhibitors. (B) Western blotting of DAPP1 immunoprecipitates after treatment of human platelets for 5 minutes with the platelet primers, thrombopoietin (200 ng/mL), insulin-like growth factor-1 (200 nM), or the agonists described in panel A. (C) DAPP1 immunoprecipitates from mouse platelets stimulated for 5 minutes with CRP (10 μg/mL) or thrombin (αT, 0.5 U/mL) were blotted for 4G10 (pY) and DAPP1. (D) DAPP1 expression in wild-type (WT) and DAPP1−/− (KO) mouse platelets. Results are representative of at least 3 independent experiments.
DAPP1-deficient mice display increased platelet activation and thrombus formation
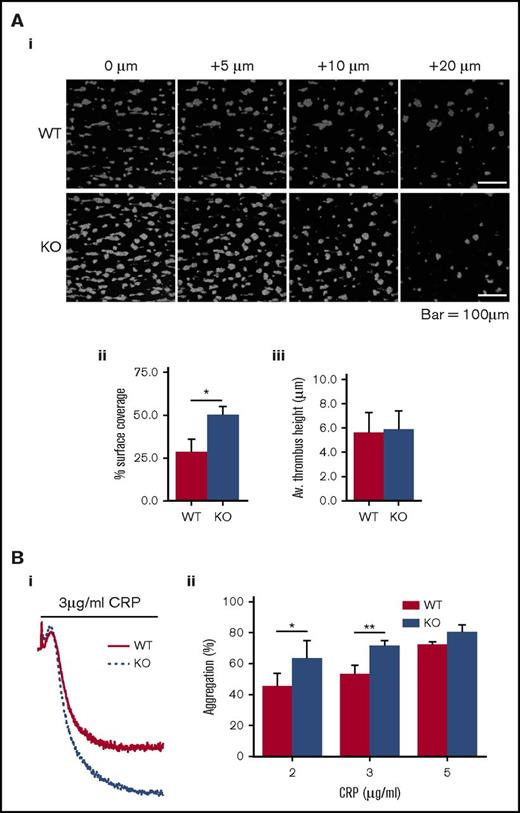
We established that mouse platelets express DAPP1, and that it undergoes thrombin- and CRP-induced tyrosine phosphorylation, as observed in human platelets (Figure 4C). We confirmed DAPP1 was absent from the platelets of DAPP1−/− mice and that these animals exhibit normal hematological parameters (Figure 4D; supplemental Table 2), and we set out to define the role of this protein in platelet activation and thrombus formation. Activation by collagen exposed after blood vessel injury is a critical early event in platelet activation,76 and so we initially performed in vitro thrombosis experiments flowing whole blood over a collagen-coated surface under noncoagulating conditions. Strikingly, we observed increased thrombus surface coverage with blood from DAPP1−/− mice compared with wild-type controls (Figure 5A). Given the essential role of GPVI in initial platelet activation in this context76 and the well-described importance of class I PI3K in this pathway,9,10,77,78 we investigated platelet function downstream of this collagen receptor by assessing CRP-induced platelet aggregation. In line with our observations for thrombus formation, GPVI-mediated aggregation was significantly enhanced in DAPP1−/− platelets (Figure 5B), suggesting that DAPP1 acts to restrain collagen-induced platelet activation.
Platelets from DAPP1−/−mice are hyperresponsive to collagen-driven functional responses. (A) Whole blood from WT or DAPP1−/− (KO) mice was loaded with DIOC6 and flowed over collagen (1000 s−1, 3 min) before fixation and imaging by confocal microscopy; (i) representative images of z-slices at indicated intervals relative to the thrombus base; (ii) histogram of surface coverage; (iii) histogram of thrombus height. n = 5 + standard error of the mean. (B) CRP-mediated platelet aggregation in WT and DAPP1−/− mouse platelets; (i) representative aggregation trace; (ii) histogram of the percentage of aggregation in response to a range of indicated CRP concentrations. n = 6 + standard error of the mean. Statistical analyses were performed by using Student t tests (A) or 2-way analysis of variance with Bonferroni post-tests (B). *P < .05; **P < .001.
Platelets from DAPP1−/−mice are hyperresponsive to collagen-driven functional responses. (A) Whole blood from WT or DAPP1−/− (KO) mice was loaded with DIOC6 and flowed over collagen (1000 s−1, 3 min) before fixation and imaging by confocal microscopy; (i) representative images of z-slices at indicated intervals relative to the thrombus base; (ii) histogram of surface coverage; (iii) histogram of thrombus height. n = 5 + standard error of the mean. (B) CRP-mediated platelet aggregation in WT and DAPP1−/− mouse platelets; (i) representative aggregation trace; (ii) histogram of the percentage of aggregation in response to a range of indicated CRP concentrations. n = 6 + standard error of the mean. Statistical analyses were performed by using Student t tests (A) or 2-way analysis of variance with Bonferroni post-tests (B). *P < .05; **P < .001.
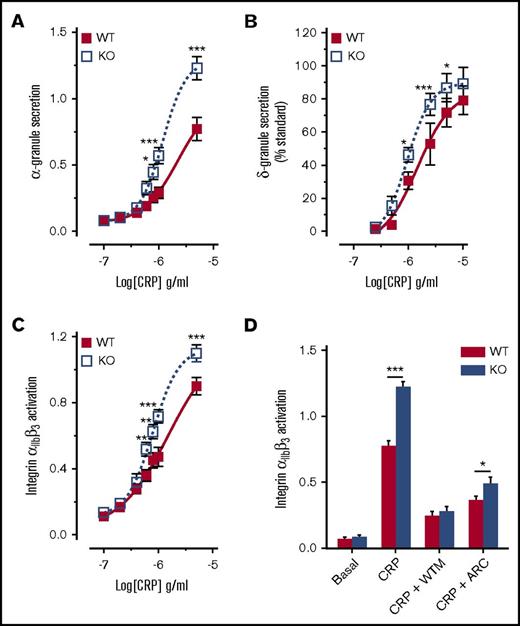
A similar negative regulatory role for DAPP1 has been reported in mast cells, where it acts to limit FcεRI-induced granule release.67 To determine whether the elevated functional responses of DAPP1−/− platelets might correspond to a similar enhancement of granule release, we conducted fluorescence-activated cell sorting (FACS) analysis and luminometry to assess platelet secretion. Compared with wild-type controls, DAPP1−/− platelets displayed significantly enhanced P-selectin exposure and ATP release in response to CRP (Figure 6A-B), confirming enhanced α and δ granule secretion, respectively. We also observed a significant increase in GPVI-mediated platelet integrin αIIbβ3 activation in the absence of DAPP1 (Figure 6C), which was blocked in the presence of the PI3K inhibitor, wortmannin (Figure 6D). Although DAPP1−/− mast cells were reported to display changes in calcium mobilization and phosphorylation of AKT and ERK,67 these parameters were not significantly altered in CRP-treated DAPP1−/− platelets (supplemental Figure 6A-B). Similarly, we observed no significant changes to the CRP-induced phosphorylation status of proximal GPVI signaling components (supplemental Figure 7), although we did detect small but significant changes in the surface expression of GPVI and GP1bα on DAPP1−/− platelets (supplemental Figure 8). In contrast to GPVI-mediated platelet function, we saw a modest decrease in PAR4-AP–induced platelet aggregation in DAPP1−/− mice, whereas α and δ granule secretion and αIIbβ3 integrin activation were unchanged in response to this agonist (supplemental Figure 9). Taken together, these results reveal that the class I PI3K effector, DAPP1, restrains platelet function downstream of GPVI, thus identifying a novel negative regulator of collagen-driven platelet activation and thrombus formation.
Platelets from DAPP1−/−mice are hyperresponsive to GPVI stimulation. (A) FACS analysis of P-selectin exposure on WT and DAPP1−/− (KO) mouse platelets in response to CRP (10 min). n = 9 + standard error of the mean. (B) ATP release by WT and DAPP1−/− mouse platelets in response to CRP. Data are expressed as peak ATP release as a percentage of a standard. n = 5 + standard error of the mean. (C) FACS analysis of integrin αIIbβ3 activation on WT and DAPP1−/− mouse platelets in response to CRP (10 min). n = 9 + standard error of the mean. (D) Integrin αIIbβ3 activation on WT and DAPP1−/− mouse platelets in response to CRP (5 µg/mL, 10 min) after preincubation for 10 minutes with either vehicle, WTM (100 nM), or AR-C66069 (ARC, 1 μM). FACS fluorescence intensities (A, C-D) were normalized to the response to maximal agonist concentration averaged per mouse pair (WT and DAPP1 KO) to preserve sample variance at the maximal concentration. Statistical analyses were performed by using 2-way analysis of variance with Bonferroni post-tests. *P < .05; **P < .001; ***P < .0001.
Platelets from DAPP1−/−mice are hyperresponsive to GPVI stimulation. (A) FACS analysis of P-selectin exposure on WT and DAPP1−/− (KO) mouse platelets in response to CRP (10 min). n = 9 + standard error of the mean. (B) ATP release by WT and DAPP1−/− mouse platelets in response to CRP. Data are expressed as peak ATP release as a percentage of a standard. n = 5 + standard error of the mean. (C) FACS analysis of integrin αIIbβ3 activation on WT and DAPP1−/− mouse platelets in response to CRP (10 min). n = 9 + standard error of the mean. (D) Integrin αIIbβ3 activation on WT and DAPP1−/− mouse platelets in response to CRP (5 µg/mL, 10 min) after preincubation for 10 minutes with either vehicle, WTM (100 nM), or AR-C66069 (ARC, 1 μM). FACS fluorescence intensities (A, C-D) were normalized to the response to maximal agonist concentration averaged per mouse pair (WT and DAPP1 KO) to preserve sample variance at the maximal concentration. Statistical analyses were performed by using 2-way analysis of variance with Bonferroni post-tests. *P < .05; **P < .001; ***P < .0001.
Discussion
Class I PI3K is an important signaling hub in human and mouse platelets, with key roles in platelet priming, activation, and stable thrombus formation, thought to be orchestrated primarily through the action of its catalytic product, PtdIns(3,4,5)P3. Direct measurements of PtdIns(3,4,5)P3 in platelets have traditionally involved the use of radiolabeled precursors and HPLC,9,79,80 yet this approach is laborious and provides no information about the fatty acyl content of this phosphoinositide.42 Although PtdIns(3,4,5)P3 has been previously measured in platelets by lipidomic MS, this measurement has lacked sensitivity, detecting this phosphoinositide only in response to a high concentration of thrombin.81 Our lipidomic analysis revealed a basal, wortmannin-sensitive level of PtdIns(3,4,5)P3 in human platelets in addition to robust thrombin- and CRP-driven PtdIns(3,4,5)P3 generation. We focused on the stearoyl/arachidonoyl species of PtdIns(3,4,5)P3, generally the most abundant molecular species in primary mammalian tissues,41,42,82,83 but we were also able to measure the less abundant C38:3 form. Conventional effectors associate with PtdIns(3,4,5)P3 primarily via its phosphorylated headgroup, and a comparison of PtdIns(3,4,5)P3-binding proteins purified by our approach and others19,20,48,84,85 suggests that most are unlikely to show absolute species specificity. However, it is possible that PtdIns(3,4,5)P3 molecular species identity contributes to the fine tuning of binding protein localization and/or function in vivo.
Although a number of PtdIns(3,4,5)P3-binding proteins have been isolated from other cell types,19,47,84-86 knowledge of these PI3K effectors in platelets prior to this study was poor, with attention primarily focused on AKT. Although our data demonstrate that AKT phosphorylation serves as a good readout for PtdIns(3,4,5)P3 generation in thrombin- and CRP-activated platelets (Figure 1), AKT is not responsible for driving all class I PI3K–regulated processes in cells and may be disconnected from class I PI3K in some contexts.87-89 Indeed, although AKT isoforms have important roles in platelets,6 other PtdIns(3,4,5)P3-binding proteins mediate key aspects of platelet biology, including platelet-specific functions.23-25,43 This highlights the need to define the individual repertoires of class I PI3K effectors in highly specialized cell types, and our study reveals for the first time the extensive network of PtdIns(3,4,5)P3-binding proteins in platelets. Strikingly, although well-characterized PtdIns(3,4,5)P3 effectors, such as AKT, BTK, and PDK1, have been shown to play roles in platelet activation and thrombus formation,6,23,24,39 the majority of proteins identified in our screen have undergone no characterization in platelets thus far, and this work provides the first insight into their function in these cells. Furthermore, we identified proteins that have received limited characterization in any tissue type, including IPCEF1, GRTP1, and TBC1D2B.
The abundance of identified proteins involved in small GTPase regulation reflects a range of GEFs and GAPs in our signalosome, a number of which have previously reported affinity for PtdIns(3,4,5)P3.46,48,51,54,66 The targets of these proteins include RHO and ARF family small GTPases, several of which play key roles in platelet function.90-92 Notably, current understanding of how these small GTPases are controlled by GEFs/GAPs in platelets is poor, yet the latter are often crucial for coupling PI3K to the regulation of cell function in other cell types. Indeed, based on work in other cells,19,53,91,93,94 several of the PtdIns(3,4,5)P3-binding GEFs/GAPs identified are likely to regulate cytoskeletal dynamics and protein trafficking in platelets in concert with other proteins identified, such as myosin 1G50 and SNX9.95 The identification of TNFAIP8 family proteins and SIN1 may permit important new insights into events such as phosphoinositide trafficking55 and mTORC2 activation,56 respectively, in platelets, whereas proteins such as FRMD4B, IPCEF1, and SKAP2 are likely to hold roles as signaling adaptors in this cell type.
The individual characterization of proteins identified in this screen by our laboratory and others will allow determination of their functional roles in the class I PI3K/PtdIns(3,4,5)P3 pathway in platelets and other cell types. Indeed, in recent work, we have identified a key role for the PtdIns(3,4,5)P3-binding RAS/RAP-GAP, RASA3, in αIIbβ3 outside-in signaling,43 and in this article, we define an important role for the SH2 and PH domain–containing adaptor protein, DAPP1, in GPVI signaling. DAPP1 has previously been shown to play both positive and negative regulatory roles, dependent on the cellular and stimulatory context.33 In B cells, DAPP1 deficiency results in impaired B-cell receptor signaling, leading to a proliferation defect in vitro,36 impaired antigen responses,68 and increased apoptosis in late-stage germinal centers in vivo,69 while DAPP1-deficient T cells display impaired in vitro proliferation and interleukin 4 (IL-4) production.96 Conversely, DAPP1-deficient B cells are hyperresponsive to IL-4 or CD40 stimulation,69 while splenic cells from trypanosome-infected DAPP1-deficient mice display increased production of the proinflammatory cytokines, IFN-γ, TNF-α, and IL-6.97 Similarly, mast cells lacking DAPP1 display enhanced degranulation and IL-6 production.67
Our work reveals that DAPP1 is regulated by PI3K and SFKs in platelets in a manner analogous to other cell types31,74,75 and acts to restrain GPVI-mediated platelet function in a negative regulatory role comparable to that observed in mast cells downstream of the high-affinity immunoglobulin E receptor, FcεRI.67 The specificity of the DAPP1 phenotype to GPVI signaling in platelets is in line with the critical importance of class I PI3K function to this pathway,9,77,78 and the ability of DAPP1 to hold a specific positive or negative role, depending on the signaling context, appears to be a common feature of such adaptor proteins in blood cells.33 Our data demonstrate that the DAPP1−/− platelet phenotype is not due to overt changes in proximal GPVI signaling, suggesting that DAPP1 may contribute to platelet function and secretion further downstream or in a parallel pathway. Previous work has demonstrated the tyrosine phosphorylation of DAPP1 to be important for its function, including roles in receptor internalization and endosomal sorting.29,33,98,99 Interestingly, we did observe small reductions in cell surface receptor expression. Although unlikely to fully explain our observed phenotype, this data may support a role for DAPP1 in platelet receptor trafficking, which could be facilitated by its dual affinity for PtdIns(3,4,5)P3 and PtdIns(3,4)P2.71 The identification of DAPP1 interacting partners and the advent of novel lipidomic approaches permitting more acute quantitative assessment of the PtdIns(3,4,5)P3/PtdIns(3,4)P2 balance in primary cells will allow greater understanding of how DAPP1 regulates cell function.
In conclusion, we have carried out an in-depth analysis of the platelet PtdIns(3,4,5)P3 signalosome by MS, yielding new insights into the molecular identity of PtdIns(3,4,5)P3 in human platelets and providing the first detailed analysis of the PtdIns(3,4,5)P3 interactome of these cells. The latter provides an important resource for future studies, facilitating work to further dissect how class I PI3Ks mediate diverse and important aspects of cell function. Indeed, it has allowed us to identify DAPP1 as a new PI3K-regulated player in GPVI-mediated platelet activation and an important negative regulator of collagen-mediated thrombus formation. Furthermore, given the challenges and limitations of directly targeting the proximal, ubiquitous, and multifunctional class I PI3Ks for therapeutic means,13,14 the characterization of downstream effectors may provide novel targets100 for the regulation of specific aspects of platelet signaling and function.
The online version of this article contains a data supplement.
Acknowledgments
The authors thank the blood donors of the School of Physiology, Pharmacology and Neuroscience (University of Bristol) and Elizabeth Aitken for mouse genotyping. The authors wish to acknowledge the assistance of Andrew Herman and the University of Bristol Faculty of Biomedical Sciences Flow Cytometry Facility. The authors also thank Asha Bayliss for helpful discussions and critical reading of the manuscript.
This work was supported by the British Heart Foundation (grants PG/12/79/29884, PG/13/11/30016, and PG/14/3/30565).
Authorship
Contribution: T.N.D. designed and performed research, collected and analyzed data, and wrote the manuscript; J.L.H. designed and performed research, collected and analyzed data, and cowrote the manuscript; K.J.H. performed proteomics analysis and edited the manuscript; K.E.A. performed lipidomics analysis and edited the manuscript; L.R.S. and P.T.H. provided lipidomics analysis, reagents, and contributed to discussion; A.J.M. provided reagents and contributed to discussion; S.F.M. performed research, contributed to discussion, and edited the manuscript; and I.H. designed and supervised research, contributed to discussion, and cowrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ingeborg Hers, School of Physiology, Pharmacology and Neuroscience, Biomedical Sciences Building, University of Bristol, Bristol BS8 1TD, United Kingdom; e-mail: i.hers@bristol.ac.uk.
References
Author notes
T.N.D. and J.L.H. contributed equally to this study.
The mass spectrometry data reported in this article have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (identifier PXD003777).


![Figure 2. Experimental workflow for proteomics experiments. (A) Pure platelet preparations were obtained from whole blood by using a multistep approach (see “Materials and methods”), and lysates were subjected to affinity capture of PtdIns(3,4,5)P3-binding proteins by using PtdIns(3,4,5)P3-coupled beads. Eluate sample complexity was reduced by sodium dodecyl sulfate-polyacrylamide gel electrophoresis separation of proteins, followed by trypsin digest, nano-HPLC, and MS analysis. Data were subject to stringent filtering and analysis using a range of bioinformatics tools. (B) Human platelet lysates were incubated with control (Ctl) or PtdIns(3,4,5)P3 [PIP3, or after preincubation with competing free PtdIns(3,4,5)P3 (PIP3+)]-coupled beads for 90 minutes at 4°C before washing, elution, and analysis by SYPRO Ruby gel staining. (C) Experiments conducted as described in panel B were subjected to liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis. Histograms demonstrate validation of the proteomics approach by quantitative analysis of known PtdIns(3,4,5)P3-binding proteins with the T3PQ method across the independent donors. Bars represent the mean of 3 independent donors + standard error of the mean. ACD, acid citrate dextrose; Ctl, control; HT, HEPES-Tyrode’s buffer; PRP, platelet-rich plasma.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/1/14/10.1182_bloodadvances.2017005173/3/m_advances005173f2.jpeg?Expires=1769223425&Signature=h8A--saIaccLDPQbYaq-PVlQrkce9~vGXHiie6~aTOZF33j-qsz2yg2wJBX0vhGrD8tlH~zwl292Pvfi~61FVmkhYXA2n3jQjqPwBNbOyIeD6qgkWTHpPlV7VS3iRkW97w~GNHbDm2IIv4mL1NbWHIYSTXtHTZNyHim5oTTm~ZgWMz3EgK5RZ4I1eX-7upHEglswAKm4ra36xWhozR1fODdN6f7z0B27cYld60F6jqyv5zlrjaEqaHxgW5UF1~DQdhzOdAIjtz3qw7IE6UC7ZHs0Miz89Yvd1D4noHc8LzeUf2lAtoOOI2M5eziY-1xQkV87HUZ2cqDEbDQw9BI5Pw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)