Key Points
IBR+VEN combination was designed to overcome resistance to IBR, but CLL and MCL samples ex vivo show resistance even to this combination.
Microenvironmental agonists (IL-10, soluble CD40L, CpG-ODN) generate resistance via NF-κB–dependent expression of Mcl-1/Bcl-xL/survivin.
Abstract
De novo resistance and rapid recurrence often characterize responses of B-cell malignancies to ibrutinib (IBR), indicating a need to develop drug combinations that block compensatory survival signaling and give deeper, more durable responses. To identify such combinations, we previously performed a combinatorial drug screen and identified the Bcl-2 inhibitor venetoclax (VEN) as a promising partner for combination with IBR in mantle cell lymphoma (MCL). We have opened a multi-institutional clinical trial to test this combination. However, analysis of primary samples from patients with MCL as well as chronic lymphocytic leukemia (CLL) revealed unexpected heterogeneous de novo resistance even to the IBR+VEN combination. In the current study, we demonstrate that resistance to the combination can be generated by microenvironmental agonists: interleukin-10 (IL-10), CD40L and, most potently, cytosine guanine dinucleotide–oligodeoxynucleotides (CpG-ODNs), which is a surrogate for unmethylated DNA and a specific agonist for Toll-like receptor 9 (TLR9) signaling. Incubation with these agonists caused robust activation of NF-κB signaling, especially alternative NF-κB, which led to enhanced expression of the antiapoptotic proteins Mcl-1, Bcl-xL, and survivin, thus decreasing dependence on Bcl-2. Inhibitors of NF-κB signaling blocked overexpression of these antiapoptotic proteins and overcame resistance. Inhibitors of Mcl-1, Bcl-xL, or survivin also overcame this resistance, and showed synergistic benefit with the IBR+VEN combination. We conclude that microenvironmental factors, particularly the TLR9 agonist, can generate de novo resistance to the IBR+VEN combination in CLL and MCL cells. This signaling pathway presents targets for overcoming drug resistance induced by extrinsic microenvironmental factors in diverse B-cell malignancies.
Introduction
The B-cell malignancies chronic lymphocytic leukemia (CLL) and mantle cell lymphoma (MCL), although distinct clinically and biologically, share common developmental themes including dependence on B-cell receptor (BCR) signaling. The advent of ibrutinib (IBR), an inhibitor of BCR signaling, represented a major advance in treatment of these malignancies.
However, responses to IBR are generally incomplete, and rarely if ever curative.1-4 The overall response rate in relapsed or refractory MCL patients is ∼70%, but with only a ∼20% complete response and median progression-free survival of ∼15 months.5,6 In CLL, overall response rate is 80% to 90% in previously treated or untreated cases, but complete response is <25% and progression-free survival at 30 months is 69% to 96%.1,2 Although responses are often durable, continuous IBR treatment is recommended as deep remission or minimal residual disease–negative status is rarely achieved. Patients failing IBR after initial response typically experience rapid disease progression and short survival.4,7-10 Thus, de novo resistance, incomplete responses, and rapid recurrence constitute disappointing outcomes for many patients.4,11,12
Relapse following initial response is sometimes due to selection for mutations in Bruton tyrosine kinase (BTK) or its substrate Phospholipase Cγ2 in CLL and MCL or RelA in MCL.13-17 However, we hypothesize that de novo resistance, partial responses, and rapid recurrence can be due in part to nongenetic, phenotypic adaptations including compensatory signaling, kinome rewiring, and other adaptive responses.18-22 Drug combinations cotargeting these phenotypic adaptations may induce broader, deeper, more durable responses.
We previously reported a combinatorial drug screen with IBR23 as an anchor drug to identify pathways that could be cotargeted with BTK. The combination of IBR and venetoclax (VEN)/ABT-199, an inhibitor of antiapoptotic protein Bcl-2,24 resulted in synergistically enhanced cytotoxicity in MCL lines. Based on these findings, we initiated a phase 1/1b trial assessing safety and efficacy of IBR+VEN in relapsed/refractory MCL (NCT02419560). Phase 1/2 studies of IBR+VEN in MCL (NCT02471391), CLL (NCT02756897), and follicular lymphoma (FL) (NCT02956382) patients have been initiated by other groups.
However, with patient samples treated ex vivo, we observed striking variability in sensitivity of CLL and MCL cells even to IBR+VEN. Although this variability could reflect genetic heterogeneity in these patients,25 we hypothesized that the de novo resistance could also be generated by variability in the in vivo environment of cancer cells.26-32 Neoplastic B cells receive growth/survival signals through interaction with components of the microenvironment such as stromal cells, cytokines, antigens, and circulating microbial/cellular DNA16,26-29,33-42 ; the latter 3 are found in lymph node (LN) and bone marrow (BM) as well as in circulation.
Here, we report that the combination of IBR+VEN generates synergistic cytotoxicity in CLL and MCL patient peripheral blood mononuclear cells (PBMCs) treated ex vivo, but with highly variable levels of de novo resistance. The cytotoxicity of IBR+VEN was greatly diminished in cells preincubated with specific agonists (soluble CD40L [sCD40L], interleukin-10 [IL-10], or cytosine guanine dinucleotide–oligodeoxynucleotides [CpG-ODNs]), characteristic of the in vivo environment. The mixture of all 3 agonists (“agonist mix”) generated almost complete resistance and induced the expression of CD69 and Ki67, characteristic of activated B cells. Preincubation with the agonist mix induced activation of the NF-κB pathway and NF-κB–dependent upregulation of the expression of antiapoptotic proteins Mcl-1, Bcl-xL, and survivin, thus reducing cell dependence on the VEN target, Bcl-2.24 Inhibition of the NF-κB pathway or of the upregulated antiapoptotic proteins overcame this resistance. These findings reveal the potential for targeting microenvironmentally activated signaling pathways to overcome adaptive and de novo resistance in CLL and MCL.
Materials and methods
Reagents, patient samples, and cell lines
Details of reagents are available in supplemental Table 1. After local institutional review board approval and in accordance with the Declaration of Helsinki, informed consent was obtained from patients. Blood from patients with circulating CLL or MCL cells was processed into PBMCs, which were used fresh, or frozen in liquid nitrogen in 90% fetal calf serum and 10% dimethyl sulfoxide (DMSO). Residual red blood cells were removed with red blood cell lysis solution (5 PRIME, Inc). For ex vivo drug screening of patient samples (Figure 1; supplemental Figures 2, 5, and 6), PBMCs were cultured in RPMI containing 10% fetal calf serum, and low levels of IL-2 (1.54 ng/mL) and CpG-ODN (0.38 μg/mL) to reduce spontaneous apoptosis as described.43,44 Cell line details are included in supplemental Materials and methods.
IBR and VEN are variably cytotoxic in CLL and MCL patient PBMCs. (A) Representative images showing cleaved PARP in CD5+/CD19+ cells in CLL (patient [Pt] 12) and MCL (Pt 22) patient PBMCs following ex vivo treatment with IBR (0.1 μM), VEN (25 nM), or IBR+VEN. The concentrations of IBR and VEN that showed 20% to 30% cytotoxicity as single agents (measured by alamarBlue) in a CLL patient PBMC were selected for ex vivo treatment of patient samples (data not shown). (B-D) PBMCs from CLL (N = 24) and MCL (N = 8) patients were treated with IBR, VEN, or IBR+VEN. The cleaved PARP in CD5+/CD19+ cells in CLL samples showing synergy (B) or no synergy (C) with IBR+VEN and in MCL samples (D) is shown. Comparable results were also obtained by analyzing the dead cell staining of CD5+/CD19+ cells in CLL and MCL patient PBMCs: samples that exhibited synergistically high cleaved PARP also showed synergistic increases in dead cell staining following ex vivo treatment with IBR and VEN (supplemental Figure 2A-C). The time to achieve maximal synergistic cytotoxicity (∼6-18 hours) was chosen as the time point used for subsequent data analysis. Synergy was calculated using the Bliss model of independence, which generates interaction scores even where 1 drug has no effect. Data are expressed as means ± SD. Statistical significance was determined by ANOVA. *P < .05, **P < .01, ****P < .0001. NS, not significant; SSC, side scatter.
IBR and VEN are variably cytotoxic in CLL and MCL patient PBMCs. (A) Representative images showing cleaved PARP in CD5+/CD19+ cells in CLL (patient [Pt] 12) and MCL (Pt 22) patient PBMCs following ex vivo treatment with IBR (0.1 μM), VEN (25 nM), or IBR+VEN. The concentrations of IBR and VEN that showed 20% to 30% cytotoxicity as single agents (measured by alamarBlue) in a CLL patient PBMC were selected for ex vivo treatment of patient samples (data not shown). (B-D) PBMCs from CLL (N = 24) and MCL (N = 8) patients were treated with IBR, VEN, or IBR+VEN. The cleaved PARP in CD5+/CD19+ cells in CLL samples showing synergy (B) or no synergy (C) with IBR+VEN and in MCL samples (D) is shown. Comparable results were also obtained by analyzing the dead cell staining of CD5+/CD19+ cells in CLL and MCL patient PBMCs: samples that exhibited synergistically high cleaved PARP also showed synergistic increases in dead cell staining following ex vivo treatment with IBR and VEN (supplemental Figure 2A-C). The time to achieve maximal synergistic cytotoxicity (∼6-18 hours) was chosen as the time point used for subsequent data analysis. Synergy was calculated using the Bliss model of independence, which generates interaction scores even where 1 drug has no effect. Data are expressed as means ± SD. Statistical significance was determined by ANOVA. *P < .05, **P < .01, ****P < .0001. NS, not significant; SSC, side scatter.
Flow cytometry and ImageStream analysis
Cell samples for phosphoprotein analysis were treated with 1 μM pervanadate, 5 nM calyculin A and washed with ice-cold phosphate-buffered saline containing pervanadate and calyculin A. Cells were stained with Live/Dead viability stain for 30 minutes. Cells for phosphoprotein and antiapoptotic or proapoptotic protein analyses by flow cytometry or ImageStream were fixed with 1.6% paraformaldehyde after Live/Dead staining, prior to surface staining. Cells were blocked with mouse immunoglobulin G (IgG) and stained for surface proteins with respective antibodies for 1 to 2 hours. Subsequently, intracellular proteins were stained after fixing in paraformaldehyde and permeabilizing with BD/eBioscience Perm/Wash buffer. Antibodies used for staining are shown in supplemental Table 1. Unconjugated antibodies were detected using phycoerythrin-conjugated anti-rabbit IgG antibody. The nucleus in fixed and permeabilized cells was stained with 7-aminoactinomycin D (7AAD). Flow cytometry and ImageStream analysis were performed using a BD FACSCalibur/LSR-Fortessa/Attune-NxT cytometer and Amnis ImageStreamX Mark II, respectively.
Cell viability assay
Cell viability (Figure 5; supplemental Figures 11 and 14) was determined by alamarBlue according to the manufacturer’s protocol (Invitrogen). Assay details are in supplemental Materials and methods.
Statistical analysis and synergy calculation
Results are presented as means ± standard deviation (SD). Statistical significance was evaluated by analysis of variance (ANOVA) or the Student t test using GraphPad Prism, version 6.0; P < .05 was considered significant. Synergy was calculated using the Bliss model of independence.45-47
Results
Leukemic B cells in CLL and MCL patient PBMCs treated ex vivo display heterogeneous de novo resistance to IBR and VEN
Cytotoxicity of IBR, VEN, and IBR+VEN was evaluated following ex vivo treatment of CLL (N = 24), MCL (N = 8) patient PBMCs, and the MCL line Mino. Apoptotic leukemic cell death was analyzed by poly ADP ribose polymerase (PARP) cleavage in CD5+/CD19+ cells by flow cytometry (supplemental Figure 1). IBR+VEN treatment caused a synergistic increase in apoptosis in 16 of 24 CLL samples as well as 7 of 8 MCL samples and in Mino cells (Figure 1) compared with single drug treatment. However, the kinetics as well as extent of cytotoxicity varied widely among patient samples, with peak synergistic cytotoxicity occurring as early as 3 hours to as late as 72 hours. Resistance to ex vivo IBR, VEN, and IBR+VEN did not correlate with previous therapy or clinical characteristics of patients (supplemental Tables 2 and 3), consistent with a previous report on CLL.48
The cytotoxicity of IBR alone was very modest and variable (supplemental Figure 3), even though 0.1 μM IBR reduced BTK phosphorylation to nearly undetectable levels (supplemental Figure 4). However, single-agent VEN treatment resulted in substantial PARP cleavage or dead cell staining, as well as enhanced cytotoxicity with IBR in a large subset of CLL or MCL patient samples (Figure 1; supplemental Figure 2). Only modest increases in cleaved PARP were detected in T lymphocytes (CD3+/CD5+ cells) from CLL (supplemental Figure 5) or MCL (supplemental Figure 6) patient PBMCs following ex vivo treatment. These data demonstrate that IBR+VEN induces synergistic cytotoxicity in leukemic B cells in a cohort of CLL and MCL patient PBMCs and a MCL line through apoptosis, with variable levels of de novo resistance.
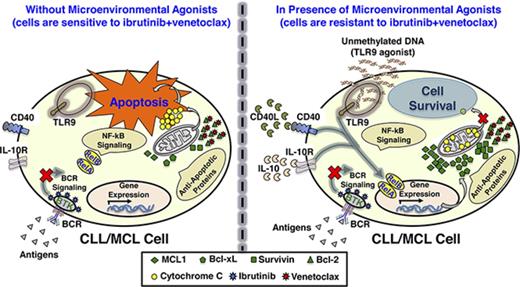
Extrinsic microenvironmental agonists generate resistance to IBR+VEN
We observed that CLL cells expressing the activation marker CD69 were less sensitive to IBR+VEN ex vivo (supplemental Figure 7), and hypothesized that extrinsic microenvironmental factors, which are capable of activating B cells,26-29,33-40 might induce drug resistance. To test this, 3 representative CLL patient PBMCs were preincubated with sCD40L, CXCL13, IL-10, IL-2, CpG-ODN, B-cell–activating factor (BAFF), or IgM, or cocultured with a follicular dendritic cell line (HK) or a bone marrow stromal cell line (HS-5)26-29,33-40 and treated with IBR+VEN as described in Figure 2A. The cleaved PARP and dead cell staining in CD5+/CD19+ cells were analyzed by flow cytometry. sCD40L, IL-10, and CpG-ODN were the most effective as single agents at inhibiting drug-induced cytotoxicity (Figure 2B; supplemental Figure 8A). The combination of these agonists (sCD40L+IL-10+CpG-ODN; “agonist mix”) resulted in robust resistance to IBR+VEN in most CLL (N = 22) and MCL (N = 8) patient PBMCs and the Mino MCL line (Figure 2C-D; supplemental Figure 8B-C). Preincubation with the agonist mix caused significantly increased expression of Ki67 and CD69 in CLL and MCL cells (Figure 2E; supplemental Figure 9), indicating induction of an activation/proliferation phenotype.16,42 High IBR and VEN concentrations were unable to completely overcome the agonist mix–induced resistance (Figure 2F), suggesting that target desensitization (eg, BTK hyperactivation or Bcl-2 overexpression) was unlikely to be responsible for drug resistance. These data show that extrinsic factors characteristic of the in vivo environment of cancer cells can generate resistance even to IBR+VEN in CLL and MCL cells.
Extrinsic microenvironmental agonists generate resistance to IBR+VEN. (A) Diagram showing the patient PBMCs cocultured with stromal cells or preincubated with soluble agonists for 12 hours. Drugs as well as a second dose of agonists were added at 12 hours and cultured for an additional 12 hours, and cleaved PARP or dead cell staining in CD5+/CD19+ cells in CLL or MCL was analyzed by flow cytometry. (B) PBMCs from CLL patients (Pts 01, 03, and 12) were cocultured with HS-5 or HK cell line at a 10:1 ratio or preincubated with sCD40L (2 μg/mL), IL-10 (0.015 μg/mL), CpG-ODN (1.5 μg/mL), CXCL13 (0.5 μg/mL), IL-2 (1.54 ng/mL), BAFF (0.25 μg/mL), or IgM (25 μg/mL) and treated with IBR (0.1 μM) + VEN (25 nM) or DMSO as well as second doses of agonists for 12 hours as shown in panel A. Cleaved PARP in CD5+/CD19+ cells was analyzed by flow cytometry. (C-D) PBMCs from 22 CLL (Pts 1-3, 6-7, 12, 14, 21, 25-28, 33, 35, 39, 43, 46, 48, 51-52, 56, and 59) and 8 MCL (Pts 22-23, 38, 45, 49, 55, 60, and 66) patients and the Mino MCL cell line were preincubated with agonist mix (sCD40L [2 μg/mL] + IL-10 [0.015 μg/mL] + CpG-ODN [1.5 μg/mL]) for 12 hours. Samples were then incubated with IBR (0.1 μM) + VEN (25 nM) or DMSO for an additional 12 hours as well as a second dose of agonist mix. Cleaved PARP in CD5+/CD19+ cells in CLL samples (C) and MCL samples/Mino cell line (D) were analyzed by flow cytometry. (E) Ki67+/CD5+/CD19+ cells in CLL and MCL patient PBMCs (Pts 06, 35, 39, 43, and 45) preincubated with or without 2 doses of agonist mix at 12-hour intervals were analyzed by flow cytometry. (F) PBMCs from CLL patients (Pt 01 and 25) were preincubated with agonist mix for 12 hours and treated with increasing concentrations of IBR (0.1, 1, and 10 μM) and VEN (6.25, 12, 25, 100, 400 nM) for 12 hours along with a second dose of agonist mix. The cleaved PARP in CD5+/CD19+ cells was analyzed by flow cytometry. Data are expressed as means ± SD. The statistical significance was determined by ANOVA. *P < .05, ***P < .001, ****P < .0001. FACS, fluorescence-activated cell sorter.
Extrinsic microenvironmental agonists generate resistance to IBR+VEN. (A) Diagram showing the patient PBMCs cocultured with stromal cells or preincubated with soluble agonists for 12 hours. Drugs as well as a second dose of agonists were added at 12 hours and cultured for an additional 12 hours, and cleaved PARP or dead cell staining in CD5+/CD19+ cells in CLL or MCL was analyzed by flow cytometry. (B) PBMCs from CLL patients (Pts 01, 03, and 12) were cocultured with HS-5 or HK cell line at a 10:1 ratio or preincubated with sCD40L (2 μg/mL), IL-10 (0.015 μg/mL), CpG-ODN (1.5 μg/mL), CXCL13 (0.5 μg/mL), IL-2 (1.54 ng/mL), BAFF (0.25 μg/mL), or IgM (25 μg/mL) and treated with IBR (0.1 μM) + VEN (25 nM) or DMSO as well as second doses of agonists for 12 hours as shown in panel A. Cleaved PARP in CD5+/CD19+ cells was analyzed by flow cytometry. (C-D) PBMCs from 22 CLL (Pts 1-3, 6-7, 12, 14, 21, 25-28, 33, 35, 39, 43, 46, 48, 51-52, 56, and 59) and 8 MCL (Pts 22-23, 38, 45, 49, 55, 60, and 66) patients and the Mino MCL cell line were preincubated with agonist mix (sCD40L [2 μg/mL] + IL-10 [0.015 μg/mL] + CpG-ODN [1.5 μg/mL]) for 12 hours. Samples were then incubated with IBR (0.1 μM) + VEN (25 nM) or DMSO for an additional 12 hours as well as a second dose of agonist mix. Cleaved PARP in CD5+/CD19+ cells in CLL samples (C) and MCL samples/Mino cell line (D) were analyzed by flow cytometry. (E) Ki67+/CD5+/CD19+ cells in CLL and MCL patient PBMCs (Pts 06, 35, 39, 43, and 45) preincubated with or without 2 doses of agonist mix at 12-hour intervals were analyzed by flow cytometry. (F) PBMCs from CLL patients (Pt 01 and 25) were preincubated with agonist mix for 12 hours and treated with increasing concentrations of IBR (0.1, 1, and 10 μM) and VEN (6.25, 12, 25, 100, 400 nM) for 12 hours along with a second dose of agonist mix. The cleaved PARP in CD5+/CD19+ cells was analyzed by flow cytometry. Data are expressed as means ± SD. The statistical significance was determined by ANOVA. *P < .05, ***P < .001, ****P < .0001. FACS, fluorescence-activated cell sorter.
NF-κB, PI3K, PKC, and MAPK pathways are activated in CLL and MCL cells preincubated with agonist mix
To understand mechanisms of resistance to IBR and VEN, we used reverse-phase protein arrays (RPPAs) to monitor signaling responses to IBR and VEN in JVM-2 and Z-138 MCL lines that were used in the drug screen that identified the IBR+VEN combination.23 We saw downregulation of phosphorylation on kinases belonging to phosphatidylinositol 3-kinase (PI3K)-AKT, MAPK, JAK-STAT, and NOTCH signaling in IBR-treated cells, cleavage of caspases and PARP in the VEN-treated samples, and dramatically enhanced cleavage of caspases and PARP in the cells treated with the IBR+VEN combination (supplemental Figure 10). Interestingly, cells recovered after longer exposures to IBR or IBR+VEN showed significantly upregulated phosphorylation on RelA/p65 S536 in the NF-κB pathway, protein kinase C (PKC) substrate myristoylated alanine-rich PKC substrate (MARCKS) S152.156, and cAMP response element binding (CREB) S133, suggesting that these phosphorylations may be markers of drug resistance. Cotargeting the NF-κB pathway with inhibitors of IκB kinase α/β (IKKα/β; BMS-345541) or proteasome (bortezomib and carfilzomib) but not the pan-PKC inhibitor sotrastaurin provided exceptional synergistic cytotoxicity with IBR+VEN in these cell lines (supplemental Figure 11A-D), at pharmacologically relevant concentrations49 (Figure 3E-I; supplemental Figure 14D). These data suggest that activation of NF-κB signaling could generate resistance to IBR+VEN.
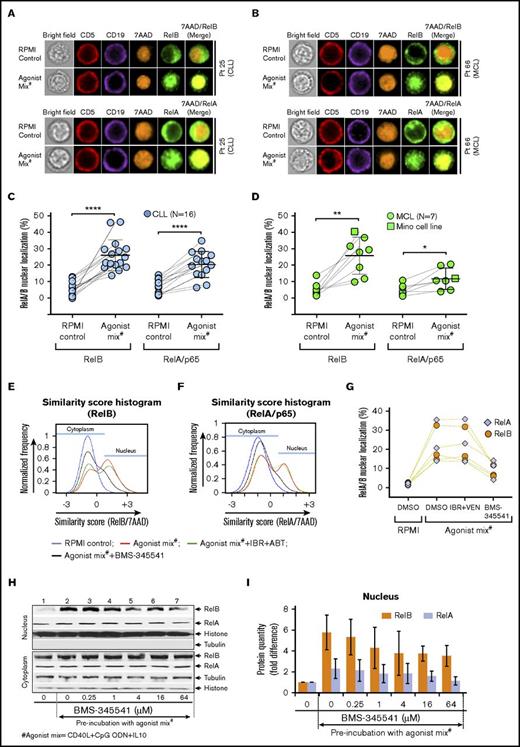
NF-κB pathway is activated in CLL and MCL cells preincubated with agonist mix. CLL and MCL patient PBMCs were preincubated with agonist mix for 12 hours. At the end of 12 hours, a second dose of agonist mix was added and incubated for another 6 hours. Cytoplasmic and nuclear localization of RelB or RelA in a minimum of 500 live/CD5+/CD19+ cells in CLL and MCL samples was analyzed in ImageStream using IDEAS ImageStream analysis software. (A-B) Representative images showing CD5 (red), CD19 (violet), RelB (green), 7AAD (orange), and 7AAD/RelB in CLL (A; top panel) and MCL (B; top panel) patient PBMCs or CD5 (red), CD19 (violet), RelA (green), 7AAD (orange), and 7AAD/RelA in CLL (A; bottom panel) and MCL (B; bottom panel) patient PBMCs. Original magnification ×60. (C-D) Percentage of nuclear localization of RelB and RelA in 16 CLL patient PBMCs (Pts 01-03, 7, 12, 21, 25-28, 33, 35, 43, 46, 51, and 52) (C) and 7 MCL patient PBMCs (Pts 23, 38, 45, 49, 55, 60, and 66) and Mino cells (D) preincubated with or without agonist mix. (E-G) CLL patient PBMCs (N = 3) were incubated with agonist mix for 12 hours and treated with BMS-345541 (16 μM) or IBR (0.1 μM) + VEN (25 nM) as well as a second dose of agonist mix for another 6 hours. The representative image of similarity score histograms showing shift in RelB (E) or RelA (F) colocalization with 7AAD. Percentage of nuclear localization of RelB and RelA in different treatment groups (G). (H-I) Mino cell line preincubated with agonist mix for 12 hours and treated with BMS-345541 (0-64 μM) and a second dose of agonist mix for another 6 hours. RelA, RelB, histone (H3), and tubulin proteins in cytoplasmic and nuclear fractions were analyzed by western blot (H). Bar graphs showing relative band densities of RelB and RelA proteins in the nuclear fractions (I). The data were average from 2 independent experiments. RelB and RelA in nuclear fractions were normalized to histone. Data are expressed as means ± SD. The statistical significance was determined by the paired Student t test. *P < .05, **P < .01, ****P < .0001.
NF-κB pathway is activated in CLL and MCL cells preincubated with agonist mix. CLL and MCL patient PBMCs were preincubated with agonist mix for 12 hours. At the end of 12 hours, a second dose of agonist mix was added and incubated for another 6 hours. Cytoplasmic and nuclear localization of RelB or RelA in a minimum of 500 live/CD5+/CD19+ cells in CLL and MCL samples was analyzed in ImageStream using IDEAS ImageStream analysis software. (A-B) Representative images showing CD5 (red), CD19 (violet), RelB (green), 7AAD (orange), and 7AAD/RelB in CLL (A; top panel) and MCL (B; top panel) patient PBMCs or CD5 (red), CD19 (violet), RelA (green), 7AAD (orange), and 7AAD/RelA in CLL (A; bottom panel) and MCL (B; bottom panel) patient PBMCs. Original magnification ×60. (C-D) Percentage of nuclear localization of RelB and RelA in 16 CLL patient PBMCs (Pts 01-03, 7, 12, 21, 25-28, 33, 35, 43, 46, 51, and 52) (C) and 7 MCL patient PBMCs (Pts 23, 38, 45, 49, 55, 60, and 66) and Mino cells (D) preincubated with or without agonist mix. (E-G) CLL patient PBMCs (N = 3) were incubated with agonist mix for 12 hours and treated with BMS-345541 (16 μM) or IBR (0.1 μM) + VEN (25 nM) as well as a second dose of agonist mix for another 6 hours. The representative image of similarity score histograms showing shift in RelB (E) or RelA (F) colocalization with 7AAD. Percentage of nuclear localization of RelB and RelA in different treatment groups (G). (H-I) Mino cell line preincubated with agonist mix for 12 hours and treated with BMS-345541 (0-64 μM) and a second dose of agonist mix for another 6 hours. RelA, RelB, histone (H3), and tubulin proteins in cytoplasmic and nuclear fractions were analyzed by western blot (H). Bar graphs showing relative band densities of RelB and RelA proteins in the nuclear fractions (I). The data were average from 2 independent experiments. RelB and RelA in nuclear fractions were normalized to histone. Data are expressed as means ± SD. The statistical significance was determined by the paired Student t test. *P < .05, **P < .01, ****P < .0001.
We then examined CLL and MCL patient samples incubated with agonist mix for activation of NF-κB, PI3K-AKT, MAPK, PKC, and JAK-STAT pathways, which are downstream of Toll-like receptor 9 (TLR9), CD40R, and IL-10R.50-54 Classical and alternative NF-κB pathway activation was determined with ImageStream, which scored nuclear translocation of NF-κB transcription factors RelA/p65 and RelB at the level of single cells. Nuclear localization of RelA and RelB in CLL and MCL cells was enhanced following preincubation with agonist mix (Figure 3A-B). Nuclear translocation of RelB was invariably enhanced in all tested CLL patient PBMCs (N = 16) (Figure 3C) as well as MCL patient PBMCs (N = 7) and Mino cells (Figure 3D) following preincubation with agonist mix. NF-κB pathway activation was also assessed by detecting RelA and RelB in cytoplasmic and nuclear fractions by subcellular fractionation and western blot analysis. Approximately 5.8 ± 1.7 fold and 2.3 ± 0.9 fold increases of RelB and RelA proteins, respectively, were detected in the nuclear fraction of agonist mix preincubated Mino cells (Figure 3H-I lanes 1-2), consistent with ImageStream data. Nuclear translocation of RelA was less pronounced compared with RelB in MCL cells (P < .01), and a similar trend was also evident in CLL cells that did not reach statistical significance (Figure 3C-D,H-I). Nuclear translocation of RelA and RelB was effectively blocked in the presence of IKKα/β inhibitor BMS-345541 in agonist mix preincubated CLL and MCL cells (Figure 3E-I).
Activation of PI3K-AKT, MAPK, and PKC pathways was evaluated by analysis of activating phosphorylations on AKT, extracellular signal-regulated kinase 1/2 (ERK1/2), or MARCKS, respectively. Preincubation with agonist mix resulted in modest increases in phosphorylation on AKT S473, ERK1/2 T202/Y204, and MARCKS S152/156 (supplemental Figure 12A). Similarly, western blots showed 2.3 ± 1.26 fold increase in AKT phosphorylation on S473 (supplemental Figure 12B-C lanes 1-2), and this phosphorylation was abolished by 2.7 μM idelalisib (supplemental Figure 12B-C lanes 3-7) comparable to published results.55 The agonist mix treatment induced a 2.4-fold increase in ERK1/2 phosphorylation on T183/Y185 (supplemental Figure 12D-E lanes 1-2), and this phosphorylation was abolished by the MEK1/2 inhibitor PD0325901 (supplemental Figure 12D-E lanes 3-4). JAK-STAT pathway activation was analyzed by measuring STAT3 protein nuclear translocation in 3 CLL patient PBMCs by ImageStream, and no significant change was evident after agonist mix treatment (data not shown). Collectively, the data demonstrate robust and consistent activation of alternative NF-κB and relatively less pronounced activation of classical NF-κB signaling in CLL and MCL cells preincubated with agonist mix. Agonist mix preincubation also induced moderate activation of PI3K-AKT, MAPK, and PKC signaling.
Expression of antiapoptotic proteins is upregulated in CLL and MCL cells preincubated with agonist mix
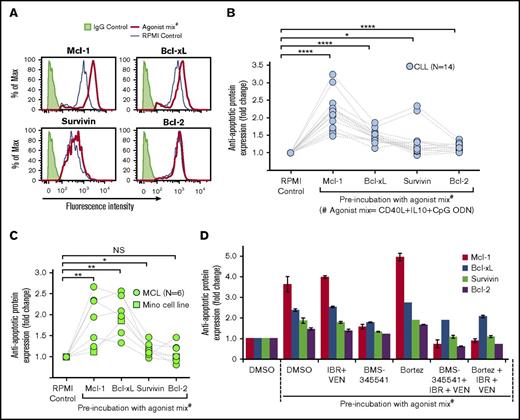
Antiapoptotic and proapoptotic protein expression was analyzed by flow cytometry in CLL and MCL patient PBMCs and Mino cells to assess their role in resistance to IBR+VEN. Significant increases occurred in expression of antiapoptotic proteins Mcl-1 and Bcl-xL, and a similar trend was seen in the expression of survivin, but none showed substantially increased Bcl-2 expression in CLL samples (Figure 4A-B). Similar results were obtained for MCL patient samples/Mino cells (Figure 4C). By contrast, agonist mix incubation only slightly altered the expression of proapoptotic proteins Noxa, Puma, BIM, and BAD (supplemental Figure 13). Overexpression of antiapoptotic proteins was not altered in the presence of IBR+VEN but was blocked by the addition of NF-κB signaling inhibitor BMS-345541, as well as by bortezomib in the presence of IBR+VEN (Figure 4D). These data show potential NF-κB–dependent overexpression of Mcl-1, Bcl-xL, and/or survivin in CLL and MCL cells incubated with agonist mix.
Antiapoptotic protein expression was upregulated in CLL and MCL cells preincubated with agonist mix. (A-C) Expression of antiapoptotic proteins Mcl-1, Bcl-xL, survivin, and Bcl-2 in 14 CLL (Pts 1, 3, 7, 12, 19, 21, 25-27, 48, 51-52, 54, and 59) and 6 MCL (Pts 22, 45, 55, 58, 60, and 66) patient PBMCs and Mino cell line incubated with 2 doses of agonist mix at 12-hour intervals was analyzed by flow cytometry. Representative histograms showing the expression of Mcl-1, Bcl-xL, survivin, and Bcl-2 in a CLL patient sample (Pt 03) treated with agonist mix (A). Antiapoptotic protein expression in CLL patient PBMCs (B) and MCL patient PBMCs/Mino cell line (C) was determined by calculating the geometric mean of fluorescence intensity (GM-FLI) of proteins in CD5+/CD19+ cells. Agonist mix treatment significantly upregulated the expression of Mcl-1 (2.14 ± 0.51 fold in CLL; 1.874 ± 0.604 fold in MCL) and Bcl-xL (1.47 ± 0.18 fold in CLL; 1.98 ± 0.44 fold in MCL), and survivin (1.3 ± 0.43 fold in CLL; 1.173 ± 0.164 fold in MCL) but not Bcl-2 (1.16 ± 0.11 fold in CLL; 1.091 ± 0.221 fold in MCL) in CLL and MCL cells. (D) A CLL patient PBMC (Pt 25) treated with agonist mix for 12 hours was incubated with BMS-345541 (BMS) (16 μM) or bortezomib (Bortez) (0.064 μM) for 9 hours along with a second dose of agonist mix, with or without IBR (0.1 μM) + VEN (25 nM) and antiapoptotic protein expression was analyzed by calculating GM-FLI of proteins in CD5+/CD19+ cells using flow cytometry. The data were normalized to DMSO control. Data are expressed as means ± SD. The statistical significance was determined by Student t test. *P < .05, **P < .01, ****P < .0001.
Antiapoptotic protein expression was upregulated in CLL and MCL cells preincubated with agonist mix. (A-C) Expression of antiapoptotic proteins Mcl-1, Bcl-xL, survivin, and Bcl-2 in 14 CLL (Pts 1, 3, 7, 12, 19, 21, 25-27, 48, 51-52, 54, and 59) and 6 MCL (Pts 22, 45, 55, 58, 60, and 66) patient PBMCs and Mino cell line incubated with 2 doses of agonist mix at 12-hour intervals was analyzed by flow cytometry. Representative histograms showing the expression of Mcl-1, Bcl-xL, survivin, and Bcl-2 in a CLL patient sample (Pt 03) treated with agonist mix (A). Antiapoptotic protein expression in CLL patient PBMCs (B) and MCL patient PBMCs/Mino cell line (C) was determined by calculating the geometric mean of fluorescence intensity (GM-FLI) of proteins in CD5+/CD19+ cells. Agonist mix treatment significantly upregulated the expression of Mcl-1 (2.14 ± 0.51 fold in CLL; 1.874 ± 0.604 fold in MCL) and Bcl-xL (1.47 ± 0.18 fold in CLL; 1.98 ± 0.44 fold in MCL), and survivin (1.3 ± 0.43 fold in CLL; 1.173 ± 0.164 fold in MCL) but not Bcl-2 (1.16 ± 0.11 fold in CLL; 1.091 ± 0.221 fold in MCL) in CLL and MCL cells. (D) A CLL patient PBMC (Pt 25) treated with agonist mix for 12 hours was incubated with BMS-345541 (BMS) (16 μM) or bortezomib (Bortez) (0.064 μM) for 9 hours along with a second dose of agonist mix, with or without IBR (0.1 μM) + VEN (25 nM) and antiapoptotic protein expression was analyzed by calculating GM-FLI of proteins in CD5+/CD19+ cells using flow cytometry. The data were normalized to DMSO control. Data are expressed as means ± SD. The statistical significance was determined by Student t test. *P < .05, **P < .01, ****P < .0001.
Inhibitors of NF-κB signaling or antiapoptotic proteins overcome drug resistance induced by the agonist mix in CLL and MCL cells
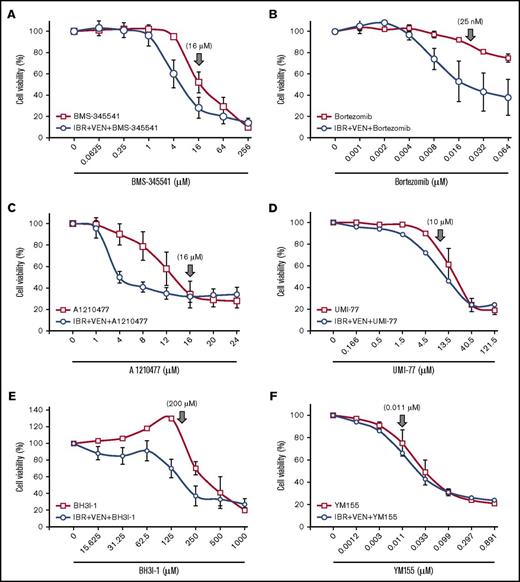
To evaluate whether agonist-induced resistance generated new therapeutic vulnerabilities, we tested whether inhibitors of the signaling pathways (supplemental Table 4) activated by agonist mix could block resistance in CLL and MCL patient samples. BMS-345541 and proteasome inhibitors bortezomib and carfilzomib, which are expected to inhibit NF-κB signaling,49 showed robust cytotoxicity in the presence and absence of IBR+VEN (Figure 5A-B; supplemental Figure 14A), at pharmacologically relevant concentrations49 (Figure 3E-I). Conversely, idelalisib and sotrastaurin showed cytotoxicity only at very high concentrations (concentrations giving greater than 90% inhibition of target kinase) (supplemental Figures 12B-C and 14B-D), likely due to off-target effects, indicating that activation of PI3K and PKC signaling by the agonists was not essential for cell survival. Similarly, although the MAPK pathway was activated in cells preincubated with agonist mix (supplemental Figure 12D-E), the MEK1/2 inhibitor PD0325901 did not alter cytotoxicity in the presence/absence of IBR+VEN (data not shown).
Inhibition of NF-κB pathway or antiapoptotic proteins overcomes IBR+VEN drug resistance in CLL and MCL cells preincubated with agonist mix. (A-F) The NF-κB pathway inhibitor BMS-345541 (A) and Bortez (B) and antiapoptotic protein inhibitor UMI-77 (Mcl-1 inhibitor) (D), BH3I-1 (Bcl-xL inhibitor) (E), and YM155 (survivin) (F) were tested in increasing concentrations with or without IBR (0.1 μM) + VEN (25 nM) in CLL (Pts 01, 03, and 12) and a MCL (Pt 45) patient PBMC preincubated with agonist mix as shown in Figure 2A. The antiapoptotic protein inhibitor A1210477 (C) was similarly tested in 4 CLL patient samples (Pts 03, 33, 35, and 46). Cell viability was determined using an alamarBlue assay. The data were normalized to DMSO treatment control or IBR+VEN in triple drug treatment. The data presented as means ± SD. ↓ indicates drug concentration at which target kinase phosphorylation/signaling pathway/apoptotic protein was maximally inhibited, based on published values.56-59
Inhibition of NF-κB pathway or antiapoptotic proteins overcomes IBR+VEN drug resistance in CLL and MCL cells preincubated with agonist mix. (A-F) The NF-κB pathway inhibitor BMS-345541 (A) and Bortez (B) and antiapoptotic protein inhibitor UMI-77 (Mcl-1 inhibitor) (D), BH3I-1 (Bcl-xL inhibitor) (E), and YM155 (survivin) (F) were tested in increasing concentrations with or without IBR (0.1 μM) + VEN (25 nM) in CLL (Pts 01, 03, and 12) and a MCL (Pt 45) patient PBMC preincubated with agonist mix as shown in Figure 2A. The antiapoptotic protein inhibitor A1210477 (C) was similarly tested in 4 CLL patient samples (Pts 03, 33, 35, and 46). Cell viability was determined using an alamarBlue assay. The data were normalized to DMSO treatment control or IBR+VEN in triple drug treatment. The data presented as means ± SD. ↓ indicates drug concentration at which target kinase phosphorylation/signaling pathway/apoptotic protein was maximally inhibited, based on published values.56-59
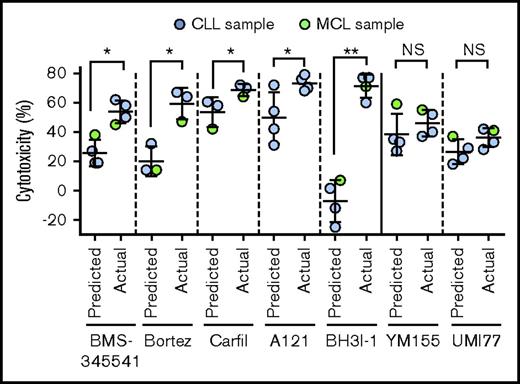
Inhibitors of Mcl-1 (A1210477 and UMI-77), Bcl-xL (BH3I-1 and ABT-737), and survivin (YM155) also induced robust cytotoxicity with or without IBR+VEN at pharmacologically relevant concentrations56-59 (Figure 5C-F; supplemental Figure 14E). The cytotoxicity 50% inhibitory concentration values for inhibitors are shown in supplemental Table 5. The NF-κB inhibitors BMS-345541, bortezomib, carfilzomib, as well as antiapoptotic protein inhibitors A1210477 and BH3I-1 showed exceptional synergistic cytotoxicity with IBR+VEN (Figure 6). These data show that inhibition of the NF-κB pathway or of the antiapoptotic proteins (Mcl-1, Bcl-xL, or survivin) leads to robust cytotoxicity in CLL and MCL cells preincubated with agonists, and each of these inhibitors is synergistically cytotoxic with IBR+VEN.
Synergistic interaction of inhibitors of NF-κB or antiapoptotic proteins with IBR+VEN combination. Synergistic interaction of inhibitors of NF-κB (BMS-345541, Bortez, and Carfilzomib) and antiapoptotic proteins (A1210477(A121), BH3I-1, UMI-77, and YM155) was determined by calculating Bliss predicted and actual cytotoxicity of individual inhibitors with IBR (0.1 μM) + VEN (25 nM) combination. The statistical significance was determined by ANOVA. *P < .05, **P < .01.
Synergistic interaction of inhibitors of NF-κB or antiapoptotic proteins with IBR+VEN combination. Synergistic interaction of inhibitors of NF-κB (BMS-345541, Bortez, and Carfilzomib) and antiapoptotic proteins (A1210477(A121), BH3I-1, UMI-77, and YM155) was determined by calculating Bliss predicted and actual cytotoxicity of individual inhibitors with IBR (0.1 μM) + VEN (25 nM) combination. The statistical significance was determined by ANOVA. *P < .05, **P < .01.
Discussion
De novo resistance to IBR, VEN, and the combination in primary CLL and MCL cells
We previously demonstrated that IBR+VEN was synergistically cytotoxic in MCL lines.23 Here, we demonstrated synergy using primary patient tumor cells from leukemic B-cell malignancies (CLL and MCL), but surprisingly observed ex vivo resistance even to this combination in many cases (Figure 1B-D; supplemental Figure 2). Similar observations were also made in CLL by Cervantes-Gomez et al.48 Extensive heterogeneity in basal phosphorylation of various signaling proteins was recently reported in patient-derived CLL/MCL/FL cells.60 Because resistance was greatest in cells expressing the activation marker CD69, characteristic of LN-resident cells, we hypothesized that cellular activation by microenvironmental interactions could generate this ex vivo resistance (supplemental Figure 7), which was also suggested by recent studies.26-32
Agonists characteristic of the in vivo environment protect CLL and MCL cells from cytotoxicity of IBR-VEN combination
Interaction of CLL/MCL cells with microenvironmental agonists can induce B-cell activation16,42 and drug resistance.26-32 Therefore, we tested the ability of various cells and agonists that are expected to be found in the cancer cell environment26-29,33-40 to protect leukemic cells from cytotoxicity of IBR+VEN. Pretreatment of CLL patient PBMCs with several of these agonists was protective (Figure 2B; supplemental Figure 8A), and the mixture of the 3 most potent (sCD40L+IL-10+CpG-ODN) generated almost complete resistance to IBR+VEN in most CLL and MCL patient samples (Figure 2C-D; supplemental Figure 8B-C). Even increasing the concentration of IBR and VEN was not enough to overcome agonist mix–induced resistance (Figure 2F), indicating that resistance was occurring not by changing the sensitivity of targets to their cognate drugs but by altering a component downstream from or parallel to the target.
Although these malignancies represent distinct diseases, with differing genetics, disease course, and sensitivity to either ibrutinib or venetoclax, they all responded to common microenvironmental agonists. These findings point to a common biology that may be targeted for therapeutic intervention in these diverse diseases. In pilot studies, we recently found that cells from FL patients also display comparable heterogeneity in sensitivity to IBR+VEN and responsiveness to the agonist mix with upregulation of NF-κB signaling and antiapoptotic proteins Mcl-1, Bcl-xL, and survivin expression (supplemental Figures 15-18). Thus, we propose that the mechanisms of drug resistance identified here draw on the developmental history of B-cell maturation, and may characterize many B-cell malignancies.
As this manuscript was under revision, Chiron et al reported that incubation of MCL PBMCs with cells expressing CD40L enhanced drug resistance of MCL cells, and sensitized the MCL cells to the protective effects of various cytokines, including IL-10, which we also identified.29 In addition, our study identified the protective role of TLR9 agonist CpG-ODN, which was not investigated by Chiron et al.29 They also reported protective effects of BAFF, which scored negatively in our assays. However, our screening was performed without the co-occurrence of supportive CD40L, which may have altered the sensitivity of our patient samples.
The most potent and consistently effective protective agonist in our assays was the TLR9 agonist CpG-ODN. This raises the interesting possibility that levels of circulating DNA could affect drug sensitivity in these B-cell malignancies. Although CpG-ODN is often thought of as a surrogate for bacterial/viral DNA, TLR9 can be activated by cellular DNA, especially mitochondrial DNA, generated during cytolysis.28 Large amounts of DNA have been detected in the plasma/serum of patients of various B-cell malignancies, including CLL,61,62 with highest levels found in relapsed patient samples.61 Elevated levels of IL-10 and sCD40L have been detected in serum of patients with CLL and other B-cell malignancies, and are associated with poor patient outcome.63,64 Although it is widely appreciated that the LN and BM environments provide a protective niche for malignant cells, our data point to mechanisms of drug resistance that can be generated within the circulation.
Wagner et al reported that ζ-chain-associated protein kinase 70–negative (ZAP70−)/immunoglobulin variable region heavy chain (IGVH) gene mutated CLL cells become apoptotic in response to CpG-ODN, in contrast to ZAP70+/IGVH unmutated, which respond proliferatively.65 However, other studies found a lack of activation/proliferation of CLL/MCL cells with CpG-ODN.66-69 We have not observed a dichotomy in spontaneous apoptosis, cellular response, or resistance to IBR+VEN in our CLL or MCL samples preincubated with agonist mix, perhaps because our studies were performed in a mixture of agonists designed to emulate some of the complexities of in vivo environments. Our results are in agreement with reports that cytokines can overcome CpG-ODN–induced apoptosis in IGVH-mutated CLL cells.34,43
Activation of NF-κB signaling in response to microenvironmental agonists is a source of resistance to IBR-VEN combination in CLL and MCL
To understand the mechanisms of resistance to IBR+VEN, we performed RPPA on MCL lines treated with IBR, VEN, and the combination (supplemental Figure 10). We observed synergistically enhanced increases both in cleaved caspases and PARP following IBR+VEN treatment, reaching saturation within 3 hours (supplemental Figure 10). The accelerated activation of the apoptosis pathway by VEN when BTK is inhibited by IBR suggests that BCR signaling is able to restrain activation of caspases by VEN, and that inhibition of BCR signaling enables increased effectiveness of VEN.70,71 IBR or IBR+VEN treatment also resulted in decreased phosphorylation of virtually all assayed signal transduction proteins, one of the few exceptions being phosphorylation on p65/RelA S536, which is known to facilitate NF-κB signaling.72 NF-κB signaling is a well-validated mechanism for resistance to drugs and other stresses,73,74 and is also highly upregulated in primary CLL/MCL cells isolated from the protective LN/BM environment.16,41,42
Our subsequent analysis of patient samples showed activation of alternative and, to a lesser extent, classical NF-κB pathways in agonist mix–treated CLL or MCL cells (Figure 3C-D,H-I). This is consistent with the analysis by Rahal et al who identified alternative NF-κB signaling as a major source of drug resistance in MCL lines.75 A recent study showed the overexpression of TLRs and several NF-κB target genes in a subset of MCL patients who also showed poor survival,76 highlighting the potential for TLR-mediated NF-κB signaling as well as its significance in MCL. A recent study by Saba et al also showed high levels of classical NF-κB signaling in LN-resident MCL cells and IBR resistance due to NF-κB signaling.16 Furthermore, Xu et al showed the overactivation of alternative NF-κB signaling in BM-resident CLL cells in a subset of patients; these cells survived better in culture and resisted fludarabine treatment ex vivo but were highly sensitive to the proteasome inhibitor MG132 that blocks NF-κB signaling.77 These findings all strengthen the concept that alternative/classical NF-κB signaling is a key node in regulating survival responses/drug effects in B-cell malignancies.
NF-κB–dependent overexpression of antiapoptotic proteins is essential for IBR+VEN resistance
The antiapoptotic proteins Mcl-1, Bcl-xL, and survivin have been identified as NF-κB target genes,78-81 and their expression is likely central to leukemic B-cell survival based on the findings of gene expression profile studies.41,42 Previous work showed higher amounts of Mcl-1 protein and Bcl-xL and survivin but not Bcl-2 messenger RNAs in CLL cells isolated from the protective environment of LNs.82 We consistently detected overexpression of Mcl-1, Bcl-xL, or survivin but not Bcl-2 protein in CLL and MCL cells preincubated with agonist mix (Figure 4A-C). The expression of these proteins was blocked by inhibitors of NF-κB (Figure 4D).
To determine whether the changes in signaling and enhanced production of antiapoptotic proteins were necessary for agonist-induced drug resistance, we tested inhibitors of signaling pathways and antiapoptotic proteins (supplemental Table 4) for cytotoxicity in CLL and MCL samples preincubated with agonist mix. BMS-345541 as well as bortezomib and carfilzomib, which inhibit NF-κB signaling49 (Figure 3E-I), generated robust cytotoxicity in CLL and MCL cells (Figure 5A-B; supplemental Figure 14A) and showed exceptional synergistic interaction with IBR+VEN (Figure 6). Similarly, inhibitors of Mcl-1, Bcl-xL, or survivin also showed significant cytotoxicity in our assays (Figure 5C-F; supplemental Figure 14E). A recent report83 described a novel Mcl-1 inhibitor that was well tolerated in vivo and effective against various cancer types, indicating the credibility of utilizing this vulnerability as a therapeutic target.
Agonist treatment generated modest activation of PI3K, MAPK, and PKC pathways (supplemental Figure 12), which are often prosurvival. However, inhibitors of these pathways did not affect cell survival at concentrations where their targets were maximally inhibited (supplemental Figures 12B-E and 14C-D). Thus, activation of these pathways by agonist mix is unlikely to be associated with drug resistance.
In summary, we have shown that CLL and MCL as well as FL cells, when exposed to agonists that are present in the in vivo microenvironment of cancer cells, become resistant to the proapoptotic activity of IBR+VEN. The agonists inhibit drug-induced apoptosis by activating NF-κB signaling. This in turn induces the synthesis of 1 or more antiapoptotic proteins that render cells less dependent on Bcl-2, which is the only known target of VEN.24 This mechanism, which can generate ex vivo phenotypic resistance, is in contrast to the central mechanism of resistance that occurs due to genetic selection, in which the IBR-inhibited BCR pathway is reactivated by mutation in pathway components.13-16 Our findings suggest that durable and complete responses to IBR, VEN, or the combination will require additional interventions that block the interacting network of survival responses available to BCR-driven and Bcl-2–driven malignancies within the in vivo environment. Furthermore, our data provide rationale for testing the inhibitors of NF-κB, Mcl-1, or TLR9 signaling in patients relapsed/refractory to IBR, VEN, or IBR+VEN.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank Susan Houchens, Kim Underwood, and other members of the University of Virginia (UVA) Hematologic Malignancies Clinical Research Team for assistance in obtaining clinical samples and information. The authors thank Eduardo M. Sotomayor at the George Washington University Cancer Center, Washington, DC, for providing HS-5 and HK cell lines. The authors are especially indebted to Daniel Gioeli and other members of the UVA Cancer Signaling Research Group for helpful comments and suggestions.
This work was supported by the UVA Cancer Center, National Institutes of Health, National Cancer Institute grant P30-CA044579, the V Foundation for Cancer Research, and the Lymphoma Research Fund of UVA. The study was also supported by a grant from the Lymphoma Research Foundation (LRF) to C.A.P. as a LRF Scholar. Clinical trial NCT02419560 is supported by a grant to UVA from AbbVie, Chicago, IL.
Authorship
Contribution: K.D.J. designed, and helped perform and analyze, most of the experiments and took the lead in writing the paper; V.L.G. performed experiments and helped in data analysis; C.A.P. and M.E.W. oversaw the clinical integration and helped in experimental design and interpretation; E.F.P. oversaw the RPPA analysis performed by J.D.W. and R.I.G.; B.J.C. and S.B. provided bioinformatics input; T.P.B. provided advice on B-cell biology and FACS analysis; M.J.A. and L.K.B. initiated the study of primary patient samples; M.J.W. and M.E.W. organized the overall project; and M.J.W. and C.A.P. oversee the overall project.
Conflict-of-interest disclosure: E.F.P. is a coinventor on US government– and university-assigned patents on the RPPA technology and can receive royalties and licensing distributions under US law. E.F.P. is also a cofounder and equity interest shareholder, consultant to Theranostics Health, Inc, the licensee for the RPPA technology discussed in this manuscript. C.A.P. receives clinical trial research support from AbbVie, via the University of Virginia Office of Sponsored Programs. M.E.W. has received clinical trial research support from Janssen and Pharmacyclics and consulting fees from Gilead Sciences. M.J.A. is an employee at Gilead Sciences and receives salary benefits. The remaining authors declare no competing financial interests.
Correspondence: Michael J. Weber, Department of Microbiology, Immunology and Cancer Biology, School of Medicine, University of Virginia, PO Box 800734, Charlottesville, VA 22908-0734; e-mail: mjw@virginia.edu.

![Figure 1. IBR and VEN are variably cytotoxic in CLL and MCL patient PBMCs. (A) Representative images showing cleaved PARP in CD5+/CD19+ cells in CLL (patient [Pt] 12) and MCL (Pt 22) patient PBMCs following ex vivo treatment with IBR (0.1 μM), VEN (25 nM), or IBR+VEN. The concentrations of IBR and VEN that showed 20% to 30% cytotoxicity as single agents (measured by alamarBlue) in a CLL patient PBMC were selected for ex vivo treatment of patient samples (data not shown). (B-D) PBMCs from CLL (N = 24) and MCL (N = 8) patients were treated with IBR, VEN, or IBR+VEN. The cleaved PARP in CD5+/CD19+ cells in CLL samples showing synergy (B) or no synergy (C) with IBR+VEN and in MCL samples (D) is shown. Comparable results were also obtained by analyzing the dead cell staining of CD5+/CD19+ cells in CLL and MCL patient PBMCs: samples that exhibited synergistically high cleaved PARP also showed synergistic increases in dead cell staining following ex vivo treatment with IBR and VEN (supplemental Figure 2A-C). The time to achieve maximal synergistic cytotoxicity (∼6-18 hours) was chosen as the time point used for subsequent data analysis. Synergy was calculated using the Bliss model of independence, which generates interaction scores even where 1 drug has no effect. Data are expressed as means ± SD. Statistical significance was determined by ANOVA. *P < .05, **P < .01, ****P < .0001. NS, not significant; SSC, side scatter.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/1/14/10.1182_bloodadvances.2016004176/3/m_advances004176f1.jpeg?Expires=1767699241&Signature=sbH504XHSctV3a0gDoNCCAAss5LhZK~y6-5S7QWnXLLHaqfYhy5e576Xvse1QqEk~4fnT8FAw8c8v8W7-Bu50sNunJVOHNJ0eGn9u5YcvfeBMjXWF5PJ4RQFeninQAWJFG6SYodM3687NkBMpmX5uJyeuBM6h9kiQ12Q1dJ9CQUc1rOLEMoeWZ90Lv9OJHI4ogWavocUcDyEnDvtuJermnKGlBVbDbkXBZkTsVmLT2ANc~HwNkFPyw~jIc-vkSwEiKiud5AUz6LkRKRDHEieVREMsHDhVaQFUSBk8RAVL0a~OY-feJy3Iors~2jYLua2SAqeOiD22PRn~nQGLS5xEA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. Extrinsic microenvironmental agonists generate resistance to IBR+VEN. (A) Diagram showing the patient PBMCs cocultured with stromal cells or preincubated with soluble agonists for 12 hours. Drugs as well as a second dose of agonists were added at 12 hours and cultured for an additional 12 hours, and cleaved PARP or dead cell staining in CD5+/CD19+ cells in CLL or MCL was analyzed by flow cytometry. (B) PBMCs from CLL patients (Pts 01, 03, and 12) were cocultured with HS-5 or HK cell line at a 10:1 ratio or preincubated with sCD40L (2 μg/mL), IL-10 (0.015 μg/mL), CpG-ODN (1.5 μg/mL), CXCL13 (0.5 μg/mL), IL-2 (1.54 ng/mL), BAFF (0.25 μg/mL), or IgM (25 μg/mL) and treated with IBR (0.1 μM) + VEN (25 nM) or DMSO as well as second doses of agonists for 12 hours as shown in panel A. Cleaved PARP in CD5+/CD19+ cells was analyzed by flow cytometry. (C-D) PBMCs from 22 CLL (Pts 1-3, 6-7, 12, 14, 21, 25-28, 33, 35, 39, 43, 46, 48, 51-52, 56, and 59) and 8 MCL (Pts 22-23, 38, 45, 49, 55, 60, and 66) patients and the Mino MCL cell line were preincubated with agonist mix (sCD40L [2 μg/mL] + IL-10 [0.015 μg/mL] + CpG-ODN [1.5 μg/mL]) for 12 hours. Samples were then incubated with IBR (0.1 μM) + VEN (25 nM) or DMSO for an additional 12 hours as well as a second dose of agonist mix. Cleaved PARP in CD5+/CD19+ cells in CLL samples (C) and MCL samples/Mino cell line (D) were analyzed by flow cytometry. (E) Ki67+/CD5+/CD19+ cells in CLL and MCL patient PBMCs (Pts 06, 35, 39, 43, and 45) preincubated with or without 2 doses of agonist mix at 12-hour intervals were analyzed by flow cytometry. (F) PBMCs from CLL patients (Pt 01 and 25) were preincubated with agonist mix for 12 hours and treated with increasing concentrations of IBR (0.1, 1, and 10 μM) and VEN (6.25, 12, 25, 100, 400 nM) for 12 hours along with a second dose of agonist mix. The cleaved PARP in CD5+/CD19+ cells was analyzed by flow cytometry. Data are expressed as means ± SD. The statistical significance was determined by ANOVA. *P < .05, ***P < .001, ****P < .0001. FACS, fluorescence-activated cell sorter.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/1/14/10.1182_bloodadvances.2016004176/3/m_advances004176f2.jpeg?Expires=1767699241&Signature=hICxDFDRfl9ehsZDtaURA~StN07cZgcBBaJhDHjRcTZipRxCdd9sPgHft99NWeAdy2Hnlngs4XLJZZjBT5LMXKLVtNWlUi4aHMYNtgPfEr2d6VpcEI2B5Gd9bsvVBqZm-Usq51sxMJSL2zO8s~hfCejKlmhlx5n19gq4xd-9LQaREPe9Xtzs1vFY9nSbMppJKYRiGTFe-BnDUH2In74yje3mDmS~u6OJi-uv3a4Bupi1tYQWvvaTQ8dn8TjCgYWRIxyWR2YZgDmxb8GpLccpWG8mLeJYwifREtY1A9WO-SnDjI4sLz3rOfOXNLP1nXQQimE1HZO7FvmxVP0-eNUZUg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)