Key Points
Low mutation load is associated with a benefit from rituximab maintenance.
The Teff signature correlates with high mutation load, is prognostic, and may distinguish immunologically distinct FL subgroups.
Abstract
Identifying follicular lymphoma (FL) patients with preexisting antitumor immunity will inform precision medicine strategies for novel cancer immunotherapies. Using clinical and genomic data from 249 FL patients, we determined the clinical impact of mutation load and an effector T-cell (Teff) gene signature as proxies for the likelihood of a functional immune response. The FL mutation load estimate varied between 0 and 33 mutations per Mb (median, 6.6), and 92% of FL patients with a high mutation load had high Teff gene expression (P = .001). The mutation load was associated with a benefit from rituximab maintenance: FL patients with low mutation loads experienced a profound benefit from rituximab maintenance (hazard ratio [HR], 0.29; 95% confidence interval [CI], 0.15-0.54; P < .001). The Teff gene signature was prognostic as a continuous predictor (P = .008), and was used to separate FL patients into 2 groups, an “inflamed” subset (Teff-high; n = 74) and an “uninflamed” subset (Teff-low; n = 75), with longer progression-free survival (PFS) in the inflamed FL subset (PFS HR, 0.39; 95% CI, 0.21-0.70; P = .002). Furthermore, the subset of inflamed FL tumors demonstrated high expression of other T-cell signatures and counterregulatory genes, which also correlate with PFS. Mutation load and Teff gene expression may help identify immunologically distinct lymphoma subsets relevant for modern immunotherapies.
Introduction
Recently discovered links between cancer genomics and immunity have reinforced the concept that some mutations trigger cytotoxic effector immune responses, and that the likelihood of an immunogenic mutation increases with mutation load.1,2 Previous studies in solid tumors showed that mutation load is a good proxy for the presence of neoantigens and linked mutation load with response to modern immunotherapy.3-6 Other studies suggest that biomarkers of T-cell biology are important for responses to checkpoint blockade.6-9 Whether these surrogates for cancer immunity are relevant in lymphoma and potentially helpful for determining which patients may benefit from modern checkpoint blockade is unknown.
Previous studies in follicular lymphoma (FL) suggest that immune infiltration is associated with outcome, although the specific immune biology contributing to outcomes is unclear.10-15 In the prerituximab era, a mixed T-cell gene signature was associated with improved outcomes, whereas an expression signature enriched for monocyte genes was associated with poor prognosis.10 Recent studies suggest, however, that the therapeutic setting may impact these associations, with macrophage infiltration correlating with worse outcomes after rituximab, cyclophosphamide, vincristine, and prednisone, but improved outcomes among patients treated with rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone.14
This study was designed to determine the clinical impact of a novel technique for estimating the mutation load of FL patients and of a 6-gene effector T-cell (Teff) signature designed to reflect functional components of the cytotoxic effector payload. Both biomarkers are tied to the proposed mechanism of anti–PD-1/PD-L1 therapies and implicated in clinical responses to modern checkpoint blockade,6-8 and have potential utility for identifying FL patient subsets that are likely to respond to cancer immunotherapies.
Methods
Characteristics of patients and the PRIMA trial
The randomized, open-label Primary Rituximab and Maintenance (PRIMA) study was undertaken in 223 centers in 25 countries.16 Patients (n = 1217) with previously untreated FL were registered in the study and received 1 of 3 nonrandomized rituximab-based immunochemotherapy induction regimens. The 1019 patients who achieved a complete or partial response were then randomly assigned to receive 2 years of rituximab maintenance therapy or observation. Pretreatment tumor biopsies were available from 249 patients (221 patients randomized to maintenance vs observation; Table 1) for FoundationOne Heme genomic profiling and 142 patients (132 patients randomized to maintenance vs observation) for RNA sequencing (RNAseq) expression profiling. Progression-free survival (PFS) from the point of randomization was defined as the primary end point. The protocol of the original PRIMA trial was approved by local or national ethics committees according to the laws of each country, and the study was undertaken in accordance with the Declaration of Helsinki.
Patient characteristics
| . | Treatment arm . | |
|---|---|---|
| Observation (n=122), n (%) . | Rituximab maintenance (n=99), n (%) . | |
| Age >60 y | 42 (34) | 34 (34) |
| Age, y | 56 (23-80) | 54 (28-76) |
| Male sex | 58 (48) | 51 (52) |
| FLIPI score | ||
| Low (0-1 risk factors) | 27 (22) | 20 (20) |
| Intermediate (2 risk factors) | 43 (35) | 39 (39) |
| High (3-5 risk factors) | 52 (43) | 40 (40) |
| Induction regimen | ||
| R-CHOP | 114 (93) | 95 (96) |
| R-CVP | 8 (7) | 4 (4) |
| R-FCM | 0 (0) | 0 (0) |
| Response to induction | ||
| Complete response | 89 (73) | 75 (76) |
| Partial response | 33 (27) | 24 (24) |
| Other | 0 (0) | 0 (0) |
| . | Treatment arm . | |
|---|---|---|
| Observation (n=122), n (%) . | Rituximab maintenance (n=99), n (%) . | |
| Age >60 y | 42 (34) | 34 (34) |
| Age, y | 56 (23-80) | 54 (28-76) |
| Male sex | 58 (48) | 51 (52) |
| FLIPI score | ||
| Low (0-1 risk factors) | 27 (22) | 20 (20) |
| Intermediate (2 risk factors) | 43 (35) | 39 (39) |
| High (3-5 risk factors) | 52 (43) | 40 (40) |
| Induction regimen | ||
| R-CHOP | 114 (93) | 95 (96) |
| R-CVP | 8 (7) | 4 (4) |
| R-FCM | 0 (0) | 0 (0) |
| Response to induction | ||
| Complete response | 89 (73) | 75 (76) |
| Partial response | 33 (27) | 24 (24) |
| Other | 0 (0) | 0 (0) |
Comprehensive genomic profiling and mutation load estimation
Comprehensive genomic profiling was performed using ≥50 ng of DNA extracted from formalin-fixed, paraffin-embedded biopsy specimens collected at study inclusion by using the FoundationOne Heme test. The pathologic diagnosis of each case was confirmed by hematoxylin and eosin–stained slides, with samples requiring a minimum of 20% tumor cells. The laboratory and computational methods employed in the FoundationOne Heme (F1H) DNA assay have been described in detail previously.6,17-19 Briefly, data were generated from the D2 bait set of the F1H test, targeting 406 genes over an ∼1.20-Mb region (1 198 160 base pairs). This next-generation sequencing platform profiles somatic mutations using high coverage (>500×) exome sequencing of 467 cancer-related genes. In addition to identifying a number of potentially deleterious mutations, the FoundationOne Heme platform was also used to estimate mutation load (per megabase) using Foundation Medicine’s mutation load algorithm.6,20 To estimate mutation load, base substitutions and indels within a 1.2-Mb region were identified by DNA sequencing; any alteration known or likely to be associated with tumorigenesis was removed, because the F1H assay is biased toward functional mutations in cancer. Synonymous mutations were counted after filtering to reduce sampling noise and as an attempt to capture mutational processes contributing to neoantigen generation.20 Noncoding alterations, alterations listed as “known somatic alterations” in the Catalogue of Somatic Mutations in Cancer database, alterations predicted to be germ line by the somatic-germ line-zygosity algorithm, known germ line alterations in the Single Nucleotide Polymorphism database, and germ line alterations occurring with ≥2 counts in the Exome Aggregation Consortium database were filtered and not counted. A breakdown of the total mutation count in each sample and the mutation load per Mb can be found in supplemental Table 1. We used the mutation load as a proxy for the probability of neoantigen formation, with a higher mutation load assumed to correlate with an increased probability of neoantigen formation. Sequencing data are available through the NCBI Sequence Read Archive under accession number PRJNA392675.
RNASeq expression profiling
Frozen biopsies collected at study inclusion were profiled by TruSeq RNAseq. To quantify the expression of immune cell subsets, we used immune signatures designed to reflect relevant antitumor immune biology and calculated signature scores by first z-score normalizing each gene’s expression values across patients, and then calculating an average z score across signature genes for each patient. Total T-cell/myeloid infiltration was estimated by using the immune response 1 (IR1) signature from Dave et al.10 Cytotoxic effector infiltration (Teff) was estimated by using a 6-gene signature (GZMA, GZMB, PRF1, IFNγ, EOMES, and CD8A), as described in previous publications.7,9 The CD4+ T-cell, regulatory T-cell (Treg), and T-follicular helper signatures were derived from the CIBERSORT LM22 matrix.21 A list of signature genes can be found in supplemental Table 2.
Statistical methods
Computational analysis of RNAseq data were performed in R (version 3.2.2, R Project for Statistical Computing). Count data were normalized by using voom, as implemented in the limma package.22 Signature genes were combined into a single score by first z-score normalizing genes across all patients, and then averaging z scores across the genes in the signature to create a single signature score for each patient. Because prior work in melanoma suggests that the vast majority of mutations are not recognized by autologous T cells,23,24 a manually selected cut-point was set to capture the highly mutated subset of FL samples with a high probability of neoantigen formation, based roughly on the inflection point in the tail of the mutation load distribution. Patients with >15 mutations/Mb (n = 19) were considered to have a high probability of neoantigen formation, and the remaining patients were stratified into mutation-low (<6.6 mutations/Mb; n = 132) or mutation-mid (≥6.6 mutations/Mb and ≤15 mutations/Mb; n = 98) groups based on the median mutation load across all patients. We used Cox regression to examine the associations with PFS, adjusting for the Follicular Lymphoma International Prognostic Index (FLIPI), age, sex, treatment arm, induction regimen, and response to initial therapy; P values are provided for exploratory purposes.
Sensitivity analysis
Given the heuristic nature of the cut-points for mutation load, we performed a sensitivity analysis to analyze the dependence of our results on specific cut-points. For both the median cut-point (6.6 mutations/Mb) and the high mutation cut-point (15 mutations/Mb), we identified a reasonable range using a bootstrapping procedure. A bootstrapped data set was formed by sampling (with replacement) 249 observations from the total data set, and for each bootstrapped data set, the median was calculated, and a mutation-high inflection point was identified manually. This process was repeated 200 times, and the minimum and maximum of the bootstrapped cut-points were used to define the range of cut-points to consider in the sensitivity analysis. The median of the bootstrapped data ranged from 5.8 to 7.5 mutations/Mb, whereas the manually selected mutation-high inflection point ranged from 11 to 20 mutations/Mb.
Results
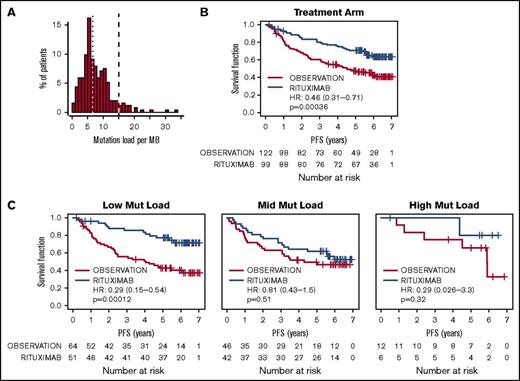
Somatic mutations were assessed in a set of 249 primary FL tumors by using the FoundationOne Heme platform. No significant association was found between mutations in individual genes and survival. However, we chose to look at the overall mutation load within the tumors, based on the hypothesis that an increased somatic mutation load is correlated with the presence of neoantigens in the tumor, increasing the likelihood of an immunogenic tumor. The mutation load among the patients in this cohort was highly variable (range, 0-33 mutations/Mb; first quartile, 4.2 mutations/Mb; median, 6.6 mutations/Mb; third quartile, 10.0 mutations/Mb; Figure 1A), but at a level comparable to lung, bladder, and head/neck malignancies.25 Overall, PFS was significantly extended in patients treated with rituximab maintenance compared with the observation arm (Figure 1B). The 3-year PFS among patients with high mutation loads was 83% compared with 66% for mid and 68% for low mutation loads, although no significant association with PFS was seen in the observation (P = .66) or rituximab (P = .13) arms, and there was no association between mutation load and age, FLIPI, or initial response. Mutation load was significantly associated, however, with benefit to rituximab maintenance: FL patients with low mutation loads experienced a profound benefit from rituximab maintenance (Figure 1C; hazard ratio [HR], 0.29; 95% confidence interval [CI], 0.15-0.54; P < .001), whereas no statistically significant benefit was seen among patients with medium (HR, 0.81; 95% CI, 0.43-1.50; P = .51) or high mutation loads (HR, 0.29; 95% CI, 0.03-3.30, P = .32). This effect of mutation load was independent of baseline factors, and no interaction with FLIPI, age, and initial response was observed (P > .05). A Cox proportional hazards model confirmed that the observed difference in the degree of benefit from rituximab maintenance significantly differed across the 3 mutation load groups (interaction P = .034). Further, the statistical significance of this effect was robust and was maintained across the full range of bootstrap-selected cut-points for the mutation-low group (P < .05 for all cut-points in 5.8-8.0 mutations/Mb).
Association of mutation load with response to rituximab therapy among FL patients. (A) The distribution of mutation load (per megabase) among untreated FL patients, grouped into mutation-high (>15 mutations/Mb, dashed line; n = 19), mutation-mid (≥6.6 mutations/Mb and ≤15 mutations/Mb; n = 85) and mutation-low (<6.6 mutations/Mb, dotted line; n = 112). (B) Kaplan-Meier estimates of PFS, stratified by treatment arm, are shown for the total biomarker-evaluated population. (C) Survival curves for patients with low, mid, and high mutation loads, stratified according to treatment arm. HRs, 95% CIs, and P values were calculated by using a Cox proportional hazards model, adjusted for age, FLIPI, sex, and response to induction therapy.
Association of mutation load with response to rituximab therapy among FL patients. (A) The distribution of mutation load (per megabase) among untreated FL patients, grouped into mutation-high (>15 mutations/Mb, dashed line; n = 19), mutation-mid (≥6.6 mutations/Mb and ≤15 mutations/Mb; n = 85) and mutation-low (<6.6 mutations/Mb, dotted line; n = 112). (B) Kaplan-Meier estimates of PFS, stratified by treatment arm, are shown for the total biomarker-evaluated population. (C) Survival curves for patients with low, mid, and high mutation loads, stratified according to treatment arm. HRs, 95% CIs, and P values were calculated by using a Cox proportional hazards model, adjusted for age, FLIPI, sex, and response to induction therapy.
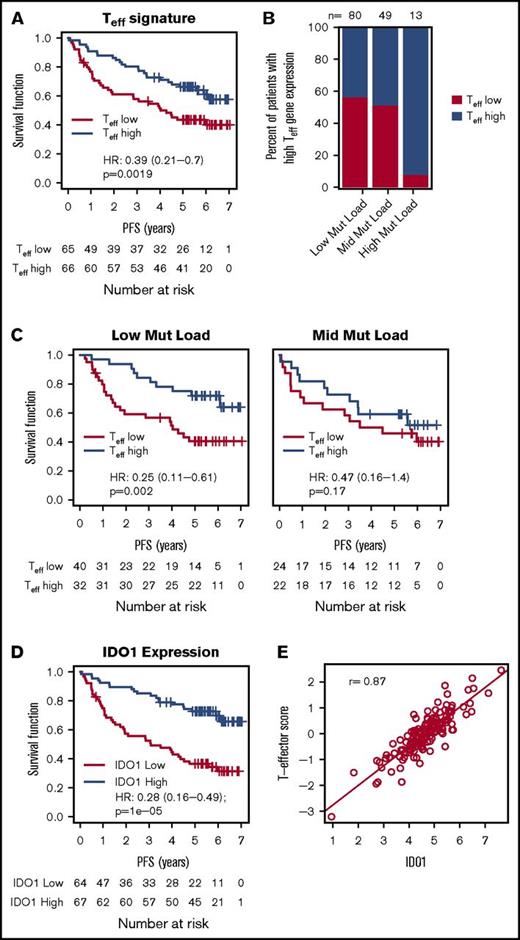
To test the hypothesis that an estimate of the functional payload (ie, interferon-γ [IFN-γ], granzyme, perforin) of tumor-infiltrating cytotoxic Teff may better approximate the likelihood of a clinically meaningful antitumor immune response, we separated FL patients into an “inflamed” subset (Teff-high; n = 74) and an “uninflamed” subset (Teff-low; n = 75) based on the aggregated expression of Teff genes. Because the Teff signature was not significantly predictive for a response to rituximab maintenance (P = .36), the 2 arms (ie, the observation arm and the rituximab maintenance arm) were combined for this analysis. The Teff signature alone was prognostic as a continuous predictor (P = .008) or as a dichotomized biomarker, with longer PFS observed in the inflamed FL subset (Figure 2A; PFS HR, 0.39; 95% CI, 0.21-0.70; P = .002). Further, the significance of this association was also insensitive to cut-points (P < .05 for all cut-points between the 25th and the 75th percentile).
Association of Teffgene expression with PFS, mutation load, and IDO1. (A) The association between the Teff gene signature and PFS in all patients treated with chemoimmunotherapy. (B) The percent of FL patients with high and low Teff scores (median cut-point) among the 3 mutation load groups. (C) The association between the Teff gene signature and PFS among mutation load groups. (D) The association between IDO1 (stratified by median IDO1 expression) and PFS among all patients treated with chemoimmunotherapy. (E) The correlation of Teff expression with levels of IDO1. HRs, 95% CIs, and P values were calculated by using a Cox proportional hazards model, adjusted for age, FLIPI, sex, and response to induction therapy.
Association of Teffgene expression with PFS, mutation load, and IDO1. (A) The association between the Teff gene signature and PFS in all patients treated with chemoimmunotherapy. (B) The percent of FL patients with high and low Teff scores (median cut-point) among the 3 mutation load groups. (C) The association between the Teff gene signature and PFS among mutation load groups. (D) The association between IDO1 (stratified by median IDO1 expression) and PFS among all patients treated with chemoimmunotherapy. (E) The correlation of Teff expression with levels of IDO1. HRs, 95% CIs, and P values were calculated by using a Cox proportional hazards model, adjusted for age, FLIPI, sex, and response to induction therapy.
Interestingly, when treated as a continuous predictor, the Teff signature did not correlate with the mutation status of any individual genes (supplemental Figure 1) or with mutation load. In contrast, 92% (12 of 13) of FL patients with high mutation loads (>15 mutations/Mb) were also inflamed compared with 44% (35 of 80) of patients with low mutation loads and 49% (24 of 49) of patients with medium mutation loads (Figure 2B; P = .001), supporting the hypothesis that high mutation loads identify an “inflamed” FL subset likely to possess immunogenic neoantigens. Sensitivity analysis showed that the association between mutation load and Teff categories was significant for a range of “mutation high” cut-points (P < .05 for cut-points between 13-19 mutations/Mb; and P > .05 for cut-points between 11-13 mutations/Mb). The low and mid mutation load FL subgroups, on the other hand, consisted of similar numbers of inflamed and uninflamed patients, potentially reflecting immunogenicity differences of individual mutations, with significantly longer PFS for inflamed patients among the low–mutation load subgroup (3-year PFS was 84.4% for inflamed patients compared with 56.6% for uninflamed patients; Figure 2C; P = .002) and a similar although statistically insignificant trend among mutation-mid patients (76.2% for inflamed patients compared with 58.3% for uninflamed patients; P = .17). The Teff signature, therefore, may identify inflamed and uninflamed FL patients and provide a useful tool for identifying immunologically distinct FL subsets with different clinical outcomes.
Teff infiltration into tumors often triggers counterregulatory mechanisms, which limit effective antitumor immune responses. Because high expression of IFN-γ (a hallmark of inflamed tumors), for instance, increases the expression of inhibitory molecules like PD-L1 and IDO1, we evaluated the impact of IDO1 on PFS. High expression of IDO1 correlated with longer PFS (Figure 2D; HR, 0.25; 95% CI, 0.14-0.45; P < .001), and there was significant overlap between IDO1-high and inflamed (Teff-high) FL tumors (Figure 2E). It is known that different T-cell subsets and other immune cells can infiltrate tumors simultaneously and may impact FL outcomes.10,15,26 Consistent with this notion, we observed that our Teff signature correlated with signatures of CD4+ T cells, Tregs, and T-follicular helper cells in addition to the IR1 signature (r > 0.70; supplemental Figure 2A), and that each of these signatures were also prognostic in this cohort (supplemental Figure 2B). These data are consistent with the interplay of pro- and anti-inflammatory immunity, wherein proinflammatory mediators like IFN-γ derived from cytotoxic effector cells drive the clinical outcome.
Discussion
Using genomic data from a large cohort of FL patients, we demonstrate an association between mutation load, immune biology, and clinical outcomes in FL. We show that mutation load is independently predictive of response to rituximab maintenance therapy, with significantly increased response among mutation-low patients. In addition, the Teff signature was shown to be prognostic, with high Teff expression correlating with improved survival.
The prognostic impact of Teff genes is consistent with prior reports of nonmalignant immune cells contributing to FL outcomes.10-13,27 The components of the Teff signature were selected to reflect a compact set of functional components of the specific cytotoxic effector payload implicated in clinical responses to modern checkpoint blockade.6-8 We found that the Teff signature, however, may also capture a more broadly defined inflamed subset of FL patients, because it is highly correlated with other T-cell messenger RNA signatures, including CD4-positive and Tregs in addition to IDO and the previously reported prognostic IR1 signature (supplemental Figure 2).10 We extend these earlier insights by demonstrating a significant correlation of the favorable IR1 prognostic signature with a simplified 6-gene Teff signature and a correlation of both signatures with favorable outcomes in a large cohort of rituximab-treated FL patients. Of note, the 41-gene IR1 signature, previously shown to be associated with tumor shrinkage and PFS among FL patients treated with pidilizumab and rituximab, contains many T-cell genes, but includes genes representing other biologies as well. Our simplified 6-gene Teff signature may capture the relevant Teff biology. We plan to evaluate the 6-gene signature in clinical trials of atezolizumab (anti–PD-L1) in FL.15 Whether a more extensive gene signature reflecting the breadth of antitumor immunity, like IR1, will outperform more targeted signatures, like the 6-gene Teff signature in the context of checkpoint blockade and chemoimmunotherapy, merits further investigation.
Mechanistic studies in mice and humans suggest that CD8 T-cell tumor infiltration triggers the recruitment of other immune cells, including immunosuppressive Tregs, IDO, and PD-L1, but the CD8 T-cells are the key mediators of antitumor immunity. These observations underlie our hypothesis that better outcomes among FL patients may be associated with high Teff gene expression.28-31 It is challenging, however, to assign the prognostic effect to the infiltration of a specific immune subset. Mechanistically, T cells with an effector phenotype are implicated in tumor killing and clearance and may provide a useful tool for identifying FL patients likely to respond to checkpoint inhibitors, as seen in solid tumors.7,8,15,32 A broader biomarker of T-cell infiltration, like CD3,27 or a composite signature capturing multiple components of antitumor immunity, like IR1,10 may also be useful for identifying immunologically distinct FL subsets, but practical considerations, like assay reproducibility and complexity, and therapeutic implications, like predicting the response to checkpoint blockade, may ultimately inform the optimal biomarker.
In addition to T cells, other nonmalignant immune cells, particularly natural killer cells, have been implicated as key mediators of rituximab’s clinical activity in NHL.11,33,34 Our Teff signature contains genes involved in both T-cell and natural killer cell effector function,35 and this overlap limits our ability to attribute the mechanism to a specific cytotoxic effector immune cell. The Teff signature, however, has been previously validated in other diseases and settings and shown to correlate with the clinical activity of anti–PD-L1 in many malignancies.7,9,15 The contribution of individual immune infiltrates to FL outcomes requires further study, and additional experiments are underway to deconvolve these highly correlated features. However, ultimately, the most useful biomarker of response in FL will be one that defines a general immune set point relevant for therapies like rituximab and anti–PD-1/PD-L1.36
The pronounced PFS benefit of rituximab maintenance among FL patients with low mutation loads treated with R-chemotherapy is novel, and the mechanism merits further interrogation. In this study, we tested a model wherein the estimated mutation load represents a proxy for the presence of neoantigens, such that the likelihood of an immunogenic mutation increases with mutation load.1-3,37,38 This model is supported by studies in melanoma, lung cancer. and bladder cancer demonstrating correlations between mutation load and response to modern T-cell targeting immunotherapies.3,6,8 One possible explanation for this effect is the vaccine hypothesis; prolonged rituximab exposure may lead to greater tumor killing, the release of novel T-cell epitopes, and the generation of new T-cell responses.39 Whether this vaccine effect is related to our findings merits further investigation. Mutation load may also represent a proxy for genomic instability or a measure of the levels of AID-induced somatic hypermutation or a surrogate for genetically heterogeneous subclones, potentially impacting resistance to cytotoxic therapy or leading to escape mutations in CD20. Additional studies are ongoing to determine the mechanism underlying this effect.
A deeper understanding of FL genomics contributing to FL immunity could lead to more informed subgrouping and tailored treatment approaches for FL patients. In this article, we demonstrate an association between mutation load, immune biology, and clinical outcomes in FL. Our data suggest that mutation load and Teff gene expression may help identify immunologically distinct lymphoma subsets relevant for precision medicine strategies in an immunotherapy era.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank the Lymphoma Study Association study team, Nadine Vailhen, Bruno Tesson, Sandra Horning, Nancy Valente, Ira Mellman, Priti Hegde, Michael Wenger, Guenter Fingerle-Rowson, Axel Boehnke, and Mark Davis for thoughtful comments and review.
Authorship
Contribution: J.M.V. and C.R.B. conceived the project, designed and performed the data analysis and interpretation, and prepared the manuscript; R.M., E.S.-G., and E.A.P. collected and assembled the data; G.M.F. applied the mutation load algorithm; G.S., R.B., L.X., F.J., P.D., P.S., and S.H. assisted in data collection, assembly, and analysis; and all authors approved the final manuscript.
Conflict-of-interest disclosure: C.R.B., R.M., R.B., E.A.P., E.S.-G., and J.M.V. are employed by Genentech and are shareholders in F. Hoffman La Roche. G.M.F. is employed by Foundation Medicine. L.X. has received honoraria from Novartis, and G.S. is a consultant for and has received from honoraria from Gilead, Janssen, Celgene, Mundipharma, Roche/Genentech, Amgen, and Novartis. The remaining authors declare no competing financial interests.
Correspondence: Jeffrey M. Venstrom, Genentech, 1 DNA Way, South San Francisco, CA 94080; e-mail: venstroj@gene.com.