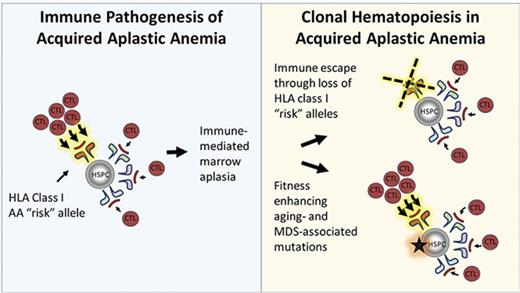
Key Points
Somatic HLA class I gene mutations are frequent in aAA and define HLA class I restricted autoimmunity in aAA.
HLA alleles targeted by inactivating mutations are overrepresented in aAA and correlate with poor therapy response and clonal evolution.
Abstract
Acquired aplastic anemia (aAA) is an acquired deficiency of early hematopoietic cells, characterized by inadequate blood production, and a predisposition to myelodysplastic syndrome (MDS) and leukemia. Although its exact pathogenesis is unknown, aAA is thought to be driven by human leukocyte antigen (HLA)–restricted T cell immunity, with earlier studies favoring HLA class II-mediated pathways. Using whole-exome sequencing (WES), we recently identified 2 patients with aAA with somatic mutations in HLA class I genes. We hypothesized that HLA class I mutations are pathognomonic for autoimmunity in aAA, but were previously underappreciated because the major histocompatibility complex (MHC) region is notoriously difficult to analyze by WES. Using a combination of targeted deep sequencing of HLA class I genes and single nucleotide polymorphism array (SNP-A) genotyping, we screened 66 patients with aAA for somatic HLA class I loss. We found somatic HLA loss in 11 patients (17%), with 13 loss-of-function mutations in HLA-A*33:03, HLA-A*68:01, HLA-B*14:02, and HLA-B*40:02 alleles. Three patients had more than 1 mutation targeting the same HLA allele. Interestingly, HLA-B*14:02 and HLA-B*40:02 were significantly overrepresented in patients with aAA compared with ethnicity-matched controls. Patients who inherited the targeted HLA alleles, regardless of HLA mutation status, had a more severe disease course with more frequent clonal complications as assessed by WES, SNP-A, and metaphase cytogenetics, and more frequent secondary MDS. The finding of recurrent HLA class I mutations provides compelling evidence for a predominant HLA class I-driven autoimmunity in aAA and establishes a novel link between immunogenetics and clonal evolution of patients with aAA.
Introduction
Acquired aplastic anemia (aAA) is an acquired bone marrow failure syndrome caused by autoimmune destruction of early hematopoietic cells1 and associated with predisposition to myelodysplastic syndrome (MDS) and leukemia.2 Although the exact pathogenesis of aAA is unknown, aAA is believed to be immune-mediated because of the clinical efficacy of T cell-directed immunosuppressive therapy (IST)3,4 and correlative studies showing lymphocyte abnormalities in patients with aAA.5-8 Several proteins have been reported as potential aAA autoantigens9-13 ; however, their roles in aAA remain unconfirmed. Earlier studies have predominantly focused on characterizing HLA class II-driven immunopathology.14-30 Despite substantial progress, more specific mechanisms of autoimmunity in aAA have not been defined.1,31-34
Recently, copy number-neutral loss of heterozygosity of chromosome arm 6p (6p CN-LOH) was identified as a recurrent somatic alteration in aAA.27 Because acquired 6p CN-LOH in aAA typically included the major histocompatibility complex (MHC) region containing multiple HLA loci,25,27 and because of genetic association of aAA with several HLA class I and II alleles,14-19,24-27,30 it was hypothesized that 6p CN-LOH emerges through immune escape of hematopoietic cells lacking certain HLA alleles.27,35 However, strong linkage disequilibrium between the HLA loci and the presence of other genes in this genomic region have made it difficult to pinpoint the causal driver of clonal hematopoiesis in patients with acquired 6p CN-LOH.
Using whole-exome sequencing (WES) to study clonal hematopoiesis in 22 patients with aAA, we recently found 2 patients with aAA with somatic loss-of-function mutations in HLA class I genes.36 We therefore wondered whether loss-of-function mutations in HLA class I genes may be common in aAA, implying a greater role of HLA class I-restricted autoimmunity than previously appreciated in this disease.1,31-34 To screen for HLA loss, we performed SNP-A genotyping and deep targeted next-generation sequencing (NGS), designed to accurately capture HLA class I genes, in 66 patients with aAA. Eleven patients (17%) had evidence of somatic HLA loss, with 13 inactivating mutations targeting 4 HLA class I alleles. Patients who inherited HLA class I alleles targeted by mutations had a more severe disease course and experienced more clonal evolution, with a bias toward somatic HLA loss in pediatric-onset aAA and toward MDS-associated changes in adult-onset aAA. Our results implicate HLA class I-driven autoimmunity in aAA pathogenesis and establish a novel link between inheriting particular HLA alleles and clonal evolution in aAA.
Methods
Additional details are provided in supplemental Methods.
Patients
Informed consent from study participants was obtained in accordance with the Declaration of Helsinki. The diagnosis and severity aAA were determined by standard criteria; congenital bone marrow failure syndromes were excluded as previously described.37-40 The 66 patients with aAA in this study included 16 patients with aAA from our previous WES cohort.36 Patients with MDS and healthy volunteers were used as controls.
Hematopathology, SNP-A, and WES analysis
Bone marrow histology was evaluated in a blinded fashion. SNP-A genotyping and comparative WES of paired bone marrow and skin fibroblast DNA were performed as previously described.25,36,39
Targeted sequencing
HLA NGS was performed as previously described40 at more than 10 000× depth. Variant calling was done with GATK Haplotype Caller,41 manually curated, and validated by WES and/or Sanger sequencing (supplemental Figure 1). Targeted sequencing of malignancy-associated genes was performed by the Penn CytoGenomics Laboratory.
Statistical analyses
The frequency of HLA alleles was compared between the largest ethnic group in our aAA cohort (white, n = 42 patients) and ethnically matched control patients.42 Response to IST was assessed 6 months after therapy according to established criteria.43 Comparisons were performed using Mann-Whitney U test for continuous variables and Fisher’s exact test for categorical variables, with a 2-tailed significance level of 0.05.
Results
Recurrent somatic loss-of-function mutations in HLA class I alleles
To characterize somatic HLA loss in aAA, we analyzed the bone marrow or peripheral blood DNA of 66 patients with aAA, using a combination of SNP-A genotyping and targeted NGS of HLA HLA-A, HLA-B, and HLA-C genes. Seventy percent of patients had pediatric-onset aAA; median age of diagnosis was 11.9 years (range, 1.5-65.6 years; supplemental Table 1; supplemental Figure 2). HLA NGS analysis was performed after a median disease duration of ∼1.1 years (range, 0-30.4 years). Twelve patients were treatment-naive.
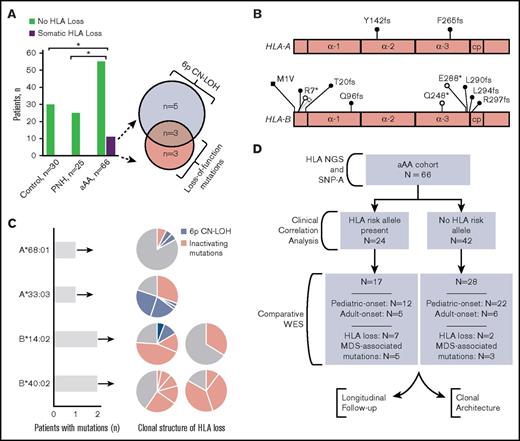
Eleven patients with aAA (17%) had somatic HLA loss; in contrast, HLA loss was not detected in 25 patients with paroxysmal nocturnal hemoglobinuria (PNH; P = .031), nor in 30 healthy control patients (P = .032; Figure 1A). Among patients with aAA, HLA loss occurred through 6p CN-LOH in 8 patients and through loss-of-function mutations in HLA-A or HLA-B genes in 6 patients; 3 patients had both acquired 6p CN-LOH and loss-of-function HLA mutations (Figure 1A-B; supplemental Table 2). Among the 6 patients with loss-of-function mutations in HLA genes, all with pediatric-onset aAA, there were a total of 13 HLA mutations, at a median of 1.5 mutations per patient (range, 1-5 mutations; Figure 1B-C). HLA mutations involved a subset of patients’ hematopoietic cells, with clone sizes ranging from 3.2% to 40.2% (Figure 1C; supplemental Table 2). Of the 13 mutations, 6 were point mutations (4 transitions and 2 transversions) and 7 were small insertions or deletions (Figure 1B); in cases amenable to clonogenic analysis, HLA mutations were confirmed to have occurred in independent clones. Similarly, in 8 patients with 6p CN-LOH, HLA loss was frequently oligoclonal, with a total of 20 6p CN-LOH events at a median of 2 clones per patient (range, 1-4 clones; Figure 1C).
Immunoediting in aAA through recurrent somatic loss of HLA class I. (A) Bar graph showing somatic HLA loss in patients with aAA compared with patients with PNH and healthy control patients. n, number of patients analyzed. *P < .05. Venn diagram shows the mechanism of HLA loss among 11 affected patients. (B) Schematic diagram showing the distribution of inactivating mutations in HLA class I alleles. α-1, α-2, and α-3 represent the α-1, α-2- and α-3 domains of HLA-A and HLA-B proteins; cp, connecting peptide. Frameshift mutations, black circles; nonsense mutations, open circles; mutated start codon, black square. (C) Bar graph showing the number of patients with mutations in a given HLA class I allele, with a corresponding pie chart plot depicting the clonal structure of HLA loss for each affected patient. The size of the pie slice corresponds to the size of each individual clone (percentage of total cells) as estimated from SNP-A analysis and percentage of mutant NGS reads. (D) A flowchart summarizing analyses performed in this manuscript.
Immunoediting in aAA through recurrent somatic loss of HLA class I. (A) Bar graph showing somatic HLA loss in patients with aAA compared with patients with PNH and healthy control patients. n, number of patients analyzed. *P < .05. Venn diagram shows the mechanism of HLA loss among 11 affected patients. (B) Schematic diagram showing the distribution of inactivating mutations in HLA class I alleles. α-1, α-2, and α-3 represent the α-1, α-2- and α-3 domains of HLA-A and HLA-B proteins; cp, connecting peptide. Frameshift mutations, black circles; nonsense mutations, open circles; mutated start codon, black square. (C) Bar graph showing the number of patients with mutations in a given HLA class I allele, with a corresponding pie chart plot depicting the clonal structure of HLA loss for each affected patient. The size of the pie slice corresponds to the size of each individual clone (percentage of total cells) as estimated from SNP-A analysis and percentage of mutant NGS reads. (D) A flowchart summarizing analyses performed in this manuscript.
Mutations were found to target a small number of specific HLA class I alleles (Figure 1C; supplemental Table 2; supplemental Table 7). The most frequently targeted were HLA-B*40:02 (8 mutations in 2 patients) and HLA-B*14:02 (3 mutations in 2 patients). In addition, 1 patient each had a loss of HLA-A*68:01 and HLA-A*33:03. Of the 3 patients harboring both 6p CN-LOH and loss-of-function HLA mutations, 2 patients lost the same HLA allele through both mechanisms (supplemental Table 2), whereas the small 6p CN-LOH clone size in the third patient precluded definitive identification of the affected haplotype.
Somatic mutations recurrently target HLA predisposition alleles
We hypothesized that recurrent loss of specific HLA alleles is a marker of their pathogenic significance in aAA. Thus, HLA alleles targeted by somatic mutations would be predicted to be more frequent in patients with aAA compared with in the general population. When we compared the prevalence of the 4 HLA alleles targeted by somatic mutations (HLA-A*33:03, HLA-A*68:01, HLA-B*14:02, or HLA-B*40:02) with that of ethnicity-matched control patients,42 HLA-B*14:02 and HLA-B*40:02 were significantly overrepresented in aAA, at 28.6% vs 5.7% (P = .000) and 9.5% vs 2.5% (P = .021) for HLA-B*14:02 and HLA-B*40:02, respectively (Table 1; supplemental Table 3). Alleles HLA-A*33:03 and HLA-B*68:01, which are more common in African, Asian, South American, and Southeast Asian ethnicities,42 did not demonstrate enrichment in our predominantly Caucasian cohort (supplemental Table 3). We confirmed our results by comparing with other, independent, control cohorts, with similar results (supplemental Table 4). In contrast, neither HLA-B*14:02 nor HLA-B*40:02 was overrepresented in Caucasian patients with MDS, also treated at our center (Table 1; supplemental Table 4).
HLA-B*14:02 and HLA*B-40:02 are enriched in patients with aAA compared with race-matched control patients
| Population . | Individuals, n . | HLA-B*14:02 . | HLA-B*40:02 . | ||||
|---|---|---|---|---|---|---|---|
| Present, n (%) . | Absent, n (%) . | P . | Present, n (%) . | Absent, n (%) . | P . | ||
| USA NMDP European Caucasian | 1 242 890 | 71 093 (5.7) | 1 171 797 (94.3) | 31 321 (2.5) | 1 211 569 (97.5) | ||
| Aplastic anemia, white | 42 | 12 (28.6) | 30 (71.4) | .000 | 4 (9.5) | 38 (90.5) | .021 |
| MDS, white | 163 | 13 (8.0) | 150 (92.0) | .233 | 4 (2.5) | 159 (97.5) | 1.000 |
| Population . | Individuals, n . | HLA-B*14:02 . | HLA-B*40:02 . | ||||
|---|---|---|---|---|---|---|---|
| Present, n (%) . | Absent, n (%) . | P . | Present, n (%) . | Absent, n (%) . | P . | ||
| USA NMDP European Caucasian | 1 242 890 | 71 093 (5.7) | 1 171 797 (94.3) | 31 321 (2.5) | 1 211 569 (97.5) | ||
| Aplastic anemia, white | 42 | 12 (28.6) | 30 (71.4) | .000 | 4 (9.5) | 38 (90.5) | .021 |
| MDS, white | 163 | 13 (8.0) | 150 (92.0) | .233 | 4 (2.5) | 159 (97.5) | 1.000 |
USA NMDP, US National Marrow Donor Program.
P values refer to comparisons between patients with aAA or MDS, as compared with the race-matched USA NMDP European Caucasian control population. Statistically significant P values are shown in bold.
To evaluate whether any other HLA class I alleles were independently associated with aAA, we extended the association analysis to all HLA class I alleles present in more than 2 patients in our patient cohort (supplemental Table 5). As expected from their genetic linkage to HLA-B*14:02 in the common Caucasian haplotypes (supplemental Table 6), alleles HLA-A*33:01 and HLA-C*08:02 were also significantly overrepresented in patients with aAA, at 7.1% vs 1.6% (P = .031) and 28.6% vs 7.7% (P = .000), respectively, as compared with ethnicity-matched control patients (supplemental Table 5). This confirmed that the only independent associations in our cohort were with HLA-B*40:02 and HLA-B*14:02 and their genetically linked loci. The ability of clonal inactivating mutations to precisely identify pathogenic HLA alleles underscores a crucial distinction between our study and genetic association approaches, which cannot distinguish indirect associations with loci in linkage disequilibrium with a true causal variant.
HLA risk alleles predispose to more severe disease course and clonal complications
We reasoned that clonal expansion of hematopoietic cells that lost a particular HLA allele necessarily reflects their increased fitness within the aAA environment. The mutant cell clones clonally expand despite continuing to express all other unmutated HLA alleles. Thus, HLA alleles targeted for somatic loss, such as the 4 alleles identified in our study (HLA-A*33:03, HLA-A*68:01, HLA-B*14:02, and HLA-B*40:02, from here on referred to as “risk alleles”) likely have an enhanced ability to either present pathogenic autoantigen or autoantigens or otherwise activate auto-reactive lymphocytes. Therefore, we hypothesized that patients with aAA carrying HLA risk alleles may have a more severe disease course and, thus, compared clinical outcomes of 24 patients carrying at least 1 of these 4 risk alleles with 42 patients without risk alleles (Figure 1D; Table 2). The 2 groups had similar baseline characteristics, including age and disease severity at diagnosis, duration of follow-up, and time to IST (supplemental Table 1; supplemental Table 8).
Clinical outcomes of patients with aplastic anemia stratified by the presence or absence of HLA risk alleles
| Clinical characteristic . | Total patients (n = 66) . | HLA risk allele present* (n = 24) . | HLA risk allele absent (n = 42) . | P . |
|---|---|---|---|---|
| Response to first-line therapy, patients | ||||
| OR (CR+PR), n (%) | 35 (76.1) | 12 (70.6) | 23 (79.3) | .722 |
| CR, n (%) | 17 (40.0) | 7 (41.2) | 10 (34.5) | |
| PR, n (%) | 18 (39.1) | 5 (29.4) | 13 (44.8) | |
| None, n (%) | 11 (23.9) | 5 (29.4) | 6 (20.7) | |
| Not evaluable, n | 20 | 7 | 13 | |
| Complications, patients | ||||
| Multiple lines of therapy or disease-related death, n (%) | 19 (38.0) | 11 (57.9) | 8 (25.8) | .036 |
| Required additional therapy, n | 18 | 10 | 8 | |
| Disease-related death after fist- line therapy, n | 1 | 1 | 0 | |
| No second-line therapy or disease-related death, n (%) | 31 (62.0) | 8 (42.1) | 23 (74.2) | |
| Not evaluable, n | 16 | 5 | 11 | |
| Patients with acquired structural chromosomal rearrangements | .005 | |||
| Abnormal, n (%) | 15 (26.8) | 11 (47.8) | 4 (12.1) | |
| Normal, n (%) | 41 (73.2) | 12 (52.2) | 29 (87.9) | |
| Not evaluable, n | 10 | 1 | 9 | |
| Patients with acquired cytogenetic abnormalities | .071 | |||
| Abnormal, n (%) | 6 (10.7) | 5 (21.7) | 1 (3.0) | |
| Normal, n (%) | 50 (89.3) | 18 (78.3) | 32 (97.0) | |
| Not evaluable, n | 10 | 1 | 9 | |
| Patients with acquired CN-LOH, n (%) | .029 | |||
| Acquired CN-LOH present | 10 (15.2) | 7 (29.2) | 3 (7.1) | |
| 6p CN-LOH | 8 (12.1) | 5 (20.8) | 3 (7.1) | |
| No CN-LOH | 56 (84.8) | 17 (70.8) | 39 (92.9) | |
| HLA loss, patients, n (%) | .013 | |||
| HLA loss present | 11 (16.7) | 8 (33.3) | 3 (7.1) | |
| No HLA loss | 55 (83.3) | 16 (66.7) | 39 (92.9) | |
| MDS transformation, patients | .013 | |||
| MDS transformation, n (%) | 4 (8.0) | 4 (22.2) | 0 (0) | |
| No MDS transformation, n (%) | 46 (92.0) | 14 (77.8) | 32 (100) | |
| Not evaluable, n | 16 | 6 | 10 |
| Clinical characteristic . | Total patients (n = 66) . | HLA risk allele present* (n = 24) . | HLA risk allele absent (n = 42) . | P . |
|---|---|---|---|---|
| Response to first-line therapy, patients | ||||
| OR (CR+PR), n (%) | 35 (76.1) | 12 (70.6) | 23 (79.3) | .722 |
| CR, n (%) | 17 (40.0) | 7 (41.2) | 10 (34.5) | |
| PR, n (%) | 18 (39.1) | 5 (29.4) | 13 (44.8) | |
| None, n (%) | 11 (23.9) | 5 (29.4) | 6 (20.7) | |
| Not evaluable, n | 20 | 7 | 13 | |
| Complications, patients | ||||
| Multiple lines of therapy or disease-related death, n (%) | 19 (38.0) | 11 (57.9) | 8 (25.8) | .036 |
| Required additional therapy, n | 18 | 10 | 8 | |
| Disease-related death after fist- line therapy, n | 1 | 1 | 0 | |
| No second-line therapy or disease-related death, n (%) | 31 (62.0) | 8 (42.1) | 23 (74.2) | |
| Not evaluable, n | 16 | 5 | 11 | |
| Patients with acquired structural chromosomal rearrangements | .005 | |||
| Abnormal, n (%) | 15 (26.8) | 11 (47.8) | 4 (12.1) | |
| Normal, n (%) | 41 (73.2) | 12 (52.2) | 29 (87.9) | |
| Not evaluable, n | 10 | 1 | 9 | |
| Patients with acquired cytogenetic abnormalities | .071 | |||
| Abnormal, n (%) | 6 (10.7) | 5 (21.7) | 1 (3.0) | |
| Normal, n (%) | 50 (89.3) | 18 (78.3) | 32 (97.0) | |
| Not evaluable, n | 10 | 1 | 9 | |
| Patients with acquired CN-LOH, n (%) | .029 | |||
| Acquired CN-LOH present | 10 (15.2) | 7 (29.2) | 3 (7.1) | |
| 6p CN-LOH | 8 (12.1) | 5 (20.8) | 3 (7.1) | |
| No CN-LOH | 56 (84.8) | 17 (70.8) | 39 (92.9) | |
| HLA loss, patients, n (%) | .013 | |||
| HLA loss present | 11 (16.7) | 8 (33.3) | 3 (7.1) | |
| No HLA loss | 55 (83.3) | 16 (66.7) | 39 (92.9) | |
| MDS transformation, patients | .013 | |||
| MDS transformation, n (%) | 4 (8.0) | 4 (22.2) | 0 (0) | |
| No MDS transformation, n (%) | 46 (92.0) | 14 (77.8) | 32 (100) | |
| Not evaluable, n | 16 | 6 | 10 |
CN-LOH, copy number-neutral loss of heterozygosity; CR, complete response; MDS, myelodysplastic syndrome (patients were diagnosed with MDS-EB-1 [n = 1], MDS-U [single lineage dysplasia and pancytopenia; n = 1], MDS-U [based on defining cytogenetic abnormality (monosomy 7) and cytopenia]; n = 2); OR, overall response; PR, partial response.
P value pertains to the comparison of the 24 patients who carry at least 1 of the 4 HLA risk alleles* (HLA-A*33:03, HLA-A*68:01, HLA-B*14:02, or HLA-B*40:02) to the 42 patients who carry none of the HLA risk alleles.
The groups did not differ in early response to first-line IST with horse anti-thymocyte globulin and cyclosporine A with 6-month overall response rates of 70.6% in the risk allele group (12 of 17 evaluable patients) compared with 79.3% in patients without risk alleles (23 of 29; P = .722; Table 2). However, patients with risk alleles experienced a more severe clinical course later in disease, with more disease-related death, or refractory, relapsed, or transformed disease requiring additional therapy: 57.9% (11 of 19) evaluable patients in the risk allele group compared with 25.8% (8 of 31) patients without risk alleles (P = .036; Table 2).
Patients with risk alleles were also more likely to develop clonal complications (Table 2). More patients developed structural chromosomal rearrangements, at 47.8% (11 of 23) of patients with risk alleles compared with 12.1% patients without risk alleles (P = .005). Patients with risk alleles were more likely to develop HLA loss, at 33.3% (8 of 24) compared with 7.1% (3 of 42) in patients without risk alleles (P = .013). Four of 18 evaluable patients with risk alleles progressed to WHO-defined MDS (22.2%) compared with 0 of 32 patients lacking risk alleles (P = .013) (Table 2); of the 4, 1 had pediatric-onset aAA refractory to IST and developed MDS with der(5)t(1;5)(q11; q11.2) 3.5 years after diagnosis. After a median follow-up of 2.8 years, pediatric patients with risk alleles were more likely to develop HLA loss (7 of 15 patients, 46.7%) compared with MDS (1 of 15 patients, 6.7%; P = .035).
HLA risk alleles predispose to acquisition of recurrent somatic mutations in an age-related manner
To more comprehensively analyze clonal hematopoiesis, we used WES to compare somatic mutations in 17 patients with aAA with risk alleles with 28 patients without risk alleles; patients were selected for WES on the basis of the availability of paired bone marrow and skin DNA (Figure 1D). The 2 groups did not differ in baseline characteristics (supplemental Table 9).
Using all markers of clonality, patients with risk alleles had a trend toward more clonal hematopoiesis, at 82.4% (14 of 17) compared with 64.3% (18 of 28; supplemental Table 9; P = .311); the analysis was not powered for statistical significance for this effect size. The difference in clonal hematopoiesis was largely explained by more HLA loss in the risk allele group at 41.1% (7 of 17) compared with 7.1% (2 of 28; P = .009). Excluding HLA and PIGA mutations, ∼50% of patients in both groups had somatic mutations with a median of 0.5 to 1 mutations per patient (range, 0-18 mutations); the majority of mutations were present in both myeloid and lymphoid lineages, indicating the involvement of an early progenitor or stem cell (supplemental Table 10).
Importantly, patients with risk alleles were more likely to carry somatic alterations previously identified as recurrent in aAA,27,36,44,45 at a frequency of 64.7% in patients with risk alleles (11 of 17) compared with 17.9% in patients without risk alleles (5 of 28; P = .004; supplemental Table 9; supplemental Table 10). Recurrent somatic events in the risk allele group included HLA loss in 7 patients and MDS-associated mutations in 5 patients, including mutations in ASXL1 (n = 5), PHF6 (n = 2), RUNX1 (n = 2), DNMT3A (n = 1), SETBP1 (n = 1), SRSF2 (n = 1), and SUZ12 (n = 1), frequently co-occurring. In contrast, among 28 patients without risk alleles, only 2 patients had HLA loss and only 3 patients had MDS-associated mutations (ASXL1 [n = 1] and BCOR [n = 2]). Mutations associated with poor prognosis and progression (ASXL1, DNMT3A, RUNX1)44,46 occurred at a higher frequency of 29.4% (5 of 17) in the risk allele group as compared with 3.5% (1 of 28) in patients without risk alleles (P = .022; supplemental Table 9; supplemental Table 10).
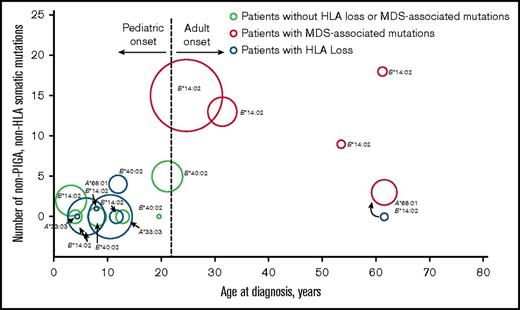
Manifestations of clonal hematopoiesis differed on the basis of age at aAA diagnosis (Figure 2). Among 17 patients with risk alleles, mutations in MDS-associated genes were found solely in adult-onset aAA (100%, 5 of 5) compared with 0% (0 of 12) of patients with pediatric-onset aAA (P = .000). The only recurrent events in pediatric-onset patients were PNH clones and HLA loss. This age-related dichotomy was most pronounced among patients with risk alleles: 6 of the 12 (50%) pediatric patients with risk alleles developed clonal HLA loss compared with only 1 of the 22 (4.5%) pediatric patients who did not carry risk alleles (P = .004). Similarly, patients with adult-onset aAA trended to more MDS-associated mutations if they had risk alleles, at 100% (5 of 5) in adult patients with risk alleles as compared with 33.3% (2 of 6) of patients without risk alleles (P = .061).
Age-related dichotomy of clonal hematopoiesis in aAA, manifested as frequent HLA loss in younger patients and as MDS-associated somatic mutations in older patients. A bubble scatter plot of somatic mutation analysis of the 17 patients with 1 of the 4 HLA risk alleles (HLA-A*33:03, HLA-A*68:01, HLA-B*14:02, or HLA-B*40:02); each patient is represented by a circle. The number of somatic nonsynonymous coding and regulatory region mutations identified by comparative WES is plotted on the y-axis, with the corresponding patient’s age at aAA diagnosis plotted on the x-axis; duration of disease at sequencing is depicted as the area of each point. PNH clones are not shown. Patients with HLA loss, as determined by the presence of either 6p CN-LOH or inactivating mutations in HLA alleles, are shown in blue. Patients with MDS-associated somatic mutations are shown in red. One patient had a transient clone of whole chromosome 6 CN-LOH early in disease course (blue circle accompanied by curved black arrow at the bottom right), which disappeared and was replaced by a dominant clone with MDS-associated mutations (red circle, indicated by a curved black arrow at the bottom right). HLA risk alleles are indicated next to each of the points; A, HLA-A; B, HLA-B.
Age-related dichotomy of clonal hematopoiesis in aAA, manifested as frequent HLA loss in younger patients and as MDS-associated somatic mutations in older patients. A bubble scatter plot of somatic mutation analysis of the 17 patients with 1 of the 4 HLA risk alleles (HLA-A*33:03, HLA-A*68:01, HLA-B*14:02, or HLA-B*40:02); each patient is represented by a circle. The number of somatic nonsynonymous coding and regulatory region mutations identified by comparative WES is plotted on the y-axis, with the corresponding patient’s age at aAA diagnosis plotted on the x-axis; duration of disease at sequencing is depicted as the area of each point. PNH clones are not shown. Patients with HLA loss, as determined by the presence of either 6p CN-LOH or inactivating mutations in HLA alleles, are shown in blue. Patients with MDS-associated somatic mutations are shown in red. One patient had a transient clone of whole chromosome 6 CN-LOH early in disease course (blue circle accompanied by curved black arrow at the bottom right), which disappeared and was replaced by a dominant clone with MDS-associated mutations (red circle, indicated by a curved black arrow at the bottom right). HLA risk alleles are indicated next to each of the points; A, HLA-A; B, HLA-B.
Most MDS-related mutations occurred in patients carrying the HLA-B*14:02 allele. These included all 4 patients with MDS transformation, 4 of the 5 patients with cytogenetic abnormalities, and 5 of the 8 patients with MDS-related mutations. The rates of MDS and cytogenetic abnormalities in patients with HLA-B*14:02 were significantly higher than in the rest of the cohort: 4 of 10 evaluable patients with HLA-B*14:02 (40%) developed MDS compared with 0 of 40 evaluable patients without HLA-B*14:02 (P = .003), and 4 of 13 evaluable patients with HLA-B*14:02 (30.8%) acquired cytogenetic abnormalities compared with 2 (4.7%) of 43 patients without HLA-B*14:02 (P = .022). Among 45 patients evaluated by WES, 5 of 11 patients with HLA-B*14:02 (45.5%) developed MDS-associated somatic mutations compared with 3 (8.8%) of 34 patients without HLA-B*14:02 (P = .014). Importantly, HLA-B*14:02 was the only risk allele represented within the relatively small number of adults in our study. Absence of the other 3 risk alleles in our adult subpopulation did not statistically deviate from their lower overall prevalence in our cohort.
HLA loss is sufficient for clonal expansion, but does not prevent further clonal evolution
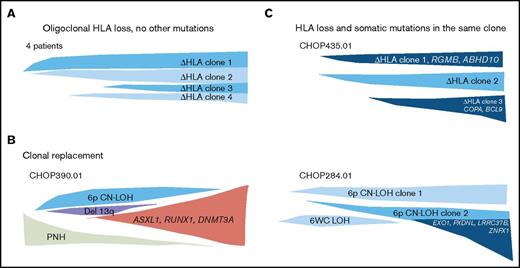
Because we did not observe co-occurrence of HLA loss and MDS-associated mutations, we asked whether HLA loss prevents further clonal evolution by abrogating HLA-restricted immune pressure on mutant cells. To evaluate this, we analyzed clonal architecture in 9 patients with HLA loss who were studied by comparative WES. None of these patients developed MDS during study follow-up. Four patients (44.4%) had no other nonsynonymous coding or regulatory region somatic mutations, suggesting HLA loss alone can act as a driver of clonal hematopoiesis. Five patients (55.6%) carried other somatic mutations (supplemental Table 10). Clonal architecture analysis showed that somatic mutations could be subclonal to HLA loss or could arise as a dominant, independent clone (Figure 3; supplemental Figure 3; supplemental Figure 4). Our results indicate that although somatic HLA loss is not associated with MDS, MDS-associated mutations, or with more general genomic instability, it also does not preclude further clonal evolution.
HLA loss is sufficient for clonal expansion, but does not prevent clonal evolution. A schematic diagram of 3 types of clonal architecture found in patients with somatic HLA loss. (A) Four patients had oligoclonal hematopoiesis with multiple independent events of HLA loss, without other nonsynonymous or regulatory region somatic mutations. (B) One patient had HLA loss and somatic mutations occurring in independent clones, where the clone bearing MDS-associated mutations expanded to replace the clone with HLA loss. (C) Two patients were found to have oligoclonal hematopoiesis with clones missing HLA alleles occurring in the same cells as somatic mutations, which were either subclonal or co-occurred with HLA loss in all cells analyzed.
HLA loss is sufficient for clonal expansion, but does not prevent clonal evolution. A schematic diagram of 3 types of clonal architecture found in patients with somatic HLA loss. (A) Four patients had oligoclonal hematopoiesis with multiple independent events of HLA loss, without other nonsynonymous or regulatory region somatic mutations. (B) One patient had HLA loss and somatic mutations occurring in independent clones, where the clone bearing MDS-associated mutations expanded to replace the clone with HLA loss. (C) Two patients were found to have oligoclonal hematopoiesis with clones missing HLA alleles occurring in the same cells as somatic mutations, which were either subclonal or co-occurred with HLA loss in all cells analyzed.
HLA risk alleles predispose to ongoing clonal evolution over time
We hypothesized that HLA risk alleles predispose to clonal complications by facilitating a sustained immune pressure, which selects for clonal expansion of cells with an increased fitness within the aAA environment. Thus, we studied clonal hematopoiesis longitudinally, using a combination of cytogenetics, SNP-A, and WES and/or targeted sequencing, as permitted by sample availability. Seven patients had serial follow-up by WES, and 11 adult patients were screened for mutations associated with hematologic malignancies by NGS.
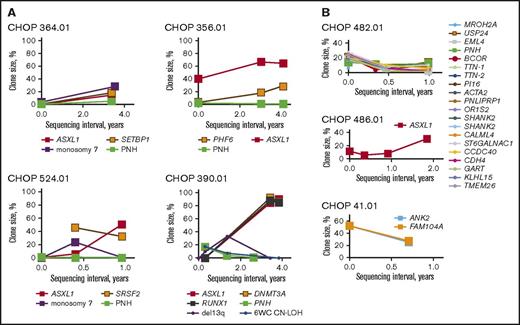
Clonal structure in risk allele patients was unstable over time, with frequent chromosomal alterations and emergence of somatic mutations (supplemental Table 11). All 5 patients with HLA risk alleles who had cytogenetic abnormalities exhibited karyotypic shift over the course of disease, including cytogenetic remission (n = 1), new chromosomal abnormalities (n = 2), and transient cytogenetic changes with clonal replacement (n = 2). Similarly, 2 of the 7 risk allele patients with acquired CN-LOH had dynamic changes, including emergence of 5q CN-LOH (n = 1) and a transient 6 whole chromosome CN-LOH (n = 1). Similarly, 1 patient without an identified risk allele, but with an acquired 6p CN-LOH clone, also showed dynamic changes with loss of 6 whole chromosome CN-LOH clone (supplemental Figure 4). Finally, serial follow-up showed that patients with risk alleles had large clone sizes and exhibited clonal expansion over time, whereas clonal dynamics in patients without risk alleles were more variable (Figure 4). Differences in duration of follow-up between the 2 groups limited our ability to draw more definitive conclusions from this comparison.
HLA risk alleles predispose to clonal expansion over time. Shown are the longitudinal analyses of frequencies of somatic alterations in patients with (A) and without (B) risk alleles. Clone size was estimated as percentage immunophenotypic PNH granulocytes for PNH, percentage abnormal metaphases for chromosomal abnormalities, percentage abnormal cells from SNP-A for 6p CN-LOH, and 2 × percentage NGS mutant reads for somatic mutations (to account for diploid status).
HLA risk alleles predispose to clonal expansion over time. Shown are the longitudinal analyses of frequencies of somatic alterations in patients with (A) and without (B) risk alleles. Clone size was estimated as percentage immunophenotypic PNH granulocytes for PNH, percentage abnormal metaphases for chromosomal abnormalities, percentage abnormal cells from SNP-A for 6p CN-LOH, and 2 × percentage NGS mutant reads for somatic mutations (to account for diploid status).
Discussion
Our results demonstrate that somatic loss-of-function mutations in HLA class I genes are common in aAA, second in frequency only to loss of PIGA. We found that inactivating mutations can identify HLA risk alleles, defined in our study as alleles, somatic loss of which increases hematopoietic cell fitness and leads to clonal expansion within an aAA environment. We identified 4 HLA risk alleles, HLA-A*33:03, HLA-A*68:01, HLA-B*14:02, and HLA-B*40:02, and have shown that patients carrying these alleles have a more severe disease course and are more likely to develop chromosomal abnormalities and other clonal changes, such as HLA loss in pediatric-onset aAA and, for HLA-B*14:02, MDS-associated mutations in adult-onset aAA. Our findings strongly support the role of HLA class I-mediated autoimmunity in aAA pathogenesis and establish a novel link between inheriting HLA risk alleles and the risk for clonal evolution.
Early studies in aAA focused on the role of HLA class II genes, particularly HLA-DR,14-19 which function in the context of antigen-presenting cells to present exogenous peptides for recognition by CD4+ T helper cells.47 In contrast, our data, as well as the data from several association studies24,26 and studies of 6p CN-LOH,27,48 suggest a pathogenic role of HLA class I-mediated autoimmunity, which normally serves to present endogenous antigens, such as those derived from viruses and intracellular bacteria, for recognition by CD8+ T cells.47 In genetic association studies, the high degree of polymorphism in the chromosome 6p genomic region, as well as its dense linkage disequilibrium, have made it exceedingly challenging to tease out the exact locations of etiologic variants in aAA. We identified several patients who had oligoclonal 6p CN-LOH concurrent with inactivating mutations in the same HLA class I allele, therefore identifying the loss of the HLA class I allele as the driving force of clonal evolution in these patients. Interestingly, a recent study employing WES in 52 patients with aAA,44 did not identify HLA gene mutations, whereas we found 13 mutations in 66 patients with aAA. One possible explanation for the discrepancy between our studies is that the MHC region is not amenable to standard WES analysis because of its high degree of polymorphism, leading to problems in accurate HLA alignment using standard methods.49 In addition, WES frequently does not achieve sufficient coverage of the MHC region to detect low-frequency mutations. Our study used more than 10 000× deep NGS optimized for analysis of the MHC region. Alternatively, our cohort included more patients with pediatric-onset aAA, in whom we find HLA mutations to be more common, whereas the study by Yoshizato and colleagues44 analyzed an older cohort enriched for patients with known MDS-associated mutations.44,50 Nevertheless, there are 2 recent case reports identifying mutations in HLA-B*40:02 in 2 East Asian patients with aAA,51,52 showing that these mutations occur in aAA, but require specialized detection strategies to be detected with high sensitivity.
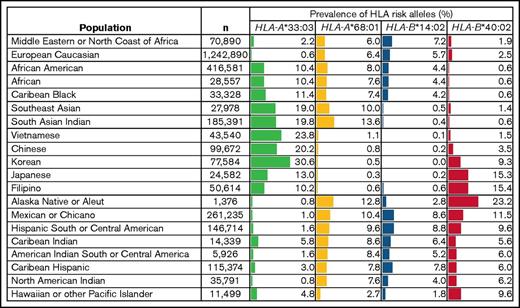
Indeed, while our manuscript was in preparation, Zaimoku and colleagues used flow cytometric methods to identify recurrent HLA-B*40:02 mutations in 28 Japanese patients with aAA carrying HLA-B*40:02 allele,53 pointing to the involvement of HLA-B*40:02 allele in aAA in Japan, which has a more restricted HLA pool and a significantly higher frequency of HLA-B*40:02 (Figure 5). Interestingly, the incidence of aAA is also 2- to 3-fold higher in East Asia.54 Although both our studies report HLA mutations in aAA, they have many differences. Unlike the HLA-B*40:02-specific approach by Zaimoku et al, we used an unbiased NGS screen to identify loss-of-function mutations in HLA class I genes in a more ethnically heterogeneous cohort of 66 American patients. Although Zaimoku et al studied adults, who commonly carry age-associated somatic mutations, our cohort was predominantly pediatric, an age range where somatic mutations are normally absent. Thus, our independent findings of HLA class I loss by different approaches, in different ethnicities and different age groups, powerfully validate each other and strongly point to the role of HLA class I-mediated autoimmunity in aAA pathogenesis. We also identified 3 HLA risk alleles beyond HLA-B*40:02 and showed that HLA-B*14:02 is of particular importance in aAA in North America. Another novel aspect of our study is the demonstration that having HLA risk alleles leads to a more complicated disease course and more clonal evolution, and that HLA-B*14:02 is linked to MDS-related somatic changes in adults, suggesting HLA allele heterogeneity likely underlies population-based differences in aAA course and complications.
Prevalence of the 4 aAA HLA risk alleles in worldwide populations. Shown is a table of the frequencies of the 4 HLA risk alleles in worldwide populations, compiled from the US National Marrow Donor Program HLA dataset.42,59 Frequencies are listed as percentage of individuals that have the allele in the population, depicted with the corresponding bar graph. n, number of individuals in each population cohort.
Prevalence of the 4 aAA HLA risk alleles in worldwide populations. Shown is a table of the frequencies of the 4 HLA risk alleles in worldwide populations, compiled from the US National Marrow Donor Program HLA dataset.42,59 Frequencies are listed as percentage of individuals that have the allele in the population, depicted with the corresponding bar graph. n, number of individuals in each population cohort.
The 4 HLA risk alleles identified in our study account for 36% of all patients in our cohort and 76% of patients with HLA loss. Although all patients with aAA are presumed to carry 1 or more HLA alleles capable of autoantigen binding and autoimmune T-cell activation, recurrent somatic loss of specific HLA alleles followed by clonal expansion of mutant clones suggests mutated alleles play a more significant role in mediating aAA pathogenesis compared with the unmutated HLA alleles within a given patient. Because of HLA allele heterogeneity in the population, loss of a specific HLA allele may not always result in the same degree of immune escape because of differences in remaining alleles. Of the 8 patients with 6p CN-LOH in our cohort, 3 carried no identified risk alleles. We hypothesize that HLA elements lost through 6p CN-LOH in these patients also contain HLA class I alleles of increased pathogenicity. Because not all pathogenic alleles were identified, our results likely underestimate deleterious effects of HLA risk alleles through inclusion of patients with yet-unidentified risk alleles in the “no risk allele” group.
The finding of increased clonal evolution in patients with HLA risk alleles was unexpected and indicates that hematopoietic cells in these patients experience a stronger selective pressure, likely related to enhanced autoantigen binding and/or stronger T-cell activation. We speculate that differences in clonal manifestations of patients with pediatric- and adult-onset aAA are related to the higher prevalence of subclinical age-associated mutations in adults.55-58 In pediatric population, where age-related mutations are rare, PIGA and HLA loss are the primary mechanisms of early bone marrow response to immune selective pressure. This may explain the lower rate of secondary MDS in pediatric-onset aAA, a population where after a median of 3.1-year follow-up in our study, only 1 (2.4%) of 41 patients with evaluable pediatric-onset aAA developed secondary MDS, whereas 19.5% (8 of 41) developed somatic HLA loss (P = .029).
Our study has limitations. aAA is rare, and the number of patients studied is relatively small; however, this is 1 of the largest aAA cohorts, with a comprehensive NGS analysis of HLA class I alleles and robust clinical correlation analysis. Our cohort is predominantly pediatric; thus, future studies will be needed to extend our analysis in adults. Because of our robust mutation validation requirements, our results underestimate HLA mutations below 5% allele frequency. Our in silico analysis did not identify a common structural element within the peptide binding cleft of the 4 HLA risk alleles. Instead, we favor existence of a small number of shared structural elements. Larger collaborative studies in different ethnic groups are necessary to validate our findings and to identify additional risk alleles for characterization of shared structural motifs.
In conclusion, we have shown that selected HLA class I alleles predispose to aAA, worsen the disease course, and increase the risk for clonal complications in an age-dependent manner. Our finding of somatic immunoediting of HLA class I alleles in aAA exposes the underlying mechanism of aAA autoimmunity, and opens new avenues for development of targeted therapies to prevent or redirect autoimmunity in aAA. Our data establish a novel link between inherited HLA alleles in patients with aAA and risk for clonal evolution and support long-term clinical follow-up of patients with aAA with HLA risk alleles, including screening for MDS-associated mutations in adult-onset aAA.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank all patients and their referring physicians for participation in our studies. The authors are grateful to Jian-Meng Fan and Carine Cattier for technical assistance, to Beverly Paul for clinical coordination, and to Philip Mason and Peter Klein and the members of Philip Mason’s and M.B.’s laboratories for helpful discussions. The authors thank the University of Pennsylvania and Children’s Hospital of Philadelphia (CHOP) hematopathology departments for pathology review, and Juan Perin for assistance with bioinformatic analysis.
This work was supported by an American Association for Cancer Research–Amgen, Inc. Fellowship in Clinical/Translational Cancer Research; Aplastic Anemia and MDS International Foundation Research Grant; National Institutes of Health, National Heart, Lung, and Blood Institute K08 HL132101 (D.V.B.); University of Pennsylvania/CHOP Institute for Translational Medicine and Therapeutics/ Clinical and Translational Research Center; Foerderer Grant; National Institutes of Health, National Heart, Lung, and Blood Institute K08 HL122306 (T.S.O.); National Institutes of Health, National Cancer Institute R01 CA105312; Buck Family Endowed Chair in Hematology (M.B.); and National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases R24DK103001 (J.A.B., S.T.C, and M.B.).
Authorship
Contribution: M.B. conceptualized the study of clonal hematopoiesis, initiated the project, and started the aAA patient registry, which is now housed at CHOP; D.V.B. initiated the study of clonal hematopoiesis and HLA loss in aAA, oversaw adult patients with aAA, designed the study, performed NGS, genetic, clinical, and statistical analyses, interpreted all data, and wrote the manuscript; T.S.O. initiated the study of HLA loss in aAA, performed clinical record review, oversees bone marrow failure patient enrollment, and leads the aAA patient registry; D.S.M. established the technology of HLA NGS at CHOP and oversaw HLA NGS; J.L.D. performed HLA typing and HLA bioinformatic analysis; H.M.X. performed WES and HLA bioinformatic analysis; N.S., J.A., H.H., and W.Y. performed Sanger validation of WES results, myeloid and lymphoid lineage analysis of mutations, and clonogenic assays; D.F. performed HLA NGS; P.N. enrolled patients into the registry, facilitated sample acquisition, and performed clinical record review; N.P. performed NGS and genetic analysis and Sanger validation; G.M.P. designed and oversaw human subjects’ research aspects of the project; Y.L. performed statistical analysis; J.A.B. established the SNP-A technology at CHOP and performed cytogenetic and SNP-A analysis at CHOP; J.K. and D.L.P. performed HLA association analysis in patients with MDS; J.J.D.M. established targeted NGS of MDS-associated mutations at the University of Pennsylvania and oversaw targeted NGS and cytogenetic analysis at the University of Pennsylvania; L.C.E. and S.T.C. provided immunology and hematopoiesis expertise; and all authors assisted in the interpretation of results, edited the manuscript, and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Daria V. Babushok, Division of Hematology-Oncology, Hospital of the University of Pennsylvania, PCAM 12 South, 3400 Civic Center Blvd, Philadelphia PA 19104; e-mail: daria.babushok@uphs.upenn.edu.