Abstract
The chronic behavior of mature lymphoid malignancies, with relapses occurring years apart in many patients, has until recently been unexplained. Patterns of relapse also differ vastly between disease entities, with some being highly curable by chemotherapy whereas others are destined to reemerge after treatment. Lately, the use of next-generation sequencing techniques has revealed essential information on the clonal evolution of lymphoid malignancies. Also, experimental xenograft transplantation point to the possible existence of an ancestral (stem) cell. Such a malignant lymphoid stem cell population could potentially evade current therapies and be the cause of chronicity and death in lymphoma patients; however, the evidence is divergent across disease entities and between studies. In this review we present an overview of genetic studies, case reports, and experimental evidence of the source of mature lymphoid malignancy and discuss the perspectives.
Introduction
The definitive cure of lymphoid malignancy has long been a challenge for clinicians and scientists. The introduction of the MOPP (nitrogen mustard, vincristine, procarbazine, and prednisone) chemotherapy regimen in the 1970s first sparked hopes about achieving this goal.1 Since then, promising new therapies have emerged; high-dose therapy with stem cell support in the late 1990s,2-4 immunochemotherapy in the 2000s,5 and, recently, small molecule inhibitors.6-8 However, long-term follow-up after state-of-the-art treatments of follicular lymphoma (FL), mantle cell lymphoma (MCL), T-cell lymphoma (TCL), hairy cell leukemia (HCL), mucosa-associated lymphoid (MALT) tissue lymphoma, and chronic lymphocytic leukemia (CLL) still reveal persistent risks of relapse and no signs of plateau in survival curves.7-15 Most indolent lymphoid entities are believed to be incurable with present therapies as opposed to aggressive lymphomas, which are highly curable with chemotherapy. In addition, combinations of novel drugs have revealed serious side effects, which hamper attempts to achieve durable remissions.16 Identification and elimination of the residual source of malignancy is indeed challenging and a crucial step for cure of these diseases.
In acute leukemia and solid tumors, major advances have been made in the understanding of cancer stem cells with unique mapping of cells and novel genetic sequencing approaches.17,18 In the field of lymphoid malignancies, the cancer stem cell theory has not been investigated until very recently, and the chronic nature of mature lymphoid disorders, often with relapses occurring years after apparently successful treatment, is so far unexplained. Several recent investigations have identified new pieces of this puzzle, with different conclusions drawn.19-24
Next-generation sequencing (NGS) of sequential paired tumor samples point to ancestral clones that initiate both the first lymphoid tumor and subsequent relapse tumor.25-28 In addition, several case stories report the onset of identical lymphoid malignancy in both donor and recipient after allogeneic hematopoietic stem cell (HSC) transplantation.29-34 Collectively, these results imply that at least some lymphomas may derive from an underlying premalignant clone, indicating the existence of quiescent lymphoma stem cells with potential for malignant transformation. The phenotype of these cells, their origin, and niche are yet to be characterized. Identification of the initiating clone will perhaps enable the design of a targeted treatment to avoid/decrease subsequent relapses.
We present here an overview of the current clinical and biological evidence (or lack thereof) of malignant lymphoid stem cells and present implications for future research.
Origin of lymphoid malignancy
Normal lymphoid development
To follow the pathogenic concepts presented in this review, a short summary of the normal formation of mature B and T cells is provided. The classic characteristic of stem cells (healthy as well as malignant) is their ability to self-renew and maintain long-term clonal growth, which is assessed by functional repopulation assays.17 These analyses involve either a series of transplantation of cells in serial (animal) recipients or in situ tracking in a patient or an animal in which the same clone/stem cell is repeatedly identified. The cells are usually characterized by the CD34+ CD38− cell surface markers, both in normal and malignant hematopoiesis. The ancestral cell, which gives rise to B, T, and natural killer (NK) cells, is considered the multilymphoid progenitor, which also can develop into monocytes, macrophages, and dendritic cells. This entity has been found in humans to be a subpopulation of the CD34+CD38− HSC compartment marked by CD90neg–loCD45RA+.35 The exact steps in the subdivision between B-, T-, or NK-cell progenitors are not clear and likely a continuous process, as shown for B cell evolution.36
After commitment to the B-cell lineage, a series of events leads to the formation of mature B cells, which function in the adaptive immune response. The early stages of B-cell development (from pro- to pre- to immature B cell) are clearly identified by consecutive ordered rearrangements of the immunoglobulin (Ig) gene segments, first the heavy chain (IgH) and later the light chain (IgL) genes, creating a wide variety of possible immunoglobulin conformations.37 When the Ig chain sequences are settled, the antigen receptor, or B-cell receptor (BCR), is expressed on the cell surface and tested for reactivity with self-antigens. Autoreactive cells can reedit the IgL genes (so-called receptor editing) and thereby gain tolerance to self-antigens. Activation of the BCR is also important for proper B-cell maturation. Loss of the BCR leads to back-differentiation (upregulation of genes characteristic of earlier stages of B-cell development) and inhibition of receptor editing.38 Once matured, the naive B cells leave the bone marrow and enter the lymph node germinal center. Here they interact with antigens and can become antibody-producing plasma cells. T cells follow similar developmental stages from lymphoid progenitors that enter the thymus to fully mature circulating T cells. Activation of the Notch receptor is essential for T-cell lineage commitment37 ; comparable to B cells, T cells follow maturation by rearranging their α-, β-, and γ-receptor genes. In the thymus they are also tested for self-tolerance and ultimately mature as either CD4 or CD8 single-positive T cells that migrate to the periphery.
Whether some cells can carry oncogenic features from a stem cell population through the extensive developmental steps to a fully (im)mature B/T cell, which becomes malignant, is controversial and will be discussed later. It is unknown if such ancestral defects lead to so-called “cancer stem cells” that potentially cause relapse of chronic lymphoid malignancies. Or if back-differentiation of malignant cells can entail stem cell properties that lead to a chronic disease state. The nomenclature of cancer stem cells can be confusing, especially because of the difficult separation of true cancer stem cells (identified by serial transplantation), “ordinary” cancer cells, and premalignant/transitory cells.17 This is further complicated by diseases (such as lymphoma) in which no cell markers of cancer stem cells have been identified.
Evidence of lymphoma-initiating cells: allogeneic transplantation of premalignant clones
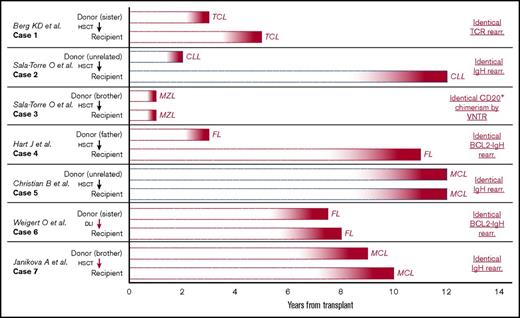
The existence of lymphoma stem cells is supported by several case stories showcasing apparent transmission of premalignant lymphoid cells from 1 individual to another. Allogeneic HSC transplantation allows transmission of not only healthy HSCs, but also disease-causing clones of both myeloid and lymphoid phenotypes, which can develop into full-blown hematological malignancy in the recipient (and donor). To our knowledge, 7 cases exist in which both recipient and donor develop the same lymphoid disease after allogeneic transplant (Figure 1).29-34 The time lapse between allogeneic transplant and disease onset ranges from 10 months to 12 years, which indicates the presence of a premalignant progenitor cell. Each case also demonstrates genetic sequence homology between the tumors of the recipient and the donor, thereby excluding a chance finding. One of the most intriguing cases (Figure 1, case 5) describes simultaneous occurrence of mantle cell lymphoma in both male donor and unrelated female recipients 12 years after allogeneic stem cell transplantation for acute myeloid leukemia. The malignant clone was found to be derived from the male donor (tumor with Y chromosome), thereby supporting the origin of a latent premalignant clone. However, it was not possible to identify the malignant clone by immunoglobulin-specific polymerase chain reaction (PCR) in a stored sample of the allogeneic transplant. In 2 other cases (Figure 1, cases 6 and 732,33 ), tumor markers were identified in earlier samples (ie, in stored bone marrow sample and in donor-leukocyte infusion, respectively).
Cases presenting onset of identical lymphoid malignancy in both donor and recipient after allogeneic hematopoietic stem cell transplantation. Seven cases of apparent transmission of a premalignant clone after transplantation of stem cell material (hematopoietic stem cell transplantation) or DLI. A red arrow from donor to recipient indicates the malignant clone was identified in stored material before onset of disease. Red bars specify transplant from related donor; blue bars specify transplant from unrelated donor. The text on the right indicates the molecular evidence of related clonal disease between the 2 patients in each case. DLI, donor leukocyte infusion; MZL, marginal zone lymphoma; TCR, T-cell receptor; rear., rearrangement; VNTR, variable number tandem repeats.
Cases presenting onset of identical lymphoid malignancy in both donor and recipient after allogeneic hematopoietic stem cell transplantation. Seven cases of apparent transmission of a premalignant clone after transplantation of stem cell material (hematopoietic stem cell transplantation) or DLI. A red arrow from donor to recipient indicates the malignant clone was identified in stored material before onset of disease. Red bars specify transplant from related donor; blue bars specify transplant from unrelated donor. The text on the right indicates the molecular evidence of related clonal disease between the 2 patients in each case. DLI, donor leukocyte infusion; MZL, marginal zone lymphoma; TCR, T-cell receptor; rear., rearrangement; VNTR, variable number tandem repeats.
The time course of lymphoma development and onset are somewhat parallel in most cases. However, in cases 2 and 4, the onset of lymphoid malignancy was delayed many years in the recipient compared with the donor. This could imply an important role of the tumor microenvironment in promoting or suppressing malignancy. Also, in case 4, a different IgH sequence was found in the 2 tumors. This makes it unlikely that the donor tumor was derived from a circulation tumor cell and supports the theory of a common lymphoma progenitor cell. Collectively, these 7 cases illustrate the prolonged natural history of lymphomagenesis and suggest the existence of cancer stem cell–like cells in these diseases. What drives or nurtures the putative lymphoma stem cells from a dormant state to overt lymphoid cancer remain to be elucidated.
Somatic mutations and CD34+ stem cells
In recent years, research has shed light on possible dormant premalignant lymphoid clones in healthy individuals. Two seminal papers found that specific somatic mutations in peripheral blood cells are detected in healthy adults and precede the onset of hematologic malignancy.20,21 The frequency increases by age; by age 70, ∼10% of healthy individuals have common aberrations. These mutations primarily occur in the genes encoding epigenetic regulators (eg, DNMT3A, TET2, ASXL1), which are mainly involved in myeloid cancers, but also occur in lymphoid malignancies.39-41 The presence of these specific somatic mutations has subsequently been termed clonal hematopoiesis of indeterminant potential (CHIP),42 which can be perceived as an analogy to monoclonal gammopathy of undetermined significance (MGUS), the premalignant state of multiple myeloma. Individuals who carry a CHIP mutation have a highly increased risk (hazard ratio, ∼12 [adjusted for age and sex]), of developing a myeloid or lymphoid cancer several years later. However, this outcome is rare and the positive predictive value of a CHIP mutation for developing hematological disease is low. MGUS also confer an increased risk of developing lymphoma, although this is more moderate, with a relative risk of 2.4 to 3.7. Additionally, individuals with MGUS have no heightened risk of developing myeloid disease.43,44 A recent study of patients undergoing autologous transplantation for lymphoma showed that CHIP mutations were highly prevalent (found in ∼30% of harvest samples) and were associated with inferior survival, but not with an increased rate of relapse.45 Hence, the importance of these mutations in clinical practice, especially regarding lymphoid malignancies, remains to be settled.
In spite of the unknown clinical importance of these findings, they imply a link between presumable stem cell mutations and subsequent lymphoid cancer. This is opposed to the currently accepted cell of origin concept, which suggests development from the various differentiated B- and T-cell subsets.46,47 Lymphoma and CLL are generally not considered to originate from mixed-linage precursors, such as identified by mixed-lineage leukemia translocations in acute leukemias.48 However, there are cases of composite lymphoma in which 2 different malignant lymphoid histologies are present in the same biopsy.49-51 Also, there is evidence of several separately developed B-cell clones (2 distinct variable-diversity-joining gene recombinations) in the same sample from patients with lymphoproliferative disorders.52 These are relatively rare events and also suggest a common origin of malignancy in the B-cell lineage.
There is now a growing amount of evidence supporting a stem cell origin of some lymphoid tumors (Table 1). Very recently, it has been shown that most patients with CLL (∼90%) harbor the same mutations in their CD34+ stem cells as in their malignant CD19+ bone marrow cells.22 The genetic changes most often shared between the stem cells and the mature malignant cells were NOTCH1 and XPO1 mutations, which were identified both by NGS and fluorescence in situ hybridization. Some common CLL-mutations (TP53, FBXW7, and SF3B1) were, however, primarily found in the mature B lymphocytes, leading to speculations of a hierarchy of early and late mutation onset. Mutations in the stem cells of CLL patients have also been found in the peripheral blood, although only in about one-half of the examined patients.19 In HCL, there is solid evidence of HSC involvement. Mutations in the BRAF V600E gene, which is found in the vast majority of bone marrow samples from HCL patients, has also been identified in the CD34+ stem cells of these patients.53 These findings are striking because both CLL and HCL have traditionally been considered lymphatic malignancies with a mature B-cell origin.
Genetic analysis of stem and progenitor cells in lymphoid disease
| Material . | CLL . | DLBCL . | FL . | MCL . | HCL . | TCL . |
|---|---|---|---|---|---|---|
| CD34+ HSC | +mut19,22 | −mut23 | — | — | +mut53 | +mut41,103 |
| CLP | +mut19 | — | +mut25,26 (in silico) | CD45+ with t(11;14)101 | +mut53 | — |
| Normal tissue (blood sample, buccal swabs) | — | +mut41 | — | — | +mut114 | +mut41,54 |
| Proposed COO86 | Naïve B cell | Activated/germinal center B cell | Centrocyte | Mantle cell | Mature B cell | Different T-cell subtypes |
| Material . | CLL . | DLBCL . | FL . | MCL . | HCL . | TCL . |
|---|---|---|---|---|---|---|
| CD34+ HSC | +mut19,22 | −mut23 | — | — | +mut53 | +mut41,103 |
| CLP | +mut19 | — | +mut25,26 (in silico) | CD45+ with t(11;14)101 | +mut53 | — |
| Normal tissue (blood sample, buccal swabs) | — | +mut41 | — | — | +mut114 | +mut41,54 |
| Proposed COO86 | Naïve B cell | Activated/germinal center B cell | Centrocyte | Mantle cell | Mature B cell | Different T-cell subtypes |
—, material has not been genetically analyzed with genetic sequencing; CLP, common lymphoid progenitor; COO, cell of origin; mut, mutation; +mut, material in the specified disease has been found to have a genetic defect also found in the lymphoid tumors; −mut, material in the specified disease has not been found to have a genetic defect comparable to that of lymphoid tumors.
In patients with TCL, mutation of the TET2 gene has especially been described in CD34+ stem cells. A paper by Quivoron et al41 identified 5 patients with TET2 mutations in both their tumor sample and matched normal DNA (Table 1), indicating a common stem cell progenitor. Colony assays of sorted CD34+ stem cells from 1 patient with TCL showed TET2 mutation in 7% of grown colonies. Another study also identified 4 patients who both had TET2 mutations in tumor sample and reference material.54 Three of these patients also carried DNMT3A mutations in their apparently normal tissue.
There is less evidence to support a stem cell origin of diffuse large B-cell lymphoma (DLBCL). In a recent study of 60 patients with DLBCL, copy-number alterations of the nontumor cells (noninfiltrated peripheral blood and bone marrow) were analyzed.24 In cells, which were expected to be nontumor control samples, the investigators observed copy-number neutral loss of heterozygosity in key lymphoma regions. These regions included 6p21 (HLA, 3% of patients), 9p24.1 (PD-L1/L2, 5% of patients), and 17p13.1 (TP53, 2.5% of patients). In another study, however, deep sequencing of genes frequently mutated in DLBCL revealed no mutations in highly purified HSCs from 6 DLBCL patients.23 Serial sequencing of diagnostic and relapse samples from the 6 DLBCL cases revealed common somatic mutations and clonal IgH variable-diversity-joining rearrangements, but none was observed in purified HSCs from the same patients. It was suggested that the cellular source of DLBCL relapses is of a more mature B-cell type than in low-grade B-cell malignancies. These observations are concordant with the fact that, compared with other types of non-Hodgkin lymphoma, de novo DLBCL rarely shows chronicity and late relapses. Indeed, 2 years after successful first-line chemotherapy for DLBCL, the relapse rate is low and overall survival is similar to the general population.55
Potential malignant stem cells of FL have not been investigated since the 1990s. Two small studies investigated highly purified CD34+ stem cells from FL patients (12 and 7 individuals, respectively) and analyzed the genetic hallmark translocation of chromosome 14 to 18, which is phenotypical for this lymphoma subtype. The vast majority of patients did not have expression of the t(14;18)-fusion gene PCR amplicon in their HSCs, and the few that did were considered a matter of contamination.56,57 To our knowledge, no other disease-related genetic aberrations have since been analyzed in HSCs of patients with indolent lymphoma (FL and MCL).
Clonal lymphoid markers in healthy individuals
Known pathogenic markers of CLL, FL, and MCL (respectively, monoclonal B-cell lymphocytosis [MBL], the t(14;18) translocation, and the t(11;14) translocation) are not isolated to detection in diseased patients, but are also to a large extent found in the general population. High-sensitivity flow cytometry has found CLL-like clones in 3.5% to 12% of healthy individuals over 40 years,58 and only persons with >500 clonal B cells per microliter have a small annual risk (1% to 2%) of requiring treatment for manifest CLL. Both MBL and CLL clones use a restricted BCR arrangement (especially VH1, VH3, and VH4 genes). This indicates that antigenic drive contributes to the transition from benign MBL to manifest CLL.59,60 In FL tumors, the t(14;18) translocation leads to expression of the fusion gene BCL2-IGH, but it is also possible to find t(14;18)-positive cells at low levels in the peripheral blood of about one-half of healthy individuals.61 It has been suggested that the additional molecular aberrations required to develop fulminant FL are caused by both a permissive microenvironment and genetic alterations such as CREBBP mutations.18,62 The characteristic t(11;14) translocation in MCL can be found in 1% to 7% of the healthy population by sensitive PCR methods.63,64 This translocation is in itself insufficient to generate MCL, and additional genetic defects are required.65 The common nature of the known pathogenic marks imply that early identification of premalignant lymphoid cells is not possible at the moment, but novel sequencing techniques may change this.
Role of the microenvironment in development and survival of lymphoid malignancy
Despite the fact that many healthy individuals have clonal lymphoid proliferation, few develop overt cancer. The spark that initiates this may be loss of repression or activation by an environmental source. This is probably the case for MALT lymphoma, which is associated with chronic infection (typically Helicobacter pylori) and thereby chronic antigenic stimulation. MALT lymphoma immunoglobulins are known to have affinity for foreign antigens from Helicobacter species, but also for self-antigens.66 On the other hand, a mouse model of this disease has been generated by inducing MALT1 overexpression (a recurrent characteristic of MALT lymphoma) in murine HSC.67 Additional genetic abrogation by TP53 deletion led to transformation to aggressive lymphoma. Thus, based on these studies, this disease entity displays a complex interplay between stem cells, genetics, autoreactivity, and environmental factors.
Chronic antigenic stimulation is also a plausible mechanism of initiation or progression toward overt cancer in other lymphoid malignancies. In CLL, leukemic B cells express a distinct restricted repertoire of antibodies and similar antigen receptors in multiple patients, which suggests a pathogenic drive by specific antigens.68-70 There is evidence that these lymphoid clones may be derived from autoreactive precursors, which have been activated by self-antigens.71 Selection of early CLL cells may be mitigated by secondary immunoglobulin rearrangements, which are caused by self-antigen–driven receptor editing.72 In addition, cell-autonomous (independent of antigen) activation of the BCR has been identified in CLL.73 Increasing amounts of research in CLL also support that key components of the microenvironment, such as macrophages, monocytes, and nurse-like cells, are essential for the survival of malignant cells.74-76
MCL also has a preferential usage of IgH genes associated with autoreactivity.77,78 A recent study showed that in vitro coculture of MCL cells with CD40L-expressing T cells and macrophages promote primary MCL cell survival and proliferation.79
In at least 25% of FL, the tumor cells have been shown to recognize and react to self-antigens.80 However, there is no evidence of shared antigen recognition between patients in FL. The role of immunity in FL has also been investigated in a small clinical trial that examined intratumoral injection of autologous dendritic cells to specific FL tumor sites.81 After injection, universal lymphoma regression was seen in some patients, which suggests an important role for T-cell response in FL.
The response rates of checkpoint inhibitor treatment (PD1/PDL1-blockade) in lymphoid malignancies have underscored the impact of the T-cell function in tumor survival. This is particularly the case in Hodgkin lymphoma, in which the majority of cells in a tumor belong to the microenvironment. Recently, it has been shown that nearly all Hodgkin lymphomas harbor genetic alterations of the PDL1 or PDL2 loci,82 which likely explains the high response rates of checkpoint blockade monotherapy in this disease.
Treatment with novel drugs has also unveiled the function of the tumor microenvironment in lymphomagenesis. Blocking of Bruton tyrosine kinase (BTK) and phosphatidylinositol 3-kinase in CLL and MCL patients induce peripheral lymphocytosis. This was shown to be triggered by impaired BCR signaling and chemokine sensitivity, which lead to decreased extracellular matrix adhesion and homing of malignant cells to the lymph nodes.83,84 Whether blocking of this mechanism will lead to fewer relapses and long-term survival is yet to be settled. New regimens combining chemotherapy and BCR inhibition, as used in the German CLL13 trial (NCT02950051), the European MCL Triangle trial (NCT02858258), and the global DBL3001 trial on non-germinal center B-cell DLBCL (NCT01855750), are likely to give the answer.
Role of clonal genetic alterations in relapse
Evidence from sequential tumor samples
When patients with lymphoid malignancies relapse after therapy, the new tumor most often shares the same histological phenotype as described in the original biopsy.85 However, the genetic aberrations in primary tumor may not be identified in the relapse sample and vice versa. Comparison of the genetics between initial tumor and subsequent relapse material has provided considerable insight in the nature of a presumed ancestral clone (Table 2). One of the key explanatory papers, by Okosun et al, focused on serial relapse samples from patients with FL over a timespan of up to 16 years.25 Sequential lymphoma samples from 10 patients were subject to whole-genome or whole-exome sequencing, which revealed that the serial tumors mainly shared mutations in genes involved in epigenetic histone modification. Single nucleotide polymorphism array of 29 matched pairs of FL and their transformed counterparts revealed a high level of shared clonality. In addition to epigenetic modifiers, mutations of the STAT6 and TNFRSF14 genes were found to originate from the founder clone. Other genetic studies of sequential tumor samples have also identified these mutations and others that are recurrent at diagnosis and relapse; an overview can be seen in Table 2.26,28,86-89 The recurrent mutations are dispersed over the whole genome, and many have been found in multiple lymphoid histological subtypes. The proteins encoded by mutated genes in paired patient samples primarily have roles in chromatin regulation (KMT2D, CREBBP, MEF2B, EZH2, EP300), the DNA-damage pathway (TP53, ATM, SF3B1), BCR signaling (CD79, TNF(R)SF14, FAS, FOXO1, EBF1), and NF-κβ pathway (CARD11, TNFAIP3, NFKBIE, MYD88). It can thus be hypothesized that survival of dormant malignant clones, under the pressure of therapy, is governed through the pathways disrupted in tumors both at diagnosis and at subsequent relapse. It is tempting to speculate that postinduction/maintenance therapy with drugs targeting these pathways may reduce the incidence of relapse.
Recurrent gene mutations in both primary and relapse tumor samples from patients with mature lymphoid malignancies reported in the literature
| Gene . | Chromosome . | Protein function . | Lymphoid histologies . | Material . | No. of studies . | Reference . |
|---|---|---|---|---|---|---|
| TP53 | 17 | DNA-damage pathway | DLBCL, MCL, FL, CLL | PRT | 9 | 26, 27, 87, 115,,,,-120 |
| KMT2D (MLL2) | 12 | Chromatin regulation | DLBCL, MCL, tFL, FL | PRT | 8 | 25, 26, 28, 86,,,-90 |
| CREBBP | 16 | Chromatin regulation | DLBCL, tFL, FL | PRT | 7 | 25,-27, 86, 87, 89, 90 |
| MEF2B | 19 | Chromatin regulation | DLBCL, tFL, MCL | PRT | 6 | 25,-27, 87,-89 |
| BCL2 | 18 | Apoptosis pathway | DLBCL, tFL, FL | PRT | 6 | 26, 27, 87, 89, 90, 121 |
| EZH2 | 7 | Chromatin regulation | DLBCL, tFL, FL | PRT | 6 | 17, 18, 87, 89, 90, 122 |
| CARD11 | 7 | NF-κB pathway | DLBCL, FL, MCL | PRT | 5 | 25,-27, 87, 88 |
| MYD88 | 3 | NF-κB pathway | DLBCL, tFL, CLL | PRT | 4 | 26, 27, 87, 119 |
| ARID1A | 1 | Chromatin regulation | DLBCL, tFL, FL | PRT | 4 | 26, 87, 89, 122 |
| TNFAIP3 | 6 | NF-κB pathway | FL, tFL, DLBCL | PRT | 4 | 25,-27, 90 |
| B2M | 12 | MHC class 1/immunology | DLBCL, FL, MCL | PRT | 3 | 26, 87, 90 |
| EP300 | 22 | Chromatin regulation | DLBCL, FL | PRT | 6 | 25,,-28, 90, 122 |
| CD79B | 17 | BCR signaling pathway | DLBCL, tFL | PRT | 4 | 25, 27, 87, 89 |
| NOTCH1 | 9 | NOTCH pathway | CLL, tFL | PRT | 4 | 26, 119, 121, 123 |
| HIST1H1 | 6 | Apoptosis pathway | tFL, FL | PRT | 4 | 25, 26, 89, 122 |
| NFKBIE | 6 | NF-κB pathway | DLBCL, CLL | PRT | 3 | 87, 119, 124 |
| STAT6 | 12 | JAK-STAT pathway | DLBCL, tFL | PRT | 3 | 25, 26, 87 |
| TNF(R)SF14 | 1 | BCR signaling pathway | DLBCL, tFL | PRT | 3 | 25, 26, 87 |
| GNA13 | 17 | Apoptosis pathway | DLBCL, tFL | PRT | 3 | 26, 27, 89 |
| ATM | 11 | DNA-damage pathway | MCL, CLL | PRT | 3 | 88, 119, 120 |
| FAS | 10 | BCR signaling pathway | DLBCL, tFL | PRT | 2 | 87, 90 |
| FOXO1 | 13 | BCR signaling pathway | DLBCL, tFL | PRT | 2 | 26, 87 |
| MGA | 15 | Suppressor of MYC pathway | tFL, CLL | PRT | 2 | 89, 118 |
| PIM1 | 6 | Apoptosis pathway | DLBCL, FL | PRT | 2 | 26, 27 |
| SF3B1 | 2 | DNA-damage pathway | CLL | PRT | 4 | 118,-120, 125 |
| EBF1 | 5 | BCR signaling pathway | tFL | PRT | 2 | 25, 26 |
| IRF8 | 16 | Interferon signaling | tFL | PRT | 2 | 26, 89 |
| RRAGC | 1 | mTORC signaling | tFL | PRT | 2 | 89, 126 |
| SOCS1 | 16 | JAK-STAT pathway | tFL | PRT | 2 | 25, 26 |
| Gene . | Chromosome . | Protein function . | Lymphoid histologies . | Material . | No. of studies . | Reference . |
|---|---|---|---|---|---|---|
| TP53 | 17 | DNA-damage pathway | DLBCL, MCL, FL, CLL | PRT | 9 | 26, 27, 87, 115,,,,-120 |
| KMT2D (MLL2) | 12 | Chromatin regulation | DLBCL, MCL, tFL, FL | PRT | 8 | 25, 26, 28, 86,,,-90 |
| CREBBP | 16 | Chromatin regulation | DLBCL, tFL, FL | PRT | 7 | 25,-27, 86, 87, 89, 90 |
| MEF2B | 19 | Chromatin regulation | DLBCL, tFL, MCL | PRT | 6 | 25,-27, 87,-89 |
| BCL2 | 18 | Apoptosis pathway | DLBCL, tFL, FL | PRT | 6 | 26, 27, 87, 89, 90, 121 |
| EZH2 | 7 | Chromatin regulation | DLBCL, tFL, FL | PRT | 6 | 17, 18, 87, 89, 90, 122 |
| CARD11 | 7 | NF-κB pathway | DLBCL, FL, MCL | PRT | 5 | 25,-27, 87, 88 |
| MYD88 | 3 | NF-κB pathway | DLBCL, tFL, CLL | PRT | 4 | 26, 27, 87, 119 |
| ARID1A | 1 | Chromatin regulation | DLBCL, tFL, FL | PRT | 4 | 26, 87, 89, 122 |
| TNFAIP3 | 6 | NF-κB pathway | FL, tFL, DLBCL | PRT | 4 | 25,-27, 90 |
| B2M | 12 | MHC class 1/immunology | DLBCL, FL, MCL | PRT | 3 | 26, 87, 90 |
| EP300 | 22 | Chromatin regulation | DLBCL, FL | PRT | 6 | 25,,-28, 90, 122 |
| CD79B | 17 | BCR signaling pathway | DLBCL, tFL | PRT | 4 | 25, 27, 87, 89 |
| NOTCH1 | 9 | NOTCH pathway | CLL, tFL | PRT | 4 | 26, 119, 121, 123 |
| HIST1H1 | 6 | Apoptosis pathway | tFL, FL | PRT | 4 | 25, 26, 89, 122 |
| NFKBIE | 6 | NF-κB pathway | DLBCL, CLL | PRT | 3 | 87, 119, 124 |
| STAT6 | 12 | JAK-STAT pathway | DLBCL, tFL | PRT | 3 | 25, 26, 87 |
| TNF(R)SF14 | 1 | BCR signaling pathway | DLBCL, tFL | PRT | 3 | 25, 26, 87 |
| GNA13 | 17 | Apoptosis pathway | DLBCL, tFL | PRT | 3 | 26, 27, 89 |
| ATM | 11 | DNA-damage pathway | MCL, CLL | PRT | 3 | 88, 119, 120 |
| FAS | 10 | BCR signaling pathway | DLBCL, tFL | PRT | 2 | 87, 90 |
| FOXO1 | 13 | BCR signaling pathway | DLBCL, tFL | PRT | 2 | 26, 87 |
| MGA | 15 | Suppressor of MYC pathway | tFL, CLL | PRT | 2 | 89, 118 |
| PIM1 | 6 | Apoptosis pathway | DLBCL, FL | PRT | 2 | 26, 27 |
| SF3B1 | 2 | DNA-damage pathway | CLL | PRT | 4 | 118,-120, 125 |
| EBF1 | 5 | BCR signaling pathway | tFL | PRT | 2 | 25, 26 |
| IRF8 | 16 | Interferon signaling | tFL | PRT | 2 | 26, 89 |
| RRAGC | 1 | mTORC signaling | tFL | PRT | 2 | 89, 126 |
| SOCS1 | 16 | JAK-STAT pathway | tFL | PRT | 2 | 25, 26 |
Mutations are ranked after the following criteria: (1) number of studies identifying mutations in the gene and (2) number of histologies in which the mutation has been reported. Mutations were included if variant allele frequency was >5% at diagnosis/relapse, recurrent in the same patient, and identified by at least 2 independent studies.
MHC, major histocompatibility complex; PRT, primary and relapse tumors; tFL, transformed FL.
It has been suggested that the development of lymphoid malignancy may follow a pattern of either early or late clonal evolution25,27,28,90 ; recently summarized by Juskevicius et al.91 Briefly, early evolution is characterized by slow divergence of several subclones, which gives rise to a heterogeneous primary tumor. Relapse of these tumors will possibly come from smaller clones already existing in the primary tumor, a so-called intrinsic resistance. On the contrary, tumors with a late clonal evolution pattern will typically have a strong driver giving rise to fast formation of a uniform clone founding a homogenous primary tumor. Therapy resistance in the late evolution tumors will likely be caused by newly acquired mutations generated under treatment. These 2 evolutionary trajectories of malignancy likely mandate different treatment regimens tailored to the genetic profile of the tumor.
Tracking residual malignant clones during and after treatment
Although the field of minimal residual disease (MRD) has seen considerably advances in the past decade, it is only recommend in a research setting in chronic lymphoid malignancies (CLL, MCL, FL) and is still highly dependent on detection of immunoglobulin rearrangements.92 Several large studies have shown substantial prognostic effects of achieving MRD negativity after treatment of chronic lymphoid malignancies93-95 (supplemental Table 1), which makes it superior for assessment of long-time survival compared with positron emission tomography-computed tomography imaging. However, in 10% to 20% of patients, it is not possible to find an appropriate MRD marker, and at least one-quarter of patients relapse despite being MRD negative after treatment.94,96 In addition, MRD-positive patients have also been reported to remain without relapse for many years.94 In DLBCL, an approach for MRD detection was recently examined. A long-term follow-up study showed that detection of clonal DNA after treatment infers a strong risk of DLBCL relapse.97 Also, in this study, a minority of patients (7 of 94) had a persistent or progressive disease despite no detectable clonal marker, underlining the continued need of alternative techniques.
All these studies have focused on immunoglobulin rearrangements, which is not a sufficient MRD strategy for all patients. It has been shown that the use of NGS for tumor-specific mutations is as least as sensitive an MRD method as the traditional quantitative PCR measurement of immunoglobulin rearrangement.98 If the progenitor or stem cell model (Figure 2) holds true for some lymphoid malignancies, we hypothesize that NGS with high sensitivity and specificity (error-corrected NGS and digital droplet PCR) will be able to detect recurrent mutations (Table 2) at an earlier time point, and also will have improved prediction of relapse compared with traditional MRD by immunoglobulin rearrangement.
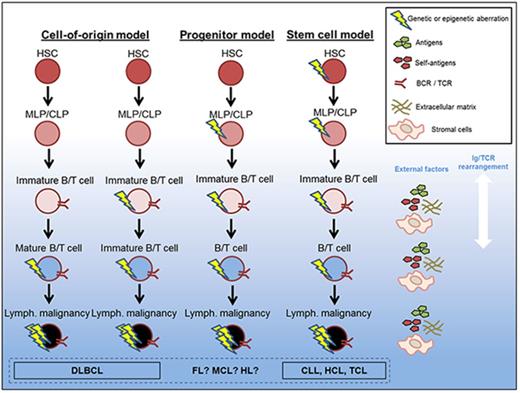
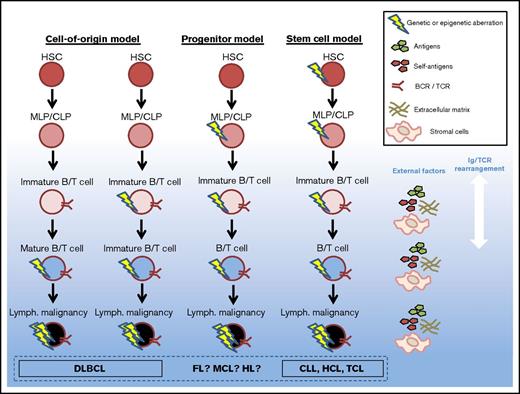
Proposed models of genetic defect origin in mature lymphoid disease. The origin of mature lymphoid malignancies is disputed; this figure represents proposed models of evolution of the malignant clones. An increasing amount of evidence points to that at least some of the mature lymphoid malignancies (CLL, HCL, and TCL) carry genetic defects inherited from HSCs. Whether this is true for all lymphoid malignancies, or if some defects originate from mature B/T cells or CLPs, is not fully elucidated. MLP, multilymphoid progenitor.
Proposed models of genetic defect origin in mature lymphoid disease. The origin of mature lymphoid malignancies is disputed; this figure represents proposed models of evolution of the malignant clones. An increasing amount of evidence points to that at least some of the mature lymphoid malignancies (CLL, HCL, and TCL) carry genetic defects inherited from HSCs. Whether this is true for all lymphoid malignancies, or if some defects originate from mature B/T cells or CLPs, is not fully elucidated. MLP, multilymphoid progenitor.
Animal models of malignant lymphoid stem cells
It is recognized that cancer stem cells are heterogeneous and often disease specific.18 This is also evidenced by a few, but very interesting, studies of serial transplantation of stem cells initiating lymphoid disease in mice. A seminal paper by Kikushige et al examined mice transplanted with sorted CD34+ stem cells from healthy donors and CLL patients, respectively.99 The mice were xenografted with highly purified stem cells from CLL patients and subsequently developed significantly more polyclonal B cells than stem cells from healthy donors. These cells subsequently matured into clones with phenotypical markers of CLL. The B-cell clones also had a striking homology of their BCR, also noted in larger patient series,69 indicating antigen drive as an essential process in malignant development, even in a xenograft model. Additionally, it was possible to take CLL-like clones from the CD34+-xenografted mice and serially transplant them into new mice, thereby indicating cancer stem cell properties. That CD34+ stem cells are the apex of CLL pathogenesis is further supported by a mouse model with long-term conditional inactivation of DNMT3A in HSCs, which develop a CLL phenotype.100
As mentioned, HCL patients have been found to harbor the BRAFV600E mutation in both mature lymphocytes, B-cell precursors, and human CD34+ stem cells.53 The same study transplanted highly purified stem cells from an untreated HCL patient into mice. After 6 months, the mice had developed an HCL immunophenotype and the BRAFV600E mutation was measurable with variant allele frequency of 4% to 9% in bone marrow from the mice, underscoring that HCL is a stem cell disease.
In MCL, a possible tumor-initiating cell has been identified. Two studies identified, respectively, CD45+ and CD133+ (both progenitor/stem cell markers) as a source of MCL outgrowth during murine serial transplantation, confirmed by the pathognomic t(11;14) translocation.101,102 However, these cell types have so far not been investigated in a clinical context and it is unknown what role these clones have in relapsed disease. As mentioned previously, a mouse model of MALT lymphoma has also been generated by overexpressing MALT1, a hallmark of this lymphoma subtype, in murine stem cells.67 In addition, sorted CD34+ stem cells from patients with MALT lymphoma and the transgenic mice showed a similar gene expression profile, which indicates a stem cell defect in this lymphoma entity.
An informative study by Scourzic et al investigated serial transplantation of stem cells with inactivated TET2 and DNMT3A, which are involved in both lymphoid and myeloid pathogenesis.103 Primary recipient mice, transplanted with defect hematopoietic stem and progenitor cells, developed both myeloid and lymphoid disease (AML, ALL), underscoring the established fact that these diseases originate from stem cells. However, when mice were serially transplanted with hematopoietic stem and progenitor cells from the primary mice and prospectively followed for ∼1 year, the majority of secondary and tertiary recipients instead developed lymphoma (T-cell angioimmunoblastic subtype). This implies that TCL originates in genetically defect stem cells.
These animal models underline the HSC as a novel part of lymphoid pathogenesis. Although the studies cumulatively provide a strong vote for the involvement of stem cells in the pathogenesis of lymphoid malignancies, further validations of these murine models are needed to consolidate the findings. The studies are few in numbers and most only describe the creation of few replicas of the animal model.
Eradication of residual malignant cells
As mentioned in the “Introduction,” late relapses occur after both immunochemotherapy and novel targeted therapies (inhibitors of BTK, phosphatidylinositol 3-kinase, and BCL2). Conversely, late relapses are rarely seen after allogeneic transplantation for lymphoma and CLL, with very few patients relapsing after 2 years and nice survival curve plateaus.104-108 This points to myeloablative therapy and graft-versus-tumor effect indeed are sufficient to eliminate residual premalignant cells. However, the side effects of allogeneic HSC transplantation restricts its use to second-line treatment (or later) and is mainly suitable for fit, younger patients with relapsed chemosensitive disease.
Autologous transplantation, a standard therapy used in first- or second-line treatment of lymphoma patients, involves transplantation of CD34+-enriched leukapheresis product after high-dose chemotherapy. To avoid residual malignant lymphoid cells in the autologous product, the possibility of transplanting highly purified CD34+ cells was examined in the past decade, but resulted in comparable effects on lymphoma response. The purification also unexpectedly led to prolonged hematologic recovery109 and increased rates of both short- and long-term infections, especially cytomegalovirus,110-113 after which method was abandoned in most centers. It can also be suspected that the purification of autologous product might not have eliminated premalignant progenitor cells.
At an earlier stage of development, there are several novel therapeutics that target stem cell–associated pathways such as Notch, Hedgehog, and Wnt.18 Some of these drugs have already gone into clinical trials, and lymphoma patients have been involved in both phase 1 and phase 2 protocols, but no results in lymphoma patients have yet been published.
Future directions
The origin of mature lymphoid malignancies and the cause of relapse after a complete remission are still largely unknown. Compared with the major myeloid histologies (AML, MDS, chronic myelomonocytic leukemia), in which the sequence of alterations leading to a malignant state is becoming unraveled,42 there is a lack of evidence on the order of events in lymphoid malignancies, and the hierarchy of mutations are not well-defined. In addition, sensitive methods for detecting residual disease in patients are at best imperfect, because a considerable fraction of patients still relapse, despite having a negative positron emission tomography-computed tomography scan and MRD test result. Treatments optimized to kill small residual malignant clones are still in their infancy and further research on this topic is warranted. In addition, it is awaited if the use of the newly developed small molecule inhibitors and antibodies are approaches to clear out residual clones. If malignant lymphoid stem cells exist, it is still uncertain whether these cells will be targeted by these novel therapies.
A more systematic and better powered investigation of HSCs in patients with lymphoid malignancies is warranted to unravel if the chronic feature of these diseases is mitigated by defect stem cells. If so, identification and characterization of these lymphoma stem cells are essential for development of novel targeted therapies that will eliminate the source of relapse.
We look forward to upcoming studies on these subjects that may change the way patients with mature lymphoid malignancies are managed. Novel insights will potentially help define treatment strategies incorporating new treatments and molecular biological analysis, thereby hopefully increasing the cure rates of these diseases.
The full-text version of this article contains a data supplement.
Acknowledgments
S.H. is supported by the Research Council at Rigshospitalet, Copenhagen. K.G. is supported by the Novo Nordisk Foundation.
Authorship
Contribution: S.H. and K.G. wrote the manuscript and prepared the figures.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Simon Husby, Epigenome Laboratory, Department of Hematology, Rigshospitalet, 3733, Building 2, 3rd Floor (Biocenter), Ole Maaløesvej 5, Copenhagen N 2200, Denmark; e-mail: simon.husby.01@regionh.dk.