Key Points
Inhibitor development in patients with nonsevere hemophilia A is associated with predicted novel binding of HLA-DRB1 with FVIII peptides.
Inhibitor development associated with high-risk F8 mutations is due, in part, to high levels of novel HLA-DRB1 FVIII peptide binding.
Visual Abstract
Professional illustration by Patrick Lane, ScEYEnce Studios.
Abstract
Development of anti-factor VIII (FVIII) inhibitory antibodies (inhibitors) is the most significant treatment complication of hemophilia A. Characteristics of the interaction between major histocompatibility complex (MHC) class II and FVIII peptides may influence FVIII antigen presentation to T cells and subsequent inhibitor development. We analyzed predicted HLA-DRB1, a subset of MHC class II, and FVIII peptide binding and its association with inhibitor development among subjects with nonsevere hemophilia A, including 20 cases (inhibitor titer ≥ 1.0 BU/mL on 2 occasions or on 1 occasion with subsequent immune tolerance induction) and 37 controls (who had received FVIII infusions and did not develop inhibitor). Using the MHC-II Binding Predictions Tool (https://www.iedb.org), the binding affinity and core binding were determined for endogenous FVIII (eFVIII) and treatment FVIII (tFVIII). A tFVIII peptide was considered novel if it was predicted to bind and present a surface to the T-cell receptor that was unique from that presented by eFVIII. Having >10 novel HLA-DRB1 allele–tFVIII peptide combinations was associated with inhibitor development (adjusted odds ratio, 4.1; 95% confidence interval, 1.1-15.0). Cases and controls with p.Arg612Cys and p.Arg2169His demonstrated a high level of novel HLA-DRB1–tFVIII peptide combinations. Assessing the likelihood that tFVIII is presented to T cells in a novel fashion may be useful for understanding and ultimately reducing the risk for inhibitor development among patients with nonsevere hemophilia, particularly those with F8 mutations other than p.Arg612Cys and p.Arg2169His.
Introduction
The development of neutralizing antibodies against factor VIII (FVIII) (inhibitors) is the most significant treatment complication affecting persons with hemophilia A. Although inhibitor development occurs in up to one third of persons with severe hemophilia (FVIII < 1 IU/dL),1 it also occurs in persons with nonsevere hemophilia A (FVIII 1-40 IU/dL), with a cumulative incidence of 13.7%.2 In contrast to severe hemophilia A in which the inhibitor risk is greatest during the first 10 days of FVIII exposure,1 the incidence of inhibitor development in nonsevere hemophilia A is consistent over the first 100 FVIII exposure days.2 Given the persistent risk over a range of FVIII exposure days, inhibitor development in nonsevere hemophilia A tends to affect older individuals (median age of 46 years).2,3 Furthermore in the setting of nonsevere hemophilia, inhibitor development often leads to a reduction in endogenous FVIII (eFVIII) activity and a marked change in clinical phenotype.4
Risk factors for inhibitor development in nonsevere hemophilia A include the intensity of treatment, the number of days, and the daily treatment dose.3,5 Surgery as an indication for treatment may also contribute;5 however, it is difficult to determine its independent effect because multiday and higher-dose FVIII treatment is routinely used in the surgical setting. Several F8 missense mutations have been reported to occur more frequently than expected in patients with nonsevere hemophilia A and inhibitor.2 Most notable is the mutation p.Arg612Cys, and others reported include p.Asp2093His, p.Arg2169His, p.Arg2178Cys, and p.Arg2248Cys.2
Because inhibitor development requires the presentation of a foreign peptide antigen to T cells by major histocompatibility complex (MHC) class II on antigen presenting cells, investigators have postulated that specific class II alleles may influence peptide binding and be associated with inhibitor development. Such an association has been seen with HLA-DRB1*0602 and HLA-DRB1*15.6 In patients with hemophilia caused by a missense mutation who are treated with FVIII-replacement products, there are potentially 2 versions of antigen (eFVIII and exogenous [treatment FVIII; tFVIII]) in circulation. It has been hypothesized that inhibitor development is influenced by differential binding between MHC class II and endogenous peptides compared with exogenous peptides that leads to a novel (ie, foreign) surface being presented to the immune system. For example, if a patient’s MHC class II does not bind eFVIII peptides but binds exogenous FVIII peptides, exogenous FVIII peptides will be seen as novel by the patient’s immune system and will be more likely to initiate an immune response. Using in silico techniques, Shepherd et al demonstrated that 15 F8 mutations were predicted to bind all 14 common HLA-DR types evaluated. These “promiscuous” binders included p.Arg612Cys and p.Arg2169His.7 The objective of this study was to evaluate whether the predicted presentation of novel surfaces based on a patient’s F8 mutation and HLA-DR type was positively associated with inhibitor development.
Methods
Participants
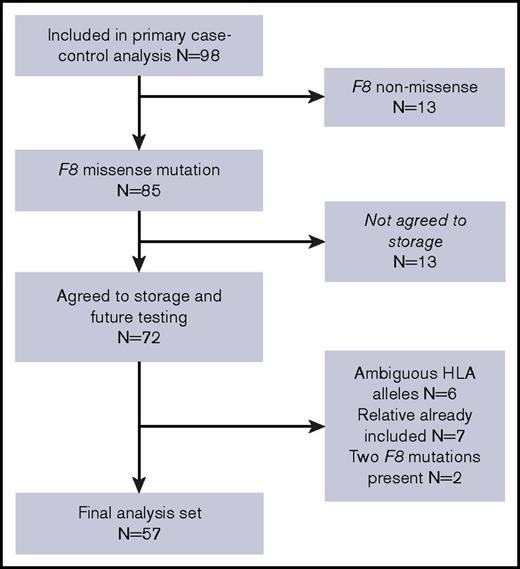
Subjects for this analysis were selected from a cohort of patients with nonsevere HA recruited as part of a case-control study.3 Subjects were included if they had agreed to participate in a repository for future research, had a single missense mutation in the F8 gene as the cause of their hemophilia, and did not have an ambiguous HLA-DRB1 type (Figure 1). If relatives were coenrolled in the study, data from the first enrolled relative were used. Case subjects had an inhibitor titer ≥1.0 BU/mL on 2 occasions or on 1 occasion with subsequent immune tolerance induction. Control subjects had prior FVIII exposure and no history of inhibitor (<0.6 BU/mL).
Laboratory testing
Blood specimens were collected, and DNA was extracted as previously described.3 DNA specimens were stored at −70°C from the time of collection between July 2007 and December 2008 until use in 2016.
F8 genotyping was performed at the Hemostasis Laboratory Branch at the Centers for Disease Control and Prevention. After exclusion of intron 22 and intron 1 inversions by polymerase chain reaction,8 all exons, intron–exon junction regions, and the 3′ untranslated regions of F8 were sequenced in both directions using a Variant-SEQr protocol on a 3730 DNA Analyzer (Applied Biosystems, Carlsbad, CA). Sequence data were analyzed with SeqScape (Applied Biosystems). F8 mutations were considered high-risk if they were previously reported to be associated with inhibitor development.2,3,9
HLA-DRB1 genotype was determined by next-generation sequencing using an Illumina MiSeq platform at Histogenetics Laboratory (Ossining, NY) to 6-digit resolution, with the first 4 digits used to describe unique HLA-DRB1 proteins (the HLA-DRB1 type).
Novel surface prediction
For both eFVIII and tFVIII, a series of 15 15-mer peptides that contained the subject’s location of the missense mutation in each amino acid position of the 15-mer peptide was modeled. Using the subject’s HLA-DRB1 type, the MHC-II Binding Predictions Tool (https://www.iedb.org) was used to predict the binding affinity of each of the 15 15-mer peptides to both of the HLA-DRB1 alleles (30 [15 peptides × 2 HLA alleles] potential binding opportunities for each subject for both eFVIII and tFVIII). The Immune Epitope Data Base (IEDB)-recommended prediction method was used to predict the binding affinity for each peptide. If the IEDB-recommended method was the consensus method,10,11 the results for each of the underlying methods was considered. A peptide was considered as binding if the predicted 50% inhibitory concentration was <1000 nmol/L. If the IEDB-recommended method included the Sturniolo method,12 a peptide was considered as binding if the percentile rank was <10.
The MHC-II Binding Predictions Tool was also used to predict the binding core for each peptide. Whether a variant would be T-cell facing was evaluated by examining the predicted binding core anchor locations for each of the HLA-DRB1 types using the SYFPEITHI database (http://www.syfpeithi.de/). For HLA-DRB1 types not included in the database, anchor locations were assumed to be the same as the locations listed for alleles with the same 2-digit resolution or P1, P4, P6, P9 if a similar type was not listed.13 If the variant was determined to be inside the binding core and not at 1 of the anchor locations, the variant was determined to be T-cell facing. If the variant was outside the binding core or at 1 of the anchor locations, the variant was determined not to be T-cell facing.
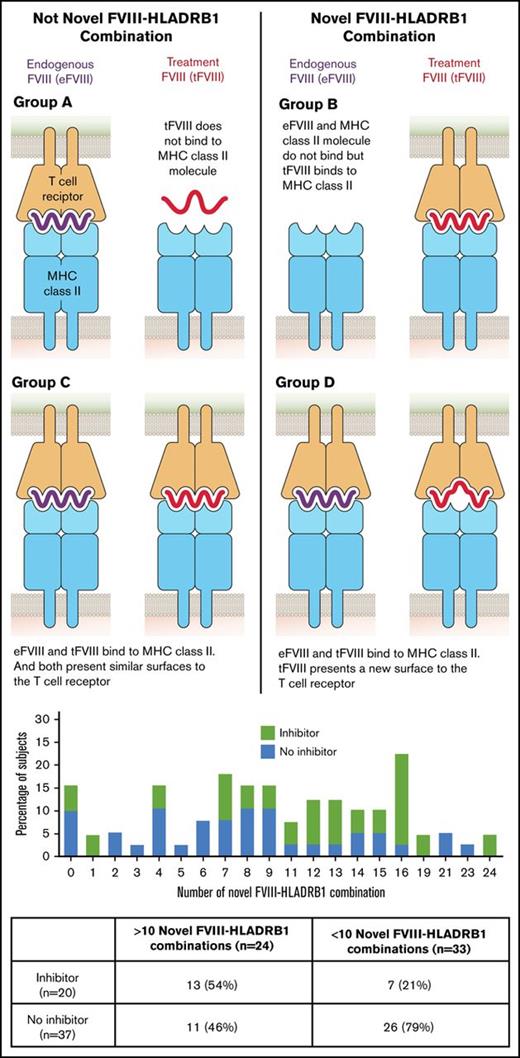
Each peptide–HLA-DRB1 interaction was categorized into 1 of 4 groups (A-D).7 Nonnovel peptides were present if eFVIII and tFVIII peptides were nonbinding (group A) or if both peptides were binding but tFVIII did not present a novel T-cell–facing variant (group C). Novel peptides were present if the eFVIII sequence was nonbinding and the tFVIII was binding (group B) or if tFVIII and eFVIII bound but tFVIII presented a novel T-cell–facing variant (group D).
Analysis
The association between inhibitor development and the number of 15-mer peptides that could present a novel peptide (group B/D; maximum = 30) was evaluated in simple and multivariable logistic-regression models. Variables, including age (<30 vs >30 years), race (white vs other), family history of inhibitor, intensive FVIII exposure, lifetime FVIII exposure days (<50 vs >50 days), product type (recombinant vs plasma derived), age at first factor exposure (age <5 vs >5 years) and baseline FVIII level (1 to <2% vs >2%), were evaluated and included if found to be a potential confounder, defined as being significantly associated with both the dependent (inhibitor status) and independent (number of group B/D peptides) variables.
Results
Twenty cases and 37 controls (56% of the original study cohort) were included in the analysis (Figure 1). The median age (case, 37 years and control, 27 years) and baseline FVIII (case, 8.0 IU/dL and control 5.5 IU/dL) were similar between groups (Table 1). Cases and controls were also similar with respect to race, ethnicity, prior number of FVIII exposure days, family history of FVIII inhibitor, FVIII product class (recombinant vs plasma derived), and being HIV or HCV antibody positive. Intensive FVIII treatment was more common in cases than in controls (P = .01).
Characteristics of study population
| Characteristic . | Controls (n = 37), n (%) . | Cases (n = 20), n (%) . | χ2 . | P . |
|---|---|---|---|---|
| Intensive FVIII treatment | 6 (16) | 10 (50) | 7.34 | .01 |
| Age, y | ||||
| <30 | 21 (57) | 7 (35) | 2.55 | .27 |
| 30-60 | 13 (35) | 10 (50) | ||
| >60 | 3 (8) | 3 (15) | ||
| Race | ||||
| White | 31 (84) | 19 (95) | 1.62 | .44 |
| African American | 5 (13) | 1 (5) | ||
| Other | 1 (3) | 0 (0) | ||
| Hispanic ethnicity | 3 (8) | 1 (5) | 0.19 | .66 |
| Family history of inhibitor | 5 (14) | 3 (15) | 0.02 | .88 |
| HCV antibody positive | 15 (41) | 7 (35) | 1.42 | .49 |
| HIV antibody positive | 1 (3) | 0 (0) | 0.74 | .69 |
| Baseline FVIII | ||||
| 1% ≤ FVIII:C < 2% | 2 (5) | 4 (20) | 3.52 | .17 |
| 2% ≤ FVIII:C ≤ 5% | 15 (41) | 5 (25) | ||
| FVIII:C > 5% | 20 (54) | 11 (55) | ||
| Age at first factor infusion, y | ||||
| Unknown | 1 (3) | 0 (0) | ||
| ≤2 | 11 (30) | 3 (15) | 4.25 | .12 |
| 3-10 | 15 (41) | 6 (30) | ||
| >10 | 10 (27) | 11 (55) | ||
| Prior FVIII exposure, d | ||||
| ≤20 | 9 (24) | 2 (10) | 4.87 | .18 |
| 21-50 | 7 (19) | 9 (45) | ||
| 51-100 | 7 (19) | 3 (15) | ||
| >100 | 14 (38) | 6 (30) | ||
| Product during prior year | ||||
| Unknown | 0 (0) | 1 (5) | ||
| Plasma derived | 8 (22) | 3 (15) | 1.37 | .50 |
| Recombinant | 24 (65) | 15 (75) | ||
| None | 5 (13) | 1 (5) | ||
| Specific missense mutations | ||||
| p.Arg612Cys | 2 (5) | 6 (30) | 6.51 | .01 |
| p.Asn1942Ser | 4 (11) | 5 (25) | 1.97 | .16 |
| p.Arg2169His | 1 (3) | 3 (15) | 3.01 | .08 |
| p.Trp2248Cys | 1 (3) | 0 (0) | 0.55 | .46 |
| Characteristic . | Controls (n = 37), n (%) . | Cases (n = 20), n (%) . | χ2 . | P . |
|---|---|---|---|---|
| Intensive FVIII treatment | 6 (16) | 10 (50) | 7.34 | .01 |
| Age, y | ||||
| <30 | 21 (57) | 7 (35) | 2.55 | .27 |
| 30-60 | 13 (35) | 10 (50) | ||
| >60 | 3 (8) | 3 (15) | ||
| Race | ||||
| White | 31 (84) | 19 (95) | 1.62 | .44 |
| African American | 5 (13) | 1 (5) | ||
| Other | 1 (3) | 0 (0) | ||
| Hispanic ethnicity | 3 (8) | 1 (5) | 0.19 | .66 |
| Family history of inhibitor | 5 (14) | 3 (15) | 0.02 | .88 |
| HCV antibody positive | 15 (41) | 7 (35) | 1.42 | .49 |
| HIV antibody positive | 1 (3) | 0 (0) | 0.74 | .69 |
| Baseline FVIII | ||||
| 1% ≤ FVIII:C < 2% | 2 (5) | 4 (20) | 3.52 | .17 |
| 2% ≤ FVIII:C ≤ 5% | 15 (41) | 5 (25) | ||
| FVIII:C > 5% | 20 (54) | 11 (55) | ||
| Age at first factor infusion, y | ||||
| Unknown | 1 (3) | 0 (0) | ||
| ≤2 | 11 (30) | 3 (15) | 4.25 | .12 |
| 3-10 | 15 (41) | 6 (30) | ||
| >10 | 10 (27) | 11 (55) | ||
| Prior FVIII exposure, d | ||||
| ≤20 | 9 (24) | 2 (10) | 4.87 | .18 |
| 21-50 | 7 (19) | 9 (45) | ||
| 51-100 | 7 (19) | 3 (15) | ||
| >100 | 14 (38) | 6 (30) | ||
| Product during prior year | ||||
| Unknown | 0 (0) | 1 (5) | ||
| Plasma derived | 8 (22) | 3 (15) | 1.37 | .50 |
| Recombinant | 24 (65) | 15 (75) | ||
| None | 5 (13) | 1 (5) | ||
| Specific missense mutations | ||||
| p.Arg612Cys | 2 (5) | 6 (30) | 6.51 | .01 |
| p.Asn1942Ser | 4 (11) | 5 (25) | 1.97 | .16 |
| p.Arg2169His | 1 (3) | 3 (15) | 3.01 | .08 |
| p.Trp2248Cys | 1 (3) | 0 (0) | 0.55 | .46 |
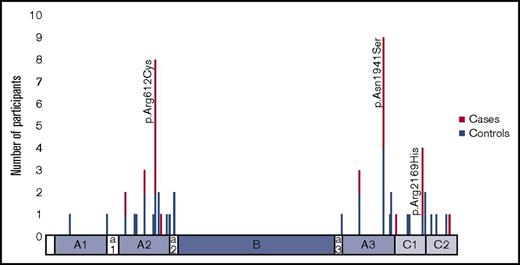
Thirty-three missense mutations were observed (Figure 2). Twenty-four were unique to 1 subject. The 3 mutations most commonly found in cases were p.Arg612Cys (n = 8), p.Asn1941Ser (n = 9), and p.Arg2169His (n = 4). These 3 mutations, in addition to p.Trp2248Cys (case n = 0; control n = 1) were considered high-risk mutations. Twenty-four unique HLA-DRB1 alleles were observed in the study population: 17 in cases and 24 in controls (Table 2). Three HLA-DRB1 alleles were found in ≥10% of cases and more often in cases than in controls (HLA-DRB1*15:01, HLA-DRB1*13:02, and HLA-DRB1*11:01), although none were statistically significantly associated with inhibitor development (referent HLA-DRB*01:01). Three alleles were found more frequently in controls than in cases (HLA-DRB1*07:01, HLA-DRB1*13:01, and HLA-DRB1*04:01), although these alleles also were not statistically significantly associated with a lack of inhibitor development (referent HLA-DRB*01:01).
Distribution of HLA-DRB1 alleles in cases and controls
| HLA type . | Controls (n = 74), n (%) . | Cases (n = 46), n (%) . | OR (95% CI) . |
|---|---|---|---|
| HLA-DRB1*01:01 | 9 (12) | 4 (10) | Ref |
| HLA-DRB1*01:02 | 0 (0) | 1 (3) | NE |
| HLA-DRB1*01:03 | 2 (3) | 0 (0) | NE |
| HLA-DRB1*03:01 | 6 (8) | 3 (8) | 1.1 (0.2-6.9) |
| HLA-DRB1*03:02 | 2 (3) | 0 (0) | NE |
| HLA-DRB1*04:01 | 6 (8) | 2 (5) | 0.8 (0.1-5.5) |
| HLA-DRB1*04:02 | 3 (4) | 0 (0) | NE |
| HLA-DRB1*04:03 | 1 (1) | 1 (3) | 2.3 (0.1-45.7) |
| HLA-DRB1*04:11 | 1 (1) | 0 (0) | NE |
| HLA-DRB1*07:01 | 14 (19) | 5 (13) | 0.8 (0.2-3.8) |
| HLA-DRB1*08:01 | 0 (0) | 1 (3) | NE |
| HLA-DRB1*08:03 | 1 (1) | 0 (0) | NE |
| HLA-DRB1*09:01 | 1 (1) | 1 (3) | 2.3 (0.1-45.7) |
| HLA-DRB1*11:01 | 3 (4) | 4 (10) | 3.0 (0.4-20.2) |
| HLA-DRB1*11:02 | 2 (3) | 0 (0) | NE |
| HLA-DRB1*11:03 | 1 (1) | 1 (3) | 2.3 (0.1-45.7) |
| HLA-DRB1*11:04 | 2 (3) | 1 (3) | 1.1 (0.1-16.3) |
| HLA-DRB1*13:01 | 3 (4) | 1 (3) | 0.8 (0.1-9.6) |
| HLA-DRB1*13:02 | 4 (5) | 5 (13) | 2.8 (0.5-16.4) |
| HLA-DRB1*13:03 | 2 (3) | 1 (3) | 1.1 (0.1-16.3) |
| HLA-DRB1*14:01 | 0 (0) | 1 (3) | NE |
| HLA-DRB1*14:54 | 2 (3) | 1 (3) | 1.1 (0.1-16.3) |
| HLA-DRB1*15:01 | 7 (9) | 7 (18) | 2.3 (0.5-10.9) |
| HLA-DRB1*15:02 | 1 (1) | 0 (0) | NE |
| HLA-DRB1*15:03 | 1 (1) | 0 (0) | NE |
| HLA type . | Controls (n = 74), n (%) . | Cases (n = 46), n (%) . | OR (95% CI) . |
|---|---|---|---|
| HLA-DRB1*01:01 | 9 (12) | 4 (10) | Ref |
| HLA-DRB1*01:02 | 0 (0) | 1 (3) | NE |
| HLA-DRB1*01:03 | 2 (3) | 0 (0) | NE |
| HLA-DRB1*03:01 | 6 (8) | 3 (8) | 1.1 (0.2-6.9) |
| HLA-DRB1*03:02 | 2 (3) | 0 (0) | NE |
| HLA-DRB1*04:01 | 6 (8) | 2 (5) | 0.8 (0.1-5.5) |
| HLA-DRB1*04:02 | 3 (4) | 0 (0) | NE |
| HLA-DRB1*04:03 | 1 (1) | 1 (3) | 2.3 (0.1-45.7) |
| HLA-DRB1*04:11 | 1 (1) | 0 (0) | NE |
| HLA-DRB1*07:01 | 14 (19) | 5 (13) | 0.8 (0.2-3.8) |
| HLA-DRB1*08:01 | 0 (0) | 1 (3) | NE |
| HLA-DRB1*08:03 | 1 (1) | 0 (0) | NE |
| HLA-DRB1*09:01 | 1 (1) | 1 (3) | 2.3 (0.1-45.7) |
| HLA-DRB1*11:01 | 3 (4) | 4 (10) | 3.0 (0.4-20.2) |
| HLA-DRB1*11:02 | 2 (3) | 0 (0) | NE |
| HLA-DRB1*11:03 | 1 (1) | 1 (3) | 2.3 (0.1-45.7) |
| HLA-DRB1*11:04 | 2 (3) | 1 (3) | 1.1 (0.1-16.3) |
| HLA-DRB1*13:01 | 3 (4) | 1 (3) | 0.8 (0.1-9.6) |
| HLA-DRB1*13:02 | 4 (5) | 5 (13) | 2.8 (0.5-16.4) |
| HLA-DRB1*13:03 | 2 (3) | 1 (3) | 1.1 (0.1-16.3) |
| HLA-DRB1*14:01 | 0 (0) | 1 (3) | NE |
| HLA-DRB1*14:54 | 2 (3) | 1 (3) | 1.1 (0.1-16.3) |
| HLA-DRB1*15:01 | 7 (9) | 7 (18) | 2.3 (0.5-10.9) |
| HLA-DRB1*15:02 | 1 (1) | 0 (0) | NE |
| HLA-DRB1*15:03 | 1 (1) | 0 (0) | NE |
NE, not evaluable; ref, referent.
A greater number of FVIII peptides bound to HLA-DRB1 alleles and formed a novel T-cell receptor (TCR) surface (category B/D) in cases than in controls (median, 12.5; interquartile range [IQR], 7.5-16 and median, 8; IQR, 4-12, respectively, P = .03) (Table 3). However, 1 case had no peptides categorized in the B/D group. The highest number of peptides that bound with a novel surface was 24 and occurred in a single case. Among subjects with and without high-risk mutations (p.Arg612Cys, p.Asn1941Ser, p.Arg2169His, p.Trp2248Cys), the proportion of subjects with >10 novel HLA-DRB1–tFVIII peptide combinations was greater in cases than in controls (Table 4). However, the proportion of controls with high-risk mutations and >10 novel HLA-DRB1–tFVIII peptide combinations was also substantial and was higher than that seen in cases with nonhigh-risk mutations.
Distribution of HLA-DRB1–tFVIII peptide binding combinations in cases and controls
| . | Controls (n = 37) . | Cases (n = 20) . | P . |
|---|---|---|---|
| Total A | 14 (7-17) | 9 (5-16) | .16 |
| Total B | 0 (0-0) | 0 (0-0) | .61 |
| Total C | 9 (6-11) | 7.5 (4.5-12.5) | .62 |
| Total D | 7 (4-11) | 11.5 (7-14.5) | .08 |
| Total A+C | 22 (18-26) | 17.5 (14-22.5) | .03 |
| Total B+D | 8 (4-12) | 12.5 (7.5-16) | .03 |
| . | Controls (n = 37) . | Cases (n = 20) . | P . |
|---|---|---|---|
| Total A | 14 (7-17) | 9 (5-16) | .16 |
| Total B | 0 (0-0) | 0 (0-0) | .61 |
| Total C | 9 (6-11) | 7.5 (4.5-12.5) | .62 |
| Total D | 7 (4-11) | 11.5 (7-14.5) | .08 |
| Total A+C | 22 (18-26) | 17.5 (14-22.5) | .03 |
| Total B+D | 8 (4-12) | 12.5 (7.5-16) | .03 |
Data are median (IQR).
Proportion of cases and controls with novel surface HLA-DRB1–tFVIII peptide binding combinations by mutation
| . | Controls with >10 novel surface HLA-DRB1–tFVIII peptide binding combinations . | Cases with >10 novel surface HLA-DRB1–tFVIII peptide binding combinations . |
|---|---|---|
| High-risk mutation | 5 (62.5) | 11 (78.6) |
| Other | 6 (20.7) | 2 (33.3) |
| . | Controls with >10 novel surface HLA-DRB1–tFVIII peptide binding combinations . | Cases with >10 novel surface HLA-DRB1–tFVIII peptide binding combinations . |
|---|---|---|
| High-risk mutation | 5 (62.5) | 11 (78.6) |
| Other | 6 (20.7) | 2 (33.3) |
Data are n (%). High-risk mutation is p.Arg162Cys, p.Asn1941Ser, p.Arg2169His, p.Trp2248Cys.
On univariate analysis, having >10 peptides that bound to HLA-DRB1 and presented a novel TCR surface was associated with inhibitor development (odds ratio [OR], 4.4; 95% confidence interval [CI], 1.4-14.0). After evaluation and adjustment for confounding, the strength of the association was unchanged (adjusted OR, 4.1; 95% CI; 1.1-15.0) (Table 5).
Effect of >10 novel surface HLA-DRB1–tFVIII peptide binding combinations on inhibitor risk
| . | OR (95% CI) . |
|---|---|
| Unadjusted model | 4.4 (1.4-14.0) |
| Adjusted model* | 4.1 (1.1-15.0) |
| . | OR (95% CI) . |
|---|---|
| Unadjusted model | 4.4 (1.4-14.0) |
| Adjusted model* | 4.1 (1.1-15.0) |
Adjusted for intensive FVIII treatment.
Likelihood Ratio Test for interaction between >10 novel peptides and intensive FVIII treatment indicated no statistically significant interaction, P = .6.
Discussion
In this analysis, prediction of novel binding of FVIII to a patient’s own HLA-DRB1 that creates a novel TCR surface was strongly associated with inhibitor development. We also observed higher levels of novel FVIII peptide binding among patients with 1 of several mutations that have previously been associated with inhibitor development. These data support the concept that 1 mechanism by which F8 mutations are associated with inhibitor development is through more frequent presentation of novel peptides due to differential binding of HLA-DRB1 by eFVIII and tFVIII.
Previous investigations of the association of FVIII peptide affinity for class II alleles with inhibitor development have used missense mutations and inhibitor status listed in databases and HLA-DRB1 alleles reported to be common in European populations.14 The study by Pashov et al evaluated patients with hemophilia A of any severity, although >92% of their mutations were associated with nonsevere hemophilia A. They used a binding score that aggregated the affinity of each possible peptide containing the missense mutation with all 10 HLA alleles; thus, 15-mer peptide binding to each of 10 HLA alleles (15 × 10 matrix) was aggregated using the lowest percentile rank of each HLA allele and averaging across the 10 alleles. A lower binding score (higher affinity) was seen in patients with a reported history of inhibitor compared with those without a reported inhibitor (OR, 7.36; 95% CI, 6.23-8.40 vs OR, 10.01; 95% CI, 9.54-10.48, P = .002).14 After adjusting for severity, the association persisted (P = .002). In contrast to that study, ours used known F8 mutations and each patient’s HLA-DRB1 alleles (15 × 2 matrix). Our finding that having a high number of HLA-DRB1–tFVIII peptide combinations that bound and presented a novel surface (group B and D) was associated with inhibitor development aligns with findings by Pashov et al.14
Two mutations frequently associated with inhibitor development, p.Arg612Cys and p.Arg2169His, were also seen in this study population. The number of HLA-DRB1–tFVIII peptide binding combinations was higher in subjects with high-risk mutations (Table 4) compared with other mutations. This is consistent with the finding of Shepherd et al that 15 F8 mutations, including p.Arg612Cys and p.Arg2169His, bound all of the 14 HLA DR alleles.7 The p.Asn1941Ser mutation was not represented in their cohort. Interestingly, in our study subjects with 1 of these 3 mutations, the number of HLA-DRB1–tFVIII peptide binding combinations was similar between cases and controls. Considering a sufficient-component cause model, a high level of HLA-DR binding can be a component of the causal mechanisms that underlie inhibitor development in patients with nonsevere hemophilia A, although it is not a necessary or sufficient component. Subjects with these mutations are at higher risk for inhibitor development, in part through the promiscuous binding of FVIII peptides containing the missense mutation with HLA-DRB1; however, other factors are required to differentiate between those who do and do not develop an inhibitor. We speculate that these factors may be environmental exposures, such as intensity of FVIII treatment or undergoing surgery, or other patient characteristics, such as other immune response characteristics. Patients with F8 mutations that do not promiscuously bind HLA-DRB1 will form a variable number of novel HLA-DRB1–tFVIII peptides that, at the present time, is not predicted based on their F8 mutation alone. It is in these patients without F8 mutations that promiscuously bind HLA-DRB1 where measurement of novel HLA-DRB1–tFVIII peptide binding may have predictive value and aid in inhibitor risk assessment.
The strength of this study is that it used patient-specific F8 genotypes and HLA-DRB1 allele combinations. In addition, relevant patient and environmental characteristics were known and could be adjusted for to reduce confounding. Despite these strengths, there are limitations. The most notable limitation is the small sample size. Specifically, only 6 cases had a mutation not encompassed by p.Arg612Cys, p.Asn1941Ser, and p.Arg2169His, thus limiting the ability to evaluate the impact of FVIII peptide binding to HLA-DR in subjects without these mutations.
In conclusion, FVIII peptide binding to HLA-DRB1 is a component, although insufficient, cause of inhibitor development in persons with nonsevere hemophilia A. HLA-DRB1 binding of FVIII and inhibitor development are more likely to occur when F8 mutations, such as p.Arg612Cys or p.Arg2169His, are present. Other risk factors are also needed as components of the causal pathway, because not all patients with 1 of these mutations develop an inhibitor when exposed to FVIII, despite a high level of novel HLA-DRB1–tFVIII peptide binding. Predicting novel HLA-DRB1–tFVIII peptide binding is likely to be useful in patients with mutations that are not known to promiscuously bind HLA-DRB1. Larger studies are needed to accurately assess the usefulness of HLA-DRB1-tFVIII peptide binding to predict inhibitor development in patients with less common missense mutations.
Acknowledgments
The authors acknowledge Althea Hammond for editorial review; Jennifer Driggers, Dorothy Ellingsen, and Christopher J. Bean for assistance with specimen preparation; and the following investigators and institutions for their enrollment of subjects in the study: Geoff Allen (Children’s Memorial Hospital, Chicago, IL), Alice Cohen (Newark Beth Israel Medical Center, Newark, NJ), Miguel Escobar (University of Texas Health Science Center and The Gulf States Hemophilia and Thrombophilia Center, Houston, TX), Cynthia Gauger (Nemours Children’s Clinic, Jacksonville, FL), John Hord (Children’s Hospital of Akron, Akron, OH), Nigel Key (University of North Carolina, Chapel Hill, NC), Rebecca Kruse-Jarres (Tulane University, New Orleans, LA), Marilyn J. Manco-Johnson (University of Colorado Denver and The Children’s Hospital, Aurora, CO), Deanna Mitchell (Helen DeVos Children’s Hospital, Grand Rapids, MI), Mark T. Reding (University of Minnesota, Minneapolis, MN), Arthur Thompson (BloodCenter Northwest, Seattle, WA), Christopher Walsh (Mount Sinai Medical Center, New York, NY), and Guy Young (Children’s Hospital Los Angeles, Los Angeles, CA).
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Authorship
Contribution: C.L.K. and A.B.P. designed the study, performed the analysis, and wrote the manuscript.
Conflict-of-interest disclosure: C.L.K. has received honoraria for participation on advisory boards from Bayer, Shire, and Genentech and for speaking engagements from Grifols and has received research support from Novo Nordisk. A.B.P. declares no competing financial interests.
Correspondence: Christine L. Kempton, 550 Peachtree St, Suite 1075, Atlanta, GA 30308; e-mail: ckempto@emory.edu.