Key Points
Immunoglobulin gene repertoire of PVRL is highly skewed, with usage of the IGHV4-34 gene in almost two thirds of cases.
PVRL differs significantly from PCNSL, which has an immunoglobulin gene repertoire resembling that of ABC-type DLBCL.
Abstract
Primary vitreoretinal lymphoma (PVRL) is a high-grade lymphoma affecting the vitreous and/or the retina. The vast majority of cases are histopathologically classified as diffuse large B-cell lymphoma (DLBCL) and considered a subtype of primary central nervous system lymphoma (PCNSL). To obtain more insight into the ontogenetic relationship between PVRL and PCNSL, we adopted an immunogenetic perspective and explored the respective immunoglobulin gene repertoire profiles from 55 PVRL cases and 48 PCNSL cases. In addition, considering that both entities are predominantly related to activated B-cell (ABC) DLBCL, we compared their repertoire with that of publicly available 262 immunoglobulin heavy variable domain gene rearrangement sequences from systemic ABC-type DLBCLs. PVRL displayed a strikingly biased repertoire, with the IGHV4-34 gene being used in 63.6% of cases, which was significantly higher than in PCNSL (34.7%) or in DLBCL (30.2%). Further repertoire bias was evident by (1) restricted associations of IGHV4-34 expressing heavy chains, with κ light chains utilizing the IGKV3-20/IGKJ1 gene pair, including 5 cases with quasi-identical sequences, and (2) the presence of a subset of stereotyped IGHV3-7 rearrangements. All PVRL IGHV sequences were highly mutated, with evidence of antigen selection and ongoing mutations. Finally, half of PVRL and PCNSL cases carried the MYD88 L265P mutation, which was present in all 4 PVRL cases with stereotyped IGHV3-7 rearrangements. In conclusion, the massive bias in the immunoglobulin gene repertoire of PVRL delineates it from PCNSL and points to antigen selection as a major driving force in their development.
Introduction
Primary intraocular lymphomas constitute rare forms of extranodular non-Hodgkin lymphoma. Upon their anatomical localization, they can be subdivided into 3 groups.1,2 The vast majority arise from the vitreous and/or the retina and, thus, are termed primary vitreoretinal lymphomas (PVRLs). Most PVRLs are high-grade diffuse large B-cell lymphomas (DLBCLs). In contrast, a minority occur in the choroid and are low-grade extranodal marginal zone B-cell lymphomas.
Because they often colocalize in the brain, the World Health Organization classification includes PVRL in the category of primary central nervous system lymphoma (PCNSL).3 Indeed, 65% to 90% of PVRL cases eventually develop central nervous system (CNS) dissemination; conversely, 15% to 25% of patients with PCNSL will present intraocular localization.2 In contrast, both tumors extremely rarely propagate outside of the CNS, with the exception of the testis,4 reflecting their dependency on an immunologically privileged microenvironment for their growth and survival. Based on their immunophenotypic and gene expression profiles, PVRL and PCNSL share features of late germinal center and activated post–germinal center B cells.5-8
The basis for the selective tropism of these lymphomatous cells for the CNS tissues remains elusive, and several, not mutually exclusive, hypotheses have been proposed. As a result of a less stringent immune surveillance, the tumor cells might survive and expand in these “immune-privileged” niches while being eliminated in the periphery.9 Homing of the malignant B cells to the CNS might also be favored by expression of the chemokine receptors CXCR4 and CXCR5 by the tumor cells and their ligands CXCL12 (SDF-1) and CXCL13 by endothelial and microglia cells.10-12 Indeed, high levels of CXCL13 in the cerebral spinal fluid correlated with response to therapy, possibly reflecting its role in lymphoma development.13 Moreover, retinal pigment epithelium cells have also been shown to express the B-cell chemokines CXCL13 and CXCL12 (SDF-1).14
Recognition of CNS-specific antigen(s) and subsequent stimulation through the B-cell receptor (BCR) might also contribute to preferential localization of the tumor cells and their expansion in CNS tissues. Antigenic stimulation is a well-recognized driving force in B lymphomagenesis,15,16 as reflected in biased immunoglobulin (IG) gene repertoires of the clonotypic BCRs in several B-cell malignancies, including DLBCL.17-20 Initially reported in small series,21-26 IG repertoire restriction in PCNSL was recently confirmed in a study including 50 cases; preferential usage of the IGHV4-34 gene was observed in 36% of cases.27 Data on PVRL are more limited and inconclusive, likely as a result of the small cohorts evaluated (<10 cases), thereby preventing any firm conclusions from being drawn.28,29 Furthermore, for some of the investigated cases, the intraocular localization was concomitant or secondary to CNS localization.28
To investigate the role of antigen selection in the ontogeny of PVRLs and its potential relevance for their unusual localization, we analyzed, in detail, the immunoglobulin heavy chain (HC) and light chain (LC) variable domain gene rearrangement sequences from 55 PVRL cases and 48 PCNSL cases, respectively. In addition, considering that these lymphomas are predominantly related to activated B-cell (ABC) DLBCL, we compared their repertoires with those of publicly available sequences of systemic DLBCLs, including 262 ABC-type and 93 germinal center B-cell (GCB) type.20 We report that the IG repertoire of PVRL is massively biased, with an overwhelming representation of the IGHV4-34 gene, and the presence of a subset of cases with highly restricted stereotyped IGHV3-7 BCR immunoglobulin. These features clearly delineate PVRL from PCNSL, which displays BCR IG repertoire features resembling those of systemic ABC-type DLBCL.
Materials and methods
Patients and samples
Fifty-five PVRL cases and 48 PCNSL cases were included in the study (supplemental Table 1). The diagnosis of PVRL was based on clinical ocular findings and extensive biological work-up of vitrectomy specimens, including cytological and immunophenotypic analyses, as well as cytokine quantification and molecular clonality assessment via IG gene rearrangement studies.30,31 PCNSLs were classified as DLBCL on the basis of morphology and immunochemistry examination, according to the World Health Organization classification.3,32
All PVRL and PCNSL samples were obtained at the time of initial presentation from HIV-negative patients. The study was approved by the CPP Ile-de-France VI Ethics Review Committee, and informed consent was obtained for all patients.
In addition, we analyzed a dataset of 398 publicly available IGHV-IGHD-IGHJ gene rearrangement sequences extracted from a recently published study including systemic DLBCL primary tumors and cell lines.20 These sequences were reanalyzed using ImMunoGeneTics (IMGT) databases and the IMGT/V-QUEST tool (http://www.imgt.org).33 Redundant, out-of-frame, truncated, or defective sequences were excluded from the analysis, resulting in 355 productive IGH sequences from 262 ABC-type and 93 GCB-type DLBCLs.
PCR amplification and sequence analysis of IG gene rearrangements
Polymerase chain reaction (PCR) amplification and sequence analysis of IG HC and LC gene rearrangements were performed as previously described.34-37 Only productive rearrangements were included in downstream analyses. Following established definitions,38,39 the V gene sequences were categorized as truly unmutated (100% germline identity), minimally mutated (97%-99.9% germline identity), or mutated (<97% germline identity). Each single-point mutation was compared with the respective germline sequence and classified as a replacement (R) or silent (S) mutation. The ratio of R/S mutations was determined within the framework regions (FRs) and complementarity-determining regions (CDRs) of the V domain. To take into account the different lengths of V regions, absolute mutation counts were normalized by the actual nucleotide length of the corresponding V region, as previously described.40 In addition, we used the BASELINe computational program (http://selection.med.yale.edu/baseline) based on a Bayesian estimation of antigen-driven selection, which takes into consideration intrinsic biases within FRs and CDRs.41,42 The IGHV4-34 rearranged sequences were searched for the N-glycosylation motifs Asn-X-Ser/Thr (N-X-S/T), where X could be any amino acid other than Pro (P), Asp (D), or Glu (E) in CDR2 and FR3, which could be created or lost as a result of the somatic hypermutation (SHM) process.43 A search for stereotyped IGHV-IGHD-IGHJ gene rearrangements was done using previously described criteria.44
Additional information is provided in supplemental Material.
MYD88 L265P mutation detection
Screening for MYD88 mutations was performed by amplifying exon 5 sequences spanning the L265P hotspot using previously published forward (GCTGTTGTTAACCCTGGGGTTGAAG) and reverse (GAACCTCAGGATGCTGGGGAAC) primers.45 PCR products were purified by a QIAquick PCR Purification Kit (Qiagen) and sequenced directly on both strands on an ABI 3730 automated sequencer.
Statistical analysis
Differences in frequencies were evaluated using descriptive statistics. Associations between variables were assessed by the χ2 test or the Fisher’s exact test of independence. Continuous variables were analyzed using analysis of variance. All tests were 2-sided, and differences were considered statistically significant for P < .05. All statistical analyses were performed using MedCalc version 18.9 software (www.medcalc.org).
Results
IGH gene repertoire
Overview.
Overall, productive IGHV-IGHD-IGHJ rearrangements from 55 PVRL and 48 PCNSL cases were analyzed; 1 PCNSL case carried double-productive rearrangements. Because most PCNSLs and PVRLs are DLBCLs with features predominantly of the ABC subtype,5-8 we also used a publicly available dataset of productive DLBCL IGHV-IGHD-IGHJ gene rearrangement sequences, including 262 cases with the ABC subtype and 93 cases with the GCB subtype. The number of cases that might represent PCNSLs within this public series is unknown; however, considering the overall low proportion of PCNSLs within the DLBCL population (<5%),3,46 their potential confounding effect is reasonably estimated to be minimal.
IGHV gene repertoire.
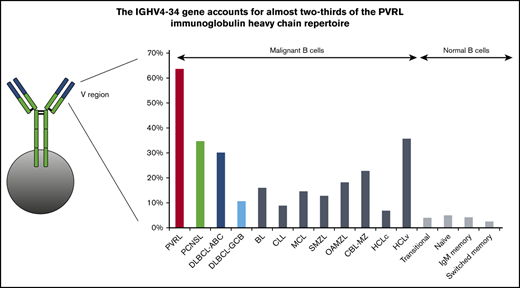
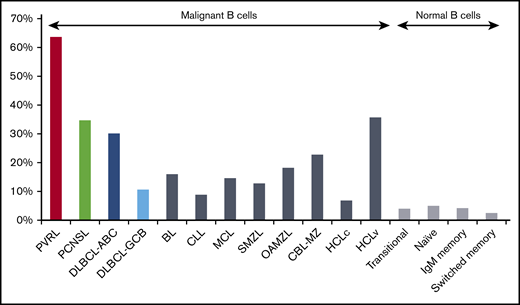
In PVRL, IGHV4 (39/55; 70.9%) and IGHV3 (14/55; 25.5%) subgroup genes accounted for 96.4% of the series (supplemental Table 2). The IGHV4-34 gene predominated (35/55; 63.6%) (Figure 1; supplemental Table 3), followed by IGHV3-7 (8/55; 14.6%). IGHV4 and IGHV3 subgroups were also predominant in PCNSL but in slightly different proportions: 51% (25/49; P = .038) and 42.9% (21/29; P = .062), respectively. These tumors also showed a bias for IGHV4-34 gene usage (17/49; 34.7%), although at a significantly lower frequency compared with PVRL (P = .003). IGHV3-7, as well as IGHV3-48, ranked second (4/49; 8.2%), at a frequency not significantly different from PVRL. IGHV4-34 and IGHV3-7 displayed biased usage in DLBCL-ABC, with proportions similar to that of PCNSL (30.2% and 7.6%, respectively) (supplemental Table 3). In contrast, they were much less predominant in DLBCL-GCB compared with PVRL, accounting for 10.8% (P < .0001) and 3.2% (P = .012), respectively, of cases, whereas the IGHV3-23 gene predominated (11.8%; P = .032). This massive IGHV4-34 bias in PVRL prompted comparisons with other B-cell malignancies,20,38,39,44,47-50 as well as normal B cells.51 As shown in Figure 2, IGHV4-34 gene frequency was by far the highest in PVRL (P < .001).
Comparative IGHV gene usage in PVRL, PCNSL, DLBCL-ABC, and DLBCL-BCB. Two genes, IGHV4-34 and IGHV3-7, account for 78% of the repertoire in PVRL. *P < .05, **P <.01, ***P < .0001. ns, not significant.
Comparative IGHV gene usage in PVRL, PCNSL, DLBCL-ABC, and DLBCL-BCB. Two genes, IGHV4-34 and IGHV3-7, account for 78% of the repertoire in PVRL. *P < .05, **P <.01, ***P < .0001. ns, not significant.
IGHV4-34 gene usage among malignant and normal B cells. The IGHV4-34 gene is used preferentially in many B-cell malignancies, but the bias in PVRL is the most important. Statistically significant differences were observed for comparisons between PVRL and all other malignant or normal B-cell populations (P < .001).20,38,39,44,47-50 BL, Burkitt lymphoma; CBL-MZ, clonal B-cell lymphocytosis of marginal zone origin; CLL, chronic lymphocytic leukemia; HCLc, hairy cell lymphoma classical type; HCLv, hairy cell lymphoma variant type; MCL, mantle cell lymphoma; OAMZL, ocular adnexa marginal zone lymphoma; SMZL, splenic marginal zone lymphoma.
IGHV4-34 gene usage among malignant and normal B cells. The IGHV4-34 gene is used preferentially in many B-cell malignancies, but the bias in PVRL is the most important. Statistically significant differences were observed for comparisons between PVRL and all other malignant or normal B-cell populations (P < .001).20,38,39,44,47-50 BL, Burkitt lymphoma; CBL-MZ, clonal B-cell lymphocytosis of marginal zone origin; CLL, chronic lymphocytic leukemia; HCLc, hairy cell lymphoma classical type; HCLv, hairy cell lymphoma variant type; MCL, mantle cell lymphoma; OAMZL, ocular adnexa marginal zone lymphoma; SMZL, splenic marginal zone lymphoma.
VH CDR3 characteristics.
IGHD genes could be identified in 54 of 55 PVRL sequences, although with some uncertainty because of the high level of somatic mutations. IGHD3 and IGHD2 subgroup genes predominated (38.2% and 21.8%, respectively), and IGHD3-10 was the most frequent gene (10/55; 18.2%). PCNSL and DLBCL-ABC displayed similar IGHD subgroup gene usage profiles, with the exception of IGHD6 subgroup genes, which were found in only 3.6% of cases of PVRL and 6.5% of cases of DLBCL-GCB (P = .467) but in 16.3% of cases of PCNSL (P = .029) and 14.9% of cases of DLBCL-ABC (P = .024) (supplemental Table 4A).
IGHJ4 was used by the majority of PVRL, PCNSL, DLBCL-ABC, and DLBCL-GCB cases (49.1%, 53.1%, 53.3% and 49.5%, respectively). It was followed by IGHJ6 in PCNSL (14.5%), DLBCL-ABC (18.4%), and DLBCL-GCB (30.1%), but it was followed by IGHJ5 in PVRL (25%), although the differences were not statistically significant, with the exception of IGHJ6 in PVRL and DLBCL-ABC (14.5% vs 30.1%; P = .034) (supplemental Table 5A). Of note, the IGHD and IGHJ gene repertoires were similar when only IGHV4-34 cases were considered (supplemental Tables 4B and 5B).
In PVRL, the VH CDR3 had a median length of 14 aa (range, 6-28), identical to that of PCNSL and DLBCL-GCB, whereas it was 15 aa long in DLBCL-ABC (supplemental Figure 1). When considering only the cases expressing IGHV4-34, the median VH CDR3 was 15 aa long in PVRL, 14 aa long in PCNSL, and 16 aa long in both DLBCL subtypes.
Stereotypy analysis.
We searched for stereotyped IGHV-IGHD-IGHJ gene rearrangements according to established criteria44 : IGHV-IGHD-IGHJ gene rearrangement sequences having (1) identical VH CDR3 length, (2) ≥50% VH CDR3 amino acid identity and 70% similarity in amino acid physicochemical properties, (3) identical offsets of shared VH CDR3 motifs, and (4) usage of IGHV genes from the same IGHV phylogenetic clan. No stereotyped cases could be identified among IGHV4-34–expressing PVRLs. In contrast, 4 of the 8 PVRL cases expressing the IGHV3-7 gene carried stereotyped VH CDR3 (Figure 3). All involved IGHV3-7/IGHJ4 gene rearrangements carrying an 11aa-long VH CDR3 with basic isoelectric point values (pI) (mean pI, 9.29), in contrast to the acidic pI values of IGHV3-7 rearrangements from nonsubset PVRL cases (mean, 5.11; P = .0018) and PCNSL cases (mean, 4.82; P =.0004) (supplemental Table 6). No stereotyped sequences could be found within the PCNSL cases, but 1 of the ABC-type DLBCLs presented similar features (Figure 3). This prompted us to search in other B-cell malignancies, including chronic lymphocytic leukemia (CLL),44 mantle cell lymphoma,38 splenic marginal zone lymphoma,39 and clonal B-cell lymphocytosis of marginal zone origin,49 where IGH stereotypy had been thoroughly investigated, but no similar restricted sequences could be found (data not shown).
HCDR3 amino acid sequence of stereotyped IGHV3-7 gene rearrangements. The 11 aa composing HCDR3 of each stereotyped IGHV3-7 case are aligned. The color reflects their origin: black, IGHV or IGHJ genes; blue, IGHD genes; red, N (or P) nucleotides; green, conserved anchor residues bordering HCDR3. Underlined amino acids resulted from replacement somatic mutations. Identical N-D-N residues for all cases in boxes. The IGHD genes contributing to HCDR3 are indicated on the right.
HCDR3 amino acid sequence of stereotyped IGHV3-7 gene rearrangements. The 11 aa composing HCDR3 of each stereotyped IGHV3-7 case are aligned. The color reflects their origin: black, IGHV or IGHJ genes; blue, IGHD genes; red, N (or P) nucleotides; green, conserved anchor residues bordering HCDR3. Underlined amino acids resulted from replacement somatic mutations. Identical N-D-N residues for all cases in boxes. The IGHD genes contributing to HCDR3 are indicated on the right.
SHM analysis.
IGHV-IGHD-IGHJ gene rearrangement sequences in PVRL displayed a heavy load of somatic mutations, with a mean IGHV germline identity of 85.9%. In fact, all sequences were mutated, with the range of IGHV germline identity spanning from 77.7% to 96.5% (median, 86.1%) (supplemental Figure 2). A particularly pronounced burden of mutations was even more evident for IGHV4-34 genes, because all but 1 sequence had IGHV germline identity <91%. PCNSL sequences were slightly less mutated (mean, 88%; P = .052 vs PVRL), with 3 cases (6.1%) belonging to the minimally mutated category (Figure 4; supplemental Table 7). In contrast, DLBCL-ABC was significantly less mutated, having a mean germline identity of 91.3% (P < .0001). In fact, 4.2% of cases were truly unmutated and 9.9% were minimally mutated, whereas only 85.9% belonged to the mutated category (Figure 4; supplemental Table 6). DLBCL-GCB was also significantly less mutated than PVRL, with a mean germline identity of 89.2% (P = .0004).
Mutational status of IGHV genes in PVRL, PCNSL, DLBCL-ABC, and DLBCL-GCB. Distribution of the SHM load in the heavy (A) and light chain (B) V domain.
Mutational status of IGHV genes in PVRL, PCNSL, DLBCL-ABC, and DLBCL-GCB. Distribution of the SHM load in the heavy (A) and light chain (B) V domain.
R/S ratios were first calculated using normalized distribution percentages, as previously described,52 to take into account the respective sequence length of each FR and CDR. The R/S ratio in PVRL IGHV rearranged sequences was higher in CDRs (3.96) compared with FRs (1.44), indicative of antigen selection pressure (supplemental Table 8). These features were also shared by PCNSL and DLBCL, as reported previously.19,27 However, using another computational methodology, which considers the inherent mutation ability of the FRs and CDRs,41,42 most of the antigen selection pressure appeared to be statistically significant only in the FRs, with a lower than expected R/S ratio indicating a negative selection (supplemental Table 9).
Recurrent amino acid changes could be observed within PVRL IGHV4-34 rearrangement sequences at certain positions (eg, G to D at VH CDR1 codon 36, S to T at VH FR2 codon 40, P to S at VH FR2 codon 45) (supplemental Figure 3). However, compared with IGHV4-34 rearrangement sequences from PCNSLs and DLBCLs, none of these preferential replacements proved to be PVRL specific (data not shown).
The IGHV4-34 germline gene harbors a motif within the VH FR1 for binding the N-acetyl-lactosamine carbohydrate epitope that is present in various exogenous and self-antigens.53-55 Although the IGHV4-34 rearrangement sequences in the present study were highly mutated, the VH FR1 motif was mostly spared from R mutations in PVRL, PCNSL, and DLBCL-ABC cases. In contrast, the AVY motif at codons 24 to 26 was significantly more mutated in DLBCL-GCB (4/10; P = .034) (Table 1).
IGHV4-34 superantigen and N-glycosylation motifs
| . | PVRL* . | PCNSL* . | DLBCL-ABC* . | DLBCL-GCB* . | P (across entities) . |
|---|---|---|---|---|---|
| IGHV4-34 super antigen motif | |||||
| Codon 6 (Q) | 1/26 (3.8) | 2/15 (13.3) | 2/79 (2.5) | 1/9 (11.1) | .237 |
| Codon 7 (W) | 0/26 (0) | 0/15 (0) | 2/79 (2.5) | 1/9 (11.1) | .256 |
| Codons 24-26 (AVY motif) | 4/35 (11.4) | 1/17 (5.9) | 7/79 (8.9) | 4/10 (40.0) | .034 |
| IGHV4-34 N-glycosylation motifs | |||||
| Disruption of CDR2 N-glycosylation motif | |||||
| Codon 57-59 (NHS) | 31/35 (88.6) | 12/17 (70.6) | 41/79 (51.9) | 41/79 (51.9) | .001 |
| Creation of a novel N-glycosylation motif in FR3 | |||||
| Codon 68-70 (NPS) | 5/35 (14.3) | 0/17 (0) | 4/79 (5.1) | 4/79 (5.1) | .198 |
| . | PVRL* . | PCNSL* . | DLBCL-ABC* . | DLBCL-GCB* . | P (across entities) . |
|---|---|---|---|---|---|
| IGHV4-34 super antigen motif | |||||
| Codon 6 (Q) | 1/26 (3.8) | 2/15 (13.3) | 2/79 (2.5) | 1/9 (11.1) | .237 |
| Codon 7 (W) | 0/26 (0) | 0/15 (0) | 2/79 (2.5) | 1/9 (11.1) | .256 |
| Codons 24-26 (AVY motif) | 4/35 (11.4) | 1/17 (5.9) | 7/79 (8.9) | 4/10 (40.0) | .034 |
| IGHV4-34 N-glycosylation motifs | |||||
| Disruption of CDR2 N-glycosylation motif | |||||
| Codon 57-59 (NHS) | 31/35 (88.6) | 12/17 (70.6) | 41/79 (51.9) | 41/79 (51.9) | .001 |
| Creation of a novel N-glycosylation motif in FR3 | |||||
| Codon 68-70 (NPS) | 5/35 (14.3) | 0/17 (0) | 4/79 (5.1) | 4/79 (5.1) | .198 |
Data are n/N (%).
Loss or gain of potential N-glycosylation sites as a consequence of SHM targeting has been described in various B-cell malignancies.56-58 The germline-encoded N-X-S/T motif within the VH CDR2 of the IGHV4-34 gene was disrupted by mutations more often in PVRL than in other types, especially systemic DLBCL (P < .001) (Table 1). In addition, a novel N-glycosylation site was created within the VH FR3 in 5 of 35 (14.3%) PVRLs, 0 of 17 (0%) PCNSLs, 4 of 79 (5.1%) DLBCLs-ABC, and 1 of 10 (10%) DLBCLs-GCB (P = .198).
Intraclonal diversity within the IG genes was assessed by cloning and sequencing multiple (16-29) independent subclones in 9 cases of PVRL, including 6 cases expressing the IGHV4-34 gene, 2 cases expressing the IGHV3-7 gene, and 1 case expressing the IGHV5-10-1 gene (supplemental Table 10). All cases demonstrated intraclonal heterogeneity, although to varying degrees, ranging from 0.05% to 0.37%, frequencies above the error rate recently determined for Platinum Taq DNA Polymerase High Fidelity (<0.015%).59 However, when considering only “confirmed” mutations (eg, those found in ≥2 subcloned sequences from a given case),40 only 6 cases displayed clear signs of intraclonal diversification, including 5 of 6 IGHV4-34–expressing cases (supplemental Figure 4).
IGK and IGL gene repertoires
Analysis of the LC gene repertoire was performed in PVRL and PCNSL. In PVRL, 31 productive LC rearrangements were obtained in 30 cases, including 23 IGKV-IGKJ and 8 IGLV-IGLJ gene rearrangement sequences. One case carried 2 productive IGKV-IGKJ and IGLV-IGLJ rearrangements. Similarly, 29 productive LC gene rearrangement sequences, including 26 IGKV-IGKJ and 3 IGLV-IGLJ gene rearrangement sequences, were obtained in 25 PCNSLs, with 4 cases bearing 2 productive IGKV-IGKJ rearrangements.
The LC gene repertoire of PVRL was also biased, although not to the same extent as for the HC. Two genes (IGKV3-20, 7 cases and IGKV3-11, 3 cases) contributed to about one third (10/31, 32.3%) of the LC gene repertoire (supplemental Table 11). Interestingly, these 2 LC genes were associated with an IGHV4-34 HC partner in all cases, resulting in about half (10/21) of the PVRL cases with IGHV4-34–encoded HC being associated with IGKV3-20 or IGK3-11 LC gene rearrangements. There was no preferential LC gene usage for IGHV3-7 PVRL cases. Of importance, all 7 IGKV3-20 gene rearrangements used the IGKJ1 gene, with 5 of these displaying very similar restricted sequences (supplemental Table 12; supplemental Figure 5).
The IGKV3-20 gene was also used preferentially in PCNSL, along with IGKV2-28/IGKV2D-28 and IGKV4-1 (4 cases each) and IGKV1-39/IGKV1D-39 (3 cases) genes (supplemental Table 11). Moreover, the association between HC and LC genes was much less pronounced than in PVRL.
All but 2 of 31 (6.5%) PVRL LC gene rearrangement sequences were mutated, with a mean germline identity of 93.3%. The PCNSL LC sequences displayed a significantly less pronounced mutational burden, having a mean germline identity of 95.5% (P = .038), with 6 of 29 (20.7%) cases being unmutated (supplemental Table 13). Interestingly, recurrent amino acid replacements were observed among some LC sequences. The most striking example involved the IGKV3-20 and IGKV3-11 gene rearrangements in PVRL; all were associated with IGHV4-34 gene rearrangements, where a serine to asparagine substitution at position 37 of CDR2 occurred in 4 of 7 cases and in 3 of 3 cases, respectively (supplemental Figure 6).
MYD88 mutations
The MYD88 L265P mutation has been shown to be a recurrent event in CNS lymphoma, with reported frequencies of up to 60%.60 We searched for the presence of this mutation in 52 PVRL cases and 28 PCNSL cases from the present series. The MYD88 L265P mutation was detected in 50% and 42.9% of cases (P = .641), respectively (Table 2). In PVRL, it was found more frequently in cases bearing IGHV4-34 (56.3%) or IGHV3-7 (62.5%) gene rearrangements than in other cases (25%), whereas the opposite was found in PCNSL, although these differences were not statistically significant (P =.153 and P = .629, respectively). Notably, within PVRL cases expressing IGHV3-7, all 4 cases carrying stereotyped VH CDR3 were MYD88 mutated, in contrast to only 1 of 4 nonstereotyped cases, although the difference was not statistically significant (P = .148), likely as a result of low patient numbers.
MYD88 L265P mutation frequency in PVRL and PCNSL according to IGHV gene usage
| IGHV gene . | PVRL* . | PCNSL* . | P . |
|---|---|---|---|
| IGHV4-34 | 18/32 (56.3) | 2/7 (28.6) | .23 |
| IGHV3-7 | 5/8 (62.5) | 1/3 (33.3) | .54 |
| IGHV3-7 subset | 4/4 (100) | .148 | |
| IGHV3-7 nonsubset | 1/4 (25) | ||
| Others | 3/12 (25) | 9/18 (50.0) | .26 |
| All | 26/52 (50) | 12/28 (42.9) | .647 |
| IGHV gene . | PVRL* . | PCNSL* . | P . |
|---|---|---|---|
| IGHV4-34 | 18/32 (56.3) | 2/7 (28.6) | .23 |
| IGHV3-7 | 5/8 (62.5) | 1/3 (33.3) | .54 |
| IGHV3-7 subset | 4/4 (100) | .148 | |
| IGHV3-7 nonsubset | 1/4 (25) | ||
| Others | 3/12 (25) | 9/18 (50.0) | .26 |
| All | 26/52 (50) | 12/28 (42.9) | .647 |
Data are n/N (%).
Discussion
The IG gene repertoire of PVRL, particularly how it compares with that of PCNSL, has remained unresolved, because of the rarity of these tumors and the very small number of reported cases.28,29 Based on a series of 55 PVRL cases, the largest series analyzed so far, we provide evidence that these lymphomas display a remarkably biased IG repertoire, considerably more pronounced than that of PCNSL.
The most striking feature involved the extremely high frequency of the IGHV4-34 gene, which was found in 63.6% of PVRLs vs only 34.7% of PCNSLs. In initial PCNSL studies, IGHV4-34 gene usage has been reported to range from 17% to 80%, but such differences might be related to the small size of the respective cohorts (5-13 cases).21-26 The largest published series (50 patients)27 found that 36% of cases expressed IGHV4-34, which was similar to our findings.
Considering that the vast majority of PVRLs and PCNSLs are classified as DLBCLs, comparison with systemic cases seemed particularly relevant. To this end, we took advantage of a recent study, for which IGH gene sequence data and histological subtype assignment were available for a large series of systemic DLBCLs.20 The overall IGHV4-34 usage frequency reached 30.2% in the ABC subtype and 10.6% for the GCB category, supporting that IGHV4-34 gene frequency is comparable in PCNSL and the ABC-subtype of DLBCL, and most likely reflects their non–germinal center cell of origin. In contrast, PVRL clearly stands out, at least from an immunogenetic perspective, displaying an overwhelming representation of the IGHV4-34 gene. Interestingly, 15 PVRL cases presented a CNS localization that was concomitant (n = 5) or occurred later in the disease course (n = 10). The IGHV4-34 gene was used by 10 of them (67%), whereas the IGHV3-7 gene was used by 2 (13%). Alternatively, 5 cases of PCNSL developed a subsequent intraocular localization, but only 1 case used the IGHV4-34 gene. These findings further indicate that antigen selection is a major force driving behind the development of PVRL.
IGVH4-34–expressing cases of PVRL also differed from their PCNSL counterparts with regard to their LC composition, with predominant usage of the IGKV3-20/IGKJ1 gene pair in the former, including 5 cases displaying almost identical VK CDR3 sequences. In addition, the second most frequently used gene, IGVK3-11, displayed some identical amino acid changes induced by somatic mutations as IGKV3-20, reflecting the probable importance of these critical residues in antigen recognition. Such a phenomenon, termed “sequence convergence,” whereby different germline genes become more similar after SHM, is reminiscent of what has been observed in broadly neutralizing anti-HIV antibodies,61 as well as in CLL with stereotyped BCRs.44
Immunoglobulins encoded by the IGHV4-34 gene are endowed with an inherent and, largely LC-independent, autoreactivity by their recognition of N-acetyl-lactosamine residues present in various autoantigens through a germline motif within the VH FR1. In healthy individuals, B cells expressing the IGHV4-34 gene constitute a sizeable fraction (5%-10%) of the normal naive repertoire, but they are usually prevented from participating in immune reactions by being kept in an anergic state.62,63 Moreover, they are censored in the germinal centers and largely excluded from the switched memory and plasma cell compartments.62,64 However, modifications introduced by SHM may allow “redemption” from autoimmunity and simultaneous acquisition of reactivity to new foreign antigens.55,63 Indeed, in patients with genetic IRAK4 or MYD88 deficiency, recombinant IGHV4-34–encoded BCRs from IgG memory cells were found to react with commensal bacteria antigens.65 Although their immunoglobulin had acquired mutations, they maintained germline NAL binding and NHS motifs, raising the possibility of a cross-reactivity between these bacterial antigens and the I/i autoantigens. In line with these findings, although IGHV4-34 sequences from PVRL and PCNSL carried a very high load of mutations, the vast majority preserved the germline N-acetyl-lactosamine-binding motif; thus, in principle, they were capable of recognizing cross-reactive foreign and self-antigens.
Additional strong evidence for the restricted nature of the IG gene repertoire in PVRL was provided by the finding of stereotyped VH CDR3 in half of IGHV3-7–expressing cases. This stereotyped VH CDR3 was also detected in 1 of the DLBCL-ABC cases, although, because of the lack of clinical information, the possibility of CNS localization cannot be excluded. Although the number of cases was very limited, the sequence similarity did not extend to the LCs, because the 2 cases available for analysis expressed different LC isotypes. The situation contrasts with the IGHV4-34 cases, for which stereotypes were not observed for the HC, whereas they were present among LCs, thereby reflecting distinct molecular requirements for antigen binding by these 2 types of “prototypic” BCRs. Another intriguing feature of this IGHV3-7 stereotypic subset involved the presence of the MYD88 L265P mutation in all 4 IGHV3-7 subset cases but only in 1 of the 4 nonsubset cases, as well as about half of the PCNSL and non–IGHV3-7 PVRL cases, in accordance with previous studies.66-69 This recalls previous observations of biased associations of specific BCR IGHV genes and particular genomic alterations in other B-cell lymphomas, including enrichment of mutations involving SF3B1 in CLL subset 2,70 MAP2K1 in IGHV4-34 hairy cell leukemia,71 KLF2 in IGHV1-2*04 splenic marginal zone lymphoma,72 and TNFAIP3 in IGHV4-34 ocular adnexa marginal zone lymphoma.73 This may reflect an exquisite synergy of immunogenic and oncogenic driving forces underlying tumorigenesis.
PVRLs displayed very high levels of mutations in their IG genes, with IGHV4-34 sequences being the most mutated. The pattern of R and S mutations was indicative of negative antigen selection in the FRs, leading to the preservation of the immunoglobulin structure. A low level of intraclonal diversification could be detected, indicative of continuing mutational activity in the tumors. These findings are consistent with a prolonged affinity-maturation process, in line with previous reports on PCNSL.21,26,27 In addition, they raise the issue of where these mutations are acquired in these tumors, because the CNS and the eye are considered immune-privileged sites.
However, a number of observations challenge the concept of the CNS being an immune-privileged site; rather, they support the notion that immune cells are present in the CNS, notably in the meninges, as well as in ectopic lymphoid structures, and participate in immune surveillance. Trafficking of B cells in the CNS and the periphery has been demonstrated in multiple sclerosis; high-throughput sequencing studies of the BCR IG repertoire have revealed shared B-cell clones in the brain parenchyma, the meningeal aggregates,74 as well as cerebral spinal fluid and peripheral blood.75,76 Whether this model can be applied to malignant B cells remains to be shown, but it provides a framework within which to explain their development in the brain and the eye.
The restrictions in the IG repertoire of PVRL reported here are highly suggestive of an antigen selection process. The nature of the antigens remains elusive, although some hints can be gained from recent studies that tested antigen reactivity of PCNSL BCR immunoglobulin, expressed as recombinant antibodies, against protein microarrays.77,78 Montesinos-Rongen et al found that these antibodies displayed polyreactivity, although not with common autoantigens but against a range of diverse proteins, including some expressed by CNS neurons. All antibodies using the IGHV4-34 gene recognized galectin-3, a carbohydrate-binding protein expressed by cells from the cerebral microenvironment.77 Because galectin-3 interacts with retinal pigment epithelial cell proteins, it raises the possibility that it might also be a target of PVRL cells.79 Alternatively, Thurner et al provided evidence that PCNSLs recognize atypically hyper–N-glycosylated forms of SAMD14 and neurabin-I, 2 proteins that are highly expressed in CNS tissue.78 However, these investigators failed to detect galectin-3 as a target of their recombinant BCRs.
Altogether, our data show that PVRLs display a very biased IG repertoire, strongly suggesting that antigen selection plays a major role in their development. Therefore, these tumors should be considered good candidates for treatment by BCR signaling pathway inhibitors. Promising results with high response rates have recently been reported with ibrutinib in PCNSL.80,81 These were confirmed in a French clinical trial testing ibrutinib as monotherapy in PCNSL, as well as PVRL.82
Data sharing requests should be sent to Frederic Davi (frederic.davi@aphp.fr).
Acknowledgments
This work was supported by the Institut Universitaire d’Ingénierie en Santé; Sorbonne Université; ERA-NET TRANSCAN-2, Project Acronym: Novel; CURAMUS (Cancer United Research Associating Medicine, University and Society, INCA-DGOS-Inserm_12560); and the Hellenic Precision Medicine Network in Oncology.
Authorship
Contribution: N.B. performed experiments and analyzed data; M.B. performed experiments; A.X., B.S., and H.M.-B. analyzed data; C.L. performed statistical analyses; N.C., C.F., T.H.C.T., S.C., C.H., A.A., P.L., C.S., V.T., K.H.-X., and B.B. provided materials and clinical data; A.X. and K.S. critically revised and contributed to the writing of the manuscript; and F.D. designed the study, supervised the research, and wrote the manuscript, which was approved by all of the authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Frederic Davi, Department of Biological Hematology, Hôpital Pitié-Salpêtrière, 47-83 Bd de l'Hôpital, 75013 Paris, France; e-mail: frederic.davi@aphp.fr.
References
Author notes
The full-text version of this article contains a data supplement.