TO THE EDITOR:
Marginal zone lymphoma (MZL) is a rare group of indolent non-Hodgkin lymphomas that originate from the marginal zone of lymphoid follicles.1 Their diagnosis remains difficult and in many cases is finally performed by the exclusion of other low-grade B-cell lymphomas. Cytogenetically, gains of chromosomes 3 and 18 have been described in all subtypes of MZLs.2 Specifically, extranodal lymphoma of the mucosa-associated lymphoid tissue is characterized by the occurrence of different translocations [t(11;18)(q21;q21), t(14;18)(q32;q21), and t(1;14)(p22;q32)].3 The deletion of 7q is the most frequent abnormality in primary splenic MZL (30%-40% of patients), and trisomy of chromosomes 3/3q is the most recurrent gain. Cytogenetics in nodal MZLs (NMZLs) is not well defined.4 Molecularly, splenic MZL and NMZL have a common genetic background, characterized by mutations in NOTCH, KLF2, and NF-κB genes.5-8 Mucosa-associated lymphoid tissue lymphomas show a heterogeneous genetic landscape in which many genes, such as TNFAIP3, KMT2C, KMT2D, CREBBP, TET2, SPEN, LRP1B, PRDM1, and EP300, may be involved.9
Here, we describe a new gene fusion IGF2BP2::LSM12 in 2 patients with MZL and translocation t(3;17)(q27;q21) in the karyotype. To our knowledge, this is the first time that this genetic alteration has been reported in cancer.
Cytogenetic tests on bone marrow (BM), peripheral blood (PB), and lymph node (LN) were performed according to the standard methods used in our laboratory. Fluorescence in situ hybridization (FISH) study using clones from human 32K bacterial artificial chromosome rearray library (BACPAC Genomics, Richmond, CA) was applied to confirm IGF2BP2::LSM12 fusion in metaphases from altered karyotypes (supplemental Figure 1). Cytoscan 750K Array Kit cytogenetic solution (Affymetrix, ThermoFisher) was used to study genetic gain, loss, and copy neutral loss of heterozygosity following the current recommendations. Optical genome mapping (OGM) was performed with the rare variant pipeline included in Bionano Solve software (version 3.5) (Bionano Genomics, San Diego, CA) and visualized in Bionano Access software (version 1.6) to identify structural variants and large copy number variants. Targeted next-generation sequencing (NGS) was performed on DNA extracted from the PB mononuclear cells or the LN. Gene libraries were prepared with a QIAseq Targeted DNA Custom Panel (Qiagen, Hilden, Germany) covering the full coding region of 15 genes involved in lymphoid malignancies (supplemental Table 1) and were sequenced with MiSeq (Illumina, San Diego, CA). Single nucleotide variants and insertions/deletions were assessed with a sensitivity of 2% of variant allele frequency. Whole-genome sequencing (WGS) was performed using DNA from BM of patient 1, allowing the analysis of single nucleotide variants, insertions/deletions, copy number variants, and rearrangements (Novogene Co, United Kingdom). All the alterations reported were reviewed using Integrative Genomics Viewer software (Broad Institute).
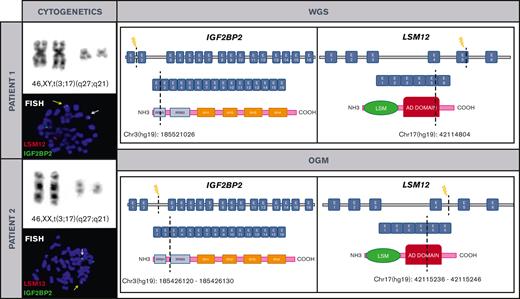
The first patient was a 45-year-old man who presented with a 4-month history of peripheral lymphadenopathy. Physical examination revealed bilateral cervical, axillary and inguinal enlarged LNs. Laboratory investigations showed a hemoglobin level of 116 g/L, leukocyte count of 6.86 × 109/L (neutrophils, 1.5 × 109/L; lymphocytes, 4.92 × 109/L), and platelet count of 60 × 109/L. A positron emission tomography scan revealed increased metabolic uptake in LNs at cervical, axillary, and inguinal regions. A PB smear examination identified atypical lymphocytes (6%). BM aspiration and biopsy showed 90% infiltration of atypical small lymphocytes. Flow cytometry (FC) analysis on BM was consistent with the diagnosis of MZL (Table 1). Karyotype of the BM revealed a single alteration t(3;17)(q27;q21). An excisional biopsy of an inguinal LN was performed, and a vaguely nodular infiltration by small lymphoid cells effacing the LN architecture was found. The immunophenotype was equivalent to that observed in the BM. Cyclin D1, SOX11, and in situ hybridization for Epstein-Barr virus were negative and proliferative index determined by Ki67 immunostaining was 10% to 20%. Cytogenetic analysis of LN cells also showed t(3;17)(q27;q21). NGS lymphoid custom panel showed the pathogenic CXCR4 frameshift mutation p.(Ser341PhefsTer3), with a variant allele frequency of 20.9%. The diagnosis of NMZL was established based on integration of the clinical, morphological, immunophenotypic, and genetic data.10 To better characterize t(3;17)(q27;q21), we performed WGS that identified a rearrangement between IGF2BP2 (cytogenetic location 3q27.2) and LSM12 (cytogenetic location 17q21.31) (supplemental Figure 2). These findings were later confirmed by FISH (Figure 1). The patient was treated with rituximab plus CHOP (cyclophosphamide, doxorubicin, vincristine, and prednisone) for 6 cycles and achieved metabolic complete remission with negative minimal residual disease measured by FC, both in PB and BM.11
Comparative table of the clinical and biological characteristics of the 2 patients
| . | Patient 1 . | Patient 2 . |
|---|---|---|
| Age, y | 47 | 76 |
| Sex | Male | Female |
| Eastern Cooperative Oncology Group scale | 0 | 0 |
| B symptoms | No | No |
| Lymph nodes affected | 2 sides of the diaphragm | 2 sides of the diaphragm |
| Splenomegaly | No | No |
| Extranodal involvement | No | Gastrointestinal infiltration |
| PB expression | Yes | Yes |
| BM involvement | Yes | Yes |
| Ann Arbor stage | IV | IV |
| Hemoglobin (g/L) | 116 | 120 |
| Platelets (× 109/L) | 60 | 234 |
| Leucocytes | ||
| Global count (× 109/L) | 6.86 | 12.7 |
| Neutrophils (× 109/L) | 1.5 | 4.2 |
| Lymphocytes (× 109/L) | 4.92 | 7.8 |
| Monoclonal gammopathy | No | IgM |
| Increased lactate dehydrogenase | No | No |
| Increased β2 microglobulin | No | No |
| HIV | Negative | Negative |
| HBV; HCV | Positive HB core antibody, negative HB surface antigen; HCV negative | Positive HB core antibody, negative HB surface antigen; HCV negative |
| Immunophenotype | CD19++, CD20++, CD79b+, CD23+, CD25+, CD5+, BCL2+++, kappa restriction CD10−, CD11c−, CD103−, CD123−, CD200−, BCL6−, cyclin D1−, SOX11− | CD19++, CD20++, CD79b+, CD5−, CD11c+, BCL2+++, kappa restriction CD10−, CD23−, CD103−, CD123−, CD200−, cyclin D1− |
| Ki67 (%) | 10-20 | 5 |
| Cytogenetics | ||
| Karyotype | 46,XY,t(3;17)(q27;q21)[10]/46,XY[10] | 46,XX,t(3;17)(q27;q21)[15]/46,XX[5] |
| FISH, IGF2BP2:LSM12 | Positive in 60% of the nuclei | Positive in 58% of the nuclei |
| FISH, IGH:CCND1 | Negative | Negative |
| FISH, BCL6 (3q27) SPLIT | Negative | Negative |
| Single nucleotide polymorphism array | No copy number alterations or loss of heterozygosis | No copy number alterations or loss of heterozygosis |
| OGM | Not done | IGF2BP2::LSM12 |
| MYD88 | Not mutated | Not mutated |
| CXCR4 | CXCR4 Ser341PhefsTer3 mutation | Not mutated |
| WGS | CXCR4 Ser341PhefsTer3 mutation IGF2BP2::LSM12 | Not done |
| . | Patient 1 . | Patient 2 . |
|---|---|---|
| Age, y | 47 | 76 |
| Sex | Male | Female |
| Eastern Cooperative Oncology Group scale | 0 | 0 |
| B symptoms | No | No |
| Lymph nodes affected | 2 sides of the diaphragm | 2 sides of the diaphragm |
| Splenomegaly | No | No |
| Extranodal involvement | No | Gastrointestinal infiltration |
| PB expression | Yes | Yes |
| BM involvement | Yes | Yes |
| Ann Arbor stage | IV | IV |
| Hemoglobin (g/L) | 116 | 120 |
| Platelets (× 109/L) | 60 | 234 |
| Leucocytes | ||
| Global count (× 109/L) | 6.86 | 12.7 |
| Neutrophils (× 109/L) | 1.5 | 4.2 |
| Lymphocytes (× 109/L) | 4.92 | 7.8 |
| Monoclonal gammopathy | No | IgM |
| Increased lactate dehydrogenase | No | No |
| Increased β2 microglobulin | No | No |
| HIV | Negative | Negative |
| HBV; HCV | Positive HB core antibody, negative HB surface antigen; HCV negative | Positive HB core antibody, negative HB surface antigen; HCV negative |
| Immunophenotype | CD19++, CD20++, CD79b+, CD23+, CD25+, CD5+, BCL2+++, kappa restriction CD10−, CD11c−, CD103−, CD123−, CD200−, BCL6−, cyclin D1−, SOX11− | CD19++, CD20++, CD79b+, CD5−, CD11c+, BCL2+++, kappa restriction CD10−, CD23−, CD103−, CD123−, CD200−, cyclin D1− |
| Ki67 (%) | 10-20 | 5 |
| Cytogenetics | ||
| Karyotype | 46,XY,t(3;17)(q27;q21)[10]/46,XY[10] | 46,XX,t(3;17)(q27;q21)[15]/46,XX[5] |
| FISH, IGF2BP2:LSM12 | Positive in 60% of the nuclei | Positive in 58% of the nuclei |
| FISH, IGH:CCND1 | Negative | Negative |
| FISH, BCL6 (3q27) SPLIT | Negative | Negative |
| Single nucleotide polymorphism array | No copy number alterations or loss of heterozygosis | No copy number alterations or loss of heterozygosis |
| OGM | Not done | IGF2BP2::LSM12 |
| MYD88 | Not mutated | Not mutated |
| CXCR4 | CXCR4 Ser341PhefsTer3 mutation | Not mutated |
| WGS | CXCR4 Ser341PhefsTer3 mutation IGF2BP2::LSM12 | Not done |
HBV, hepatitis B virus; HCV, hepatitis C virus.
Cytogenetic and molecular characterization of t(3;17)(q27;q21)/IGF2BP2::LSM12 in both patients. Chromosomes and metaphase-FISH of both patients show t(3;17)(q27;q21)/IGF2BP2::LSM12 fusion (left). Dashed line indicates the DNA and domain structure breakpoints (LSM12: NM_152344 and IGF2BP2: NM_006548.6) revealed after WGS and OGM in both patients. Breakpoints at IGF2BP2 affect RNA recognition motif 1. The RNA recognition motifs have high affinity for RNA, determining the binding capacities of the RNA-binding proteins (RBPs) to RNA, play a central role in the stability of IGF2BP-RNA complexes, and coordinate the interactions between the complex and other RBPs. Breakpoints at LSM12 affect anticodon binding domain (AD). Its function consists of binding the anticodon of transfer RNA (right). COOH, carboxylic acid; E, exon; NH3, ammonia.
Cytogenetic and molecular characterization of t(3;17)(q27;q21)/IGF2BP2::LSM12 in both patients. Chromosomes and metaphase-FISH of both patients show t(3;17)(q27;q21)/IGF2BP2::LSM12 fusion (left). Dashed line indicates the DNA and domain structure breakpoints (LSM12: NM_152344 and IGF2BP2: NM_006548.6) revealed after WGS and OGM in both patients. Breakpoints at IGF2BP2 affect RNA recognition motif 1. The RNA recognition motifs have high affinity for RNA, determining the binding capacities of the RNA-binding proteins (RBPs) to RNA, play a central role in the stability of IGF2BP-RNA complexes, and coordinate the interactions between the complex and other RBPs. Breakpoints at LSM12 affect anticodon binding domain (AD). Its function consists of binding the anticodon of transfer RNA (right). COOH, carboxylic acid; E, exon; NH3, ammonia.
We reviewed the cytogenetic database of MZLs from our institution and identified another patient with t(3;17)(q27;q21) detected in PB. This patient was a 76-year-old woman who was referred to our hematology department in 2016 because of lymphocytosis. She did not have any medical history of interest and was asymptomatic. Physical examination was unremarkable. Laboratory investigations showed a hemoglobin level of 120 g/L, a leukocyte count of 12.79 × 109/L (neutrophils, 4.2 × 109/L and lymphocytes, 7.8 × 109/L), and a platelet count of 234 × 109/L. The PB smear analysis showed 52% atypical small lymphocytes, and immunophenotype determined by FC was consistent with the diagnosis of MZL (Table 1). Karyotype of the PB showed the t(3;17)(q27;q21), and IGF2BP2::LSM12 fusion gene was confirmed by FISH. OGM was performed to clarify specific breakpoints involved in IGF2BP2::LSM12 fusion (Figure 1; supplemental Figure 3). NGS lymphoid custom panel did not detect mutations. BM biopsy showed 20% infiltration of atypical small lymphocytes with the same immunophenotype as described for the blood; cyclin D1 and SOX11 were negative, and proliferative index determined by Ki-67 immunostaining was 5%. Body computed tomography showed slightly increased LN on both sides of the diaphragm (size, <1.5 cm). The diagnosis of monoclonal B-cell lymphocytosis of marginal zone origin was established. The patient followed a watch-and-wait strategy, but 5 years after diagnosis, a computed tomography scan showed nodal progression without splenomegaly. An upper gastroduodenal endoscopy showed diffuse duodenal infiltration by small lymphoid cells with histology and immunophenotype consistent with MZL.
Here, we describe 2 patients with MZL with a novel genetic alteration t(3;17)(q27;q21)/IGF2BP2::LSM12. From the clinical perspective, both patients had disseminated LN involvement, BM infiltration, and PB expression. Clinical behavior was indolent, with slow growing lymphadenopathy in both patients. To our knowledge, this is the first time that a pathogenic relationship between IGF2BP2, LSM12, and MZL has been reported.5,12-17 The insulin-like growth factor 2 messenger RBPs (IGF2BP1, IGF2BP2, and IGF2BP3) belong to a family of RBPs that modulate important aspects of cell function in cancer biology, such as cell polarization, migration, metabolism, proliferation, and differentiation.18,IGF2BP2 has been reported as an oncogene because its overexpression has been associated with aggressive phenotype and poor prognosis in various solid tumors, acute myeloid leukemia, and diffuse large B-cell lymphomas.19-21,LSM12 is also an RBP that interacts with target messenger RNA to regulate gene posttranscriptional expression, which could affect the expression and function of proto-oncogenes and tumor suppressor genes.22 Its implication in oncogenesis has been minimally described. LSM12 has been identified as a fusion partner with BRCA1 in a pantumor survey of 346 cases of BRCA-associated tumors in males.23 In colorectal cancer, LSM12 overexpression negatively correlates with tumor immune infiltration levels, which may promote colorectal cancer transfer.24 In pancreatic adenocarcinoma, LSM12 seems to be downregulated according to a recent study.25
In summary, we report a novel genetic alteration t(3;17)(q27;q21)/IGF2BP2::LSM12 in 2 patients with MZL. Further investigations are needed to better understand the implications of this genetic alteration in lymphomagenesis.
Acknowledgments: This study was supported, in part, by Instituto de Salud Carlos III and cofunded by the European Union (grant FIS-FEDER PI19/00034), GILEAD (grants GLD18/00117, 2017SGR205, and PT20/00023), and Xarxa de Banc de Tumors de Catalunya sponsored by Pla Director d’Oncologia de Catalunya.
Contribution: R.D.-F. performed the research, analyzed and interpreted the clinical and biological results, and wrote the manuscript; C.F.-R. performed next-generation sequencing, analyzed the NGS and WGS data, and wrote the manuscript; M.L. and N.G.-G. collected and analyzed the clinical and biological data; A.F. and L.C. performed the cytologic, immunophenotypic, and histologic diagnostic; A.S. and M.S. designed the research, performed the research, analyzed and interpreted the results, and wrote the manuscript; and all authors reviewed and approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Antonio Salar, Department of Hematology, Hospital del Mar, Passeig Maritim, 25-29, 08003 Barcelona, Spain; e-mail: asalar@parcdesalutmar.cat.
References
Author notes
Data are available on request from the corresponding author, Antonio Salar (asalar@parcdesalutmar.cat).
The full-text version of this article contains a data supplement.
R.D.-F., C.F.-R., and A.S. contributed equally to this study.
M.S. and A.S. are joint senior authors.