Key Points
Detectable pre-HCT MRD, even at a low level of <10–4, and any detectable post-HCT MRD increase the risk of post-HCT relapse.
In B-cell ALL, detection of IgH clonotypes after HCT, but not non-IgH clonotypes, is associated with relapse.
Abstract
Measurable residual disease (MRD) is an adverse prognostic factor in adult patients with acute lymphoblastic leukemia (ALL) undergoing hematopoietic cell transplant (HCT). Next-generation sequencing (NGS) can detect MRD with a sensitivity of 10–6, but the prognostic value of NGS-based MRD in adult patients with ALL undergoing HCT remains minimally studied. To evaluate the prognostic value of NGS-based MRD in adult patients with ALL undergoing HCT, patients aged ≥18 years with ALL who underwent allogeneic HCT at Stanford University or Oregon Health & Science University between January 2014 and April 2021 and were evaluated for MRD using the NGS-based clonoSEQ assay were included in this study. MRD was assessed before HCT (MRDpre) and up to 1 year after HCT (MRDpost). Patients were followed up for leukemia relapse and survival for up to 2 years after HCT. In total, 158 patients had a trackable clonotype for MRD monitoring. The cumulative incidence of relapse was increased at all levels of MRDpre, including in patients who had low MRDpre of <10–4 (hazard ratio [HR], 3.56; 95% confidence interval [95% CI], 1.39-9.15). In multivariable analysis, MRDpre level remained significantly prognostic; however, detectable MRDpost was the strongest predictor of relapse (HR, 4.60; 95% CI, 3.01-7.02). In exploratory analyses limited to patients with B-cell ALL, the detection of post-HCT immunoglobulin H (IgH) MRD clonotypes, rather than non-IgH MRD clonotypes, was associated with relapse. In this analysis across 2 large transplant centers, we found that the detection of MRD by NGS at a level of 10–6 offers significant prognostic value in adults with ALL undergoing HCT.
Introduction
Measurable residual disease (MRD) monitoring after induction and consolidation and before and after HCT is the standard of care in the management of acute lymphoblastic leukemia (ALL).1-3 Detection of MRD is associated with an increased risk of relapse and death across subtypes and therapies, including before allogeneic hematopoietic cell transplant (HCT).4,5 Traditionally, MRD has been clinically defined at a sensitivity of 10–4 and is detected using either multiparameter flow cytometry or polymerase chain reaction (PCR) assays, with regulatory bodies historically using a conservative estimate of 10–3 for drug approval indications.6 Next-generation sequencing (NGS) of unique immunoglobulin (Ig) and/or T-cell receptor (TCR) clonotypes allows for a routine sensitivity of at least 10–6 and is now widely available7 and increasingly used across the United States to evaluate ALL treatment response. Studies have demonstrated that patients with ALL with MRD-negative (MRD–) results at 10–4 via multiparameter flow cytometry but MRD-positive via NGS have an increased risk of relapse compared with patients who test MRD– via both methods,8-10 highlighting the enhanced prognostic value of more sensitive NGS-based MRD. In addition, the generally close correlation between peripheral blood (PB) and bone marrow (BM) reported using NGS-based MRD is attractive for noninvasive disease monitoring.11-13
In adult patients with ALL undergoing HCT, the clinical significance of NGS-based MRD detected below 10–4 remains underreported. In a small retrospective study (n = 29) using banked samples from adults with ALL who underwent HCT, Logan et al demonstrated that NGS MRD of the PB or BM at <10–4 was highly associated with post-HCT outcomes.12 Data from larger cohorts of children undergoing HCT provide mixed results regarding the prognostic value of MRD <10–4. For example, a large study of 616 children with ALL who underwent HCT examined the role of quantitative PCR at levels <10–4 and showed that post-HCT MRD (MRDpost) at any level, including <10–4, was strongly associated with post-HCT relapse, whereas low levels of pre-HCT MRD (MRDpre) were less prognostic.14 In contrast, in another study of 56 children with ALL, both pre- and post-HCT NGS MRD strongly correlated with post-HCT relapse.9 Furthermore, the prognostic significance of various clonotypes (eg, IgH vs Igκ/Igλ) detected using NGS MRD remains unknown. For example, the uniqueness of non-IgH sequences may be less unique and, therefore, less prognostic of disease relapse.
To provide additional insights into the potential value of NGS-based MRD in adult ALL, we constructed a large cohort of patients drawn from 2 large US transplant centers and evaluated the association between NGS-based MRD and HCT outcomes. We hypothesized that both detectable MRDpre and MRDpost would be associated with worse outcomes, even for MRD <10–4.
Methods
Patient cohort and study definitions
This retrospective study included adults (≥18 years) with B-cell or T-cell ALL who underwent HCT at Stanford University or Oregon Health & Science University (OHSU) between January 2014 and April 2021, with planned MRD monitoring using the NGS-based clonoSEQ assay (Adaptive Biotechnologies, Seattle, WA).7,15 MRDpre (the MRD sample directly preceding HCT without interval therapy) and MRDpost were obtained from the BM and/or PB and were assessed per the treating physician’s routine clinical practice for up to 1 year after HCT.
We followed up with patients for leukemia relapse and/or death for up to 2 years after HCT. Relapse was defined as morphologic (≥5% blasts in PB or BM) or clinical (initiation of therapy for recurrence of disease as deemed by the treating physician). Leukemia-specific mortality was defined as the time from HCT to death due to relapse or last follow-up; nonrelapse mortality was considered a competing event.
For the primary analyses, MRD positivity was defined as the presence of a detectable IgH clonotype in B-cell ALL or the presence of a detectable TCRβ or TCRγ clonotype in T-cell ALL; conversely, MRD-negativity was defined as the absence of detectable IgH (B-cell) or TCRβ/TCRγ (T-cell) clonotypes that had previously been identified as representing the leukemic blast population. Non-IgH (ie, Igκ/Igλ) clonotypes detected after HCT in B-cell ALL were evaluated only in the exploratory analyses. NGS MRD results were normalized to residual clonal cells per million nucleated cells and characterized as undetectable (0), low (<10–4), high (≥10–4 to ≤10–3), or very high (>10–3). Because the clonoSEQ B-cell ALL assay has a limit of detection to capture malignancy-associated clonotypes, we considered MRD below the limit of detection to be low-positive. Because most MRDpre assessments were performed on BM and most MRDpost assessments were performed on PB, when patients had simultaneous samples from both sources, we used the BM result for MRDpre and the PB result for MRDpost.
Statistical analyses
Concordance between PB and BM MRD was performed using the Pearson correlation method for patients who had samples from both sources collected within 30 days of each other. Univariable Fine-Gray subdistribution hazard models were used to estimate the effect of MRDpre level on the cumulative incidences of relapse and leukemia-specific mortality in our cohort; death due to and not due to relapse were competing risks, respectively. Univariable cause-specific Cox proportional hazards models with time-dependent variables were used to evaluate the effect of MRDpost on relapse and leukemia-specific mortality. To analyze the association of both MRDpre and MRDpost levels with relapse and leukemia-specific mortality, we constructed multivariable cause-specific Cox proportional hazards models, with MRDpre as a fixed covariate and MRDpost as a time-varying covariate. A multivariable cause-specific Cox proportional hazards model with PB-based MRDpre as a fixed covariate and PB-based MRDpost as a time-varying covariate was constructed for sensitivity analyses. Exploratory analyses limited to patients with B-cell ALL only described the outcomes of patients with detectable IgH vs non-IgH (Igκ/λ) clonotypes after HCT. Analyses were performed using SAS/STAT software version9.4 (SAS Institute Inc., Cary, NC). This study was approved by the institutional review boards of Stanford University and OHSU and was conducted in accordance with the Declaration of Helsinki.
Results
Patient cohort
Among 174 patients with ALL who underwent HCT and had a diagnostic sample sent to Adaptive Biotechnologies for clonality evaluation, 158 (91%) had at least 1 dominant sequence identified for tracking and MRD monitoring, 15 (9%) failed dominant sequence identification because of polyclonality, and 1 (1%) had only a dominant TCRδ clonotype, which was not tracked via the clonoSEQ assay. Of the 15 patients without a dominant sequence, 7 (47%) had B-cell ALL, and 8 (53%) had T-cell ALL. Similar to the rest of our cohort, most patients without a dominant sequence were in first complete remission at the time of the HCT and underwent myeloablative conditioning. Of the patients without a dominant sequence, only 1 patient relapsed during follow-up, 3 patients died of nonleukemia causes, and 12 patients were alive and in remission at the last follow-up.
Of the 158 patients with a trackable clonotype, 122 (77%) underwent MRDpre assessment, and 139 (88%) underwent ≥1 MRDpost assessment; 111 (70%) underwent both MRDpre and ≥1 MRDpost assessments and formed the analytic cohort for multivariable survival models (supplemental Figures 1 and 2). In the 11 patients who underwent MRDpre but not MRDpost assessment, the reasons were death shortly after HCT (n = 9), relocation to a different state (n = 1), and physician decision to monitor MRD with BCR-ABL PCR instead of NGS in Ph+ B-cell ALL (n = 1). The source of MRD was equally distributed between PB and BM, with a greater proportion of MRDpre assessed using BM (69%) and a greater proportion of MRDpost assessed using PB (75%) samples. In 81 patients with 120 paired PB and BM MRD samples, a high concordance was observed between PB and BM MRD (Pearson coefficient = 0.78; P < .001; supplemental Table 1).
The median age of the patients was 44 years (range, 19-70 years), 62 (56%) were male, and 95 (86%) had B-cell ALL. (Table 1). Most (83%) of patients underwent myeloablative HCT, most commonly from matched unrelated donors (39%), and 80 (72%) and 24 (24%) were in first or second complete remission at the time of HCT, respectively. The 2-year incidence of relapse, nonrelapse mortality, and leukemia-specific mortality after HCT was 21%, 12%, and 11%, respectively, and the 2-year overall survival was 77%.
Demographics of study cohort
| . | MRDpre assessed (N = 122) . | MRDpost assessed (N = 139) . | MRDpre and MRDpost assessed (N = 111) . | |||
|---|---|---|---|---|---|---|
| N . | % . | N . | % . | N . | % . | |
| Age (y) | ||||||
| Median (range) | 44 (19-77) | 43 (18-71) | 44 (19-70) | |||
| 18-39 | 50 | 41 | 60 | 43 | 44 | 40 |
| 40-59 | 46 | 38 | 50 | 36 | 43 | 39 |
| 60+ | 26 | 21 | 29 | 21 | 24 | 22 |
| Sex | ||||||
| Male | 68 | 56 | 79 | 57 | 62 | 56 |
| Female | 54 | 44 | 60 | 43 | 49 | 44 |
| Race/ethnicity | ||||||
| Non-Hispanic White | 54 | 44 | 65 | 47 | 48 | 43 |
| Hispanic/Latino | 33 | 27 | 38 | 27 | 30 | 27 |
| African American | 6 | 5 | 5 | 4 | 5 | 5 |
| Asian | 18 | 15 | 19 | 14 | 18 | 16 |
| Other | 11 | 9 | 12 | 9 | 10 | 9 |
| Immunophenotype | ||||||
| B-cell | 104 | 85 | 119 | 86 | 95 | 86 |
| T-cell | 18 | 15 | 20 | 14 | 16 | 14 |
| Molecular subtype (B-cell ALL only) | ||||||
| Ph+∗ | 28 | 27 | 30 | 25 | 24 | 25 |
| Ph-like† | 13 | 13 | 13 | 11 | 13 | 14 |
| MLLr/KMT2a rearrangement∗ | 5 | 5 | 6 | 5 | 5 | 5 |
| Site of involvement | ||||||
| BM only | 101 | 83 | 116 | 84 | 94 | 85 |
| Extramedullary only | 1 | 1 | 3 | 2 | 1 | 1 |
| BM and extramedullary | 20 | 16 | 20 | 14 | 16 | 14 |
| Disease status at HCT | ||||||
| CR1 | 86 | 71 | 101 | 73 | 80 | 72 |
| CR2+ | 32 | 26 | 33 | 24 | 27 | 24 |
| Active | 4 | 3 | 5 | 4 | 4 | 4 |
| Donor type | ||||||
| Matched related donor | 39 | 32 | 45 | 32 | 37 | 33 |
| Unrelated donor | ||||||
| Matched unrelated donor | 46 | 38 | 55 | 40 | 43 | 39 |
| Mismatched unrelated donor | 9 | 7 | 10 | 7 | 8 | 7 |
| Haploidentical | 11 | 9 | 11 | 8 | 9 | 8 |
| Umbilical cord blood | 17 | 14 | 18 | 13 | 14 | 13 |
| Conditioning regimen | ||||||
| Myeloablative | 102 | 84 | 115 | 83 | 91 | 82 |
| Reduced intensity | 20 | 16 | 24 | 17 | 20 | 18 |
| 2-y outcomes | ||||||
| Cumulative incidence of relapse | 23 | 19 | 33 | 24 | 23 | 21 |
| Nonrelapse mortality | 22 | 18 | 14 | 10 | 13 | 12 |
| Leukemia-specific mortality | 12 | 10 | 20 | 14 | 12 | 11 |
| Overall survival | 88 | 72 | 105 | 76 | 86 | 77 |
| Follow-up time from HCT (mo) | ||||||
| Median (range) | 24 (12-24) | 24 (12-24) | 24 (12-24) | |||
| . | MRDpre assessed (N = 122) . | MRDpost assessed (N = 139) . | MRDpre and MRDpost assessed (N = 111) . | |||
|---|---|---|---|---|---|---|
| N . | % . | N . | % . | N . | % . | |
| Age (y) | ||||||
| Median (range) | 44 (19-77) | 43 (18-71) | 44 (19-70) | |||
| 18-39 | 50 | 41 | 60 | 43 | 44 | 40 |
| 40-59 | 46 | 38 | 50 | 36 | 43 | 39 |
| 60+ | 26 | 21 | 29 | 21 | 24 | 22 |
| Sex | ||||||
| Male | 68 | 56 | 79 | 57 | 62 | 56 |
| Female | 54 | 44 | 60 | 43 | 49 | 44 |
| Race/ethnicity | ||||||
| Non-Hispanic White | 54 | 44 | 65 | 47 | 48 | 43 |
| Hispanic/Latino | 33 | 27 | 38 | 27 | 30 | 27 |
| African American | 6 | 5 | 5 | 4 | 5 | 5 |
| Asian | 18 | 15 | 19 | 14 | 18 | 16 |
| Other | 11 | 9 | 12 | 9 | 10 | 9 |
| Immunophenotype | ||||||
| B-cell | 104 | 85 | 119 | 86 | 95 | 86 |
| T-cell | 18 | 15 | 20 | 14 | 16 | 14 |
| Molecular subtype (B-cell ALL only) | ||||||
| Ph+∗ | 28 | 27 | 30 | 25 | 24 | 25 |
| Ph-like† | 13 | 13 | 13 | 11 | 13 | 14 |
| MLLr/KMT2a rearrangement∗ | 5 | 5 | 6 | 5 | 5 | 5 |
| Site of involvement | ||||||
| BM only | 101 | 83 | 116 | 84 | 94 | 85 |
| Extramedullary only | 1 | 1 | 3 | 2 | 1 | 1 |
| BM and extramedullary | 20 | 16 | 20 | 14 | 16 | 14 |
| Disease status at HCT | ||||||
| CR1 | 86 | 71 | 101 | 73 | 80 | 72 |
| CR2+ | 32 | 26 | 33 | 24 | 27 | 24 |
| Active | 4 | 3 | 5 | 4 | 4 | 4 |
| Donor type | ||||||
| Matched related donor | 39 | 32 | 45 | 32 | 37 | 33 |
| Unrelated donor | ||||||
| Matched unrelated donor | 46 | 38 | 55 | 40 | 43 | 39 |
| Mismatched unrelated donor | 9 | 7 | 10 | 7 | 8 | 7 |
| Haploidentical | 11 | 9 | 11 | 8 | 9 | 8 |
| Umbilical cord blood | 17 | 14 | 18 | 13 | 14 | 13 |
| Conditioning regimen | ||||||
| Myeloablative | 102 | 84 | 115 | 83 | 91 | 82 |
| Reduced intensity | 20 | 16 | 24 | 17 | 20 | 18 |
| 2-y outcomes | ||||||
| Cumulative incidence of relapse | 23 | 19 | 33 | 24 | 23 | 21 |
| Nonrelapse mortality | 22 | 18 | 14 | 10 | 13 | 12 |
| Leukemia-specific mortality | 12 | 10 | 20 | 14 | 12 | 11 |
| Overall survival | 88 | 72 | 105 | 76 | 86 | 77 |
| Follow-up time from HCT (mo) | ||||||
| Median (range) | 24 (12-24) | 24 (12-24) | 24 (12-24) | |||
CR, complete remission.
Among those with B-cell ALL.
Among those with Ph-negative B-cell ALL.
MRD and post-HCT relapse: univariable analyses
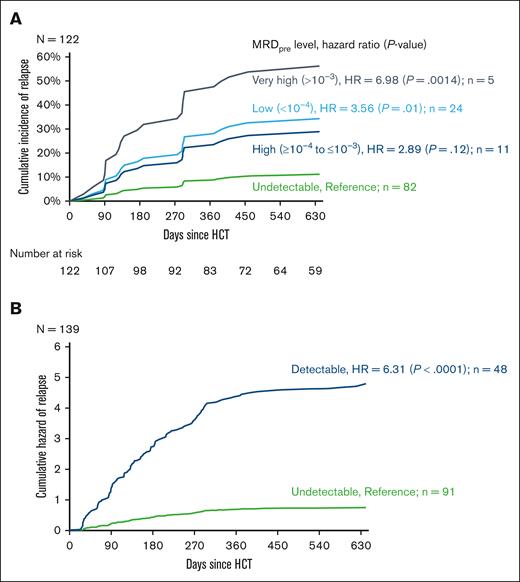
The median time between the MRDpre assessment and HCT was 24 days (range, 3-124 days). MRDpre was undetectable, low, high, and very high in 82 (67%), 24 (20%), 11 (9%), and 5 (4%), respectively. The cumulative incidence of post-HCT relapse increased per the MRDpre level (Figure 1A; Table 2), including an increase in the risk of relapse among patients with low (relative to undetectable) MRDpre (hazard ratio [HR], 3.56; 95% confidence interval [CI], 1.39-9.15; P = .01). There were no significant differences in leukemia-specific mortality based on the MRDpre level (Table 2). Given the similarity in relapse risk between patients with low and high MRDpre, these 2 groups were subsequently combined and defined as “MRDpre moderate” (<10–4 to ≤10–3) in multivariable survival models.
Risk of relapse based on MRDpre and MRDpost levels. (A) Cumulative incidence of relapse based on the MRDpre level. (B) Cumulative hazard of relapse based on the MRDpost level.
Risk of relapse based on MRDpre and MRDpost levels. (A) Cumulative incidence of relapse based on the MRDpre level. (B) Cumulative hazard of relapse based on the MRDpost level.
Univariable analysis of the cumulative incidence of relapse and leukemia-specific mortality based on MRDpre and MRDpost levels
| MRDpre level (N = 122) . | Cumulative incidence of relapse . | Leukemia-specific mortality . | ||
|---|---|---|---|---|
| HR (95% CI) . | P value . | HR (95% CI) . | P value . | |
| Undetectable | 1 | 1 | ||
| Low (<10–4) | 3.56 (1.39-9.15) | .01 | 2.90 (0.80-10.52) | .10 |
| High (>10–4 to ≤10–3) | 2.89 (0.75-11.20) | .12 | 3.37 (0.61-18.69) | .17 |
| Very high (>10–3) | 6.98 (2.12-22.91) | .0014 | 3.34 (0.41-27.55) | .26 |
| MRDpre level (N = 122) . | Cumulative incidence of relapse . | Leukemia-specific mortality . | ||
|---|---|---|---|---|
| HR (95% CI) . | P value . | HR (95% CI) . | P value . | |
| Undetectable | 1 | 1 | ||
| Low (<10–4) | 3.56 (1.39-9.15) | .01 | 2.90 (0.80-10.52) | .10 |
| High (>10–4 to ≤10–3) | 2.89 (0.75-11.20) | .12 | 3.37 (0.61-18.69) | .17 |
| Very high (>10–3) | 6.98 (2.12-22.91) | .0014 | 3.34 (0.41-27.55) | .26 |
| MRDpostlevel (N = 139) . | HR (95% CI) . | P value . | HR (95% CI) . | P value . |
|---|---|---|---|---|
| Undetectable | 1 | 1 | ||
| Detectable | 6.31 (4.56-8.74) | <.0001 | 7.74 (4.86-12.32) | <.0001 |
| MRDpostlevel (N = 139) . | HR (95% CI) . | P value . | HR (95% CI) . | P value . |
|---|---|---|---|---|
| Undetectable | 1 | 1 | ||
| Detectable | 6.31 (4.56-8.74) | <.0001 | 7.74 (4.86-12.32) | <.0001 |
The median time between HCT and the first MRDpost assessment was 32 days (range, 20-337 days), with a median monitoring frequency of every 43 days over the first year after HCT (supplemental Figure 2). MRDpost was detectable via NGS at any level in 48 (33%) patients. Patients with detectable MRDpost had a significantly increased risk of post-HCT relapse (HR, 6.31; 95% CI, 4.56-8.74; P < .0001; Figure 1B) and a significantly increased risk of leukemia-specific mortality (HR, 7.71; 95% CI, 4.86-12.31; P < .0001; Table 2).
MRD and post-HCT relapse: multivariable analyses
The results of the multivariable cause-specific Cox proportional hazards model using MRDpre as a fixed covariate and MRDpost as a time-varying covariate are shown in Figure 2. We found that after adjusting for MRDpost results, moderate (HR, 1.81; 95% CI, 1.16-2.82; P = .01) and very high (HR, 3.38; 95% CI, 1.84-6.19; P < .0001) MRDpre remained significantly associated with an increased risk of post-HCT relapse. Similarly, after adjusting for MRDpre results, detectable MRDpost at any time point remained highly associated with subsequent post-HCT relapse (HR, 4.60; 95% CI, 3.01-7.02; P < .0001). Sensitivity analyses limited to the PB MRD results demonstrated similar associations between MRDpre, MRDpost, and clinical relapse (supplemental Table 2). Multivariable models evaluating the effect of MRDpre and MRDpost on leukemia-specific mortality showed a significant association only with detectable MRDpost and leukemia-specific mortality (HR, 9.85; 95% CI, 4.98-19.54; P < .0001).
Risk of post-HCT relapse and leukemia-specific mortality based on MRDpre and MRDpost levels. HRs for (A) post-HCT relapse and (B) post-HCT leukemia-specific mortality from a cause-specific Cox proportional hazards model, with MRDpre as a fixed covariate and MRDpost as a time-varying covariate.
Risk of post-HCT relapse and leukemia-specific mortality based on MRDpre and MRDpost levels. HRs for (A) post-HCT relapse and (B) post-HCT leukemia-specific mortality from a cause-specific Cox proportional hazards model, with MRDpre as a fixed covariate and MRDpost as a time-varying covariate.
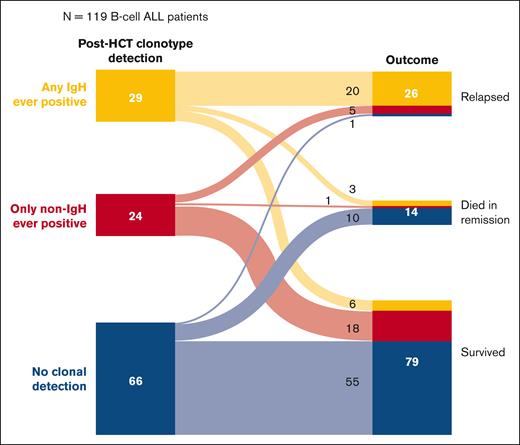
Effect of detectable non-IgH clonotypes after HCT in B-cell ALL
In contrast to the aforementioned analyses, to explore the potential prognostic significance associated with the detection of non-IgH (ie, Igκ/Igλ) MRD clonotypes in B-cell ALL, we examined the outcomes of patients with B-cell ALL included in our cohort based on MRDpost results, including IgH and non-IgH clonotypes (Figure 3). We found that of the 29 patients with solely IgH MRD clonotypes detected after HCT, 20 (61%) relapsed and only 6 (21%) survived. However, among the 24 patients with only non-IgH (Igκ/λ) MRD clonotypes detected after HCT, only 5 (21%) relapsed and 18 (75%) survived.
Description of outcomes in patients with B-cell ALL based on the specific MRD clonotype (IgH vs non-IgH) detected at any time point after HCT.
Description of outcomes in patients with B-cell ALL based on the specific MRD clonotype (IgH vs non-IgH) detected at any time point after HCT.
Discussion
In this retrospective, real-world study of 174 adult ALL HCT recipients treated across 2 large transplant centers, we demonstrated the prognostic value of MRD monitoring using highly sensitive NGS technology. We found that detectable MRDpre at levels as low as <10–4 were associated with an increased risk of post-HCT relapse. Conversely, patients entering HCT with undetectable MRDpre and those whose MRD remained undetectable via NGS monitoring for the first year after HCT had a low likelihood of relapsing by 2 years after HCT.
We were particularly interested in the association between very low levels of detectable MRDpre via NGS and the risk of post-HCT relapse because the clearance of pre-HCT MRD has growing clinical relevance, and it was previously unclear whether elimination of MRD to a level of less than 10–4 was necessary in the pre-HCT setting. The majority of our patients underwent myeloablative HCT, suggesting that the strength of the conditioning regimen was not sufficient to overcome MRD positivity. The use of blinatumomab to reduce MRD before HCT is becoming increasingly common3,6 and was recently shown in the ECOG-ACRIN E1910 phase 3 trial to improve overall survival in patients with MRD– when added to consolidation chemotherapy after intensive induction chemotherapy.16 Although these results may ultimately reduce the need for HCT in adult ALL, our data suggest that the optimal pre-HCT goal should be undetectable MRD at a level of 10–6. The use of blinatumomab to clear pretransplant MRD has not been studied in a randomized clinical trial, and it is unlikely that such a trial will ever be undertaken. Interestingly, we did not observe a significant association between detectable MRDpre and leukemia-specific mortality after HCT. This may be due to the relatively small number of leukemia-related deaths, limited follow-up of 2 years after HCT, and recent findings that after HCT relapse in ALL (particularly B-cell ALL) is more salvageable now than in the past.17
Although the association between MRDpre and relapse was significant, detectable MRDpost at any level remained highly associated with subsequent relapse and leukemia-specific mortality, regardless of the MRDpre results. These data should further encourage close monitoring of MRD in the post-HCT setting, regardless of the patient’s MRDpre results because MRD results are becoming increasingly actionable.17 However, which of the sequences initially identified in the malignant clone have prognostic significance in the post-HCT setting is important. To our knowledge, we have shown for the first time that the specific MRD clonotype matters: detectable IgH MRD clonotypes after HCT were highly associated with relapse in our primary results, whereas our exploratory findings demonstrated that detectable non-IgH (ie, Igκ/λ) clonotypes in the absence of detectable IgH clonotypes were not clinically significant. This may reflect the fact that Ig light chain sequences are, on average, less “unique” than Ig heavy chain sequences and are, therefore, more likely to have a background level contributed by independent nonmalignant clones in the same patient. The clonoSEQ assay accounts for these differences in clonotype uniqueness by adjusting the limit of detection threshold. Additional larger studies should be conducted to confirm this effect, and post-HCT relapse prevention trials should incorporate NGS MRD as a trigger for subsequent therapy.
Because this was an observational study in which MRD testing was performed in accordance with clinical practice parameters, our patient cohort included MRD obtained from both PB and BM. One advantage of the NGS-based MRD assay is its ability to use PB as a source for MRD monitoring. We previously demonstrated high concordance of MRD results in PB and BM using NGS-based MRD (n = 69 patients; 126 paired samples),11 and, here, we show similar findings using data from patients who underwent transplant at 2 centers (n = 81 patients; 120 paired samples). In this study, most of the MRDpost samples were obtained from PB, thereby strengthening the evidence related to NGS-based PB MRD and outcomes in ALL. Monitoring MRD by using PB is clearly less invasive and more cost-effective and can allow for more frequent monitoring when necessary.
Our study cohort comprised a heterogeneous population consisting of patients with both B-cell and T-cell ALL receiving various conditioning regimens and different transplant sources. However, despite this heterogeneity, we detected strong associations between MRD levels and the risk of relapse. Although of great interest, our a priori study objectives did not include subgroup or multivariable analyses incorporating age, cytogenetics, Ph-like phenotype, immunophenotype, or conditioning regimen, which may have additional clinical utility. In addition, our limited sample size did not allow for post hoc subgroup analyses. Although we did not prospectively assign the frequency or duration of NGS-based MRD monitoring because this was a retrospective analysis, most patients underwent MRD assessment within 45 days of HCT, and the most common monitoring interval was approximately every 1 or 2 months during the first year after HCT. Although most relapses occurred within the first year after HCT, the optimal duration of MRD monitoring for patients at high risk (those with detectable MRDpre and/or detectable MRDpost) was unknown. In the 12 patients with B-cell ALL with detectable MRDpost but who did not relapse, the reasons included early transplant-related mortality (n = 3), clearance of MRD with onset of graft-versus-host disease (n = 4), initiation of post-HCT tyrosine kinase inhibitor therapy in Ph+ B-cell ALL (n = 3), NGS MRD detected only below the limit of detection (n = 1), and relapse after the study cut off (n = 1).
In conclusion, we demonstrated the prognostic value of the highly sensitive NGS-based MRD in adults with ALL undergoing HCT. Our data suggest that MRD clearance to an undetectable level at the 10–6 or lower threshold is optimal before HCT, and that detectable MRD (specifically, IgH clonotypes in B-cell ALL or TCR clonotypes in T-cell ALL) after HCT via NGS of the PB or BM should raise concerns for impending clinical relapse.
Acknowledgments
This study was funded by the National Institutes of Health, National Cancer Institute (NCI) grant P01CA049605 and is partially supported by the Biostatistics Shared Resource of the NCI-sponsored Stanford Cancer Institute (P30CA124435).
Authorship
Contribution: J.T.L. and L.M. supervised and provided the concept and designed the study; E.C.L., S.E.D., J.M.G.S., S.T., A.Z., K.M., J.T.L., and L.M. were responsible for the acquisition, analysis, and interpretation of data; E.C.L. and L.M. drafted the manuscript; A.Z. and K.M. performed the statistical analysis; and all the authors critically reviewed the manuscript.
Conflict-of-interest disclosure: J.T.L. reports research funding from AbbVie and Amgen, and consultancy for Pfizer, Adaptive, Kite, and Takeda. L.M. reports research funding from Adaptive, Astellas, Jasper, Kite, and Bristol Myers Squibb; provides consulting and is a member of advisory boards for Adaptive, Amgen, CTI Biopharma, Kite, Medexus, and Pfizer; and receives honoraria from Adaptive and UpToDate. The remaining authors declare no competing financial interests.
Correspondence: Lori Muffly, 300 Pasteur Dr H0144, Stanford, CA 94305; e-mail: lmuffly@stanford.edu.
References
Author notes
∗E.C.L., S.E.D., and J.M.G.S. are joint first authors.
†J.T.L. and L.M. are joint senior authors.
Data are available on request from the corresponding author, Lori Muffly (lmuffly@stanford.edu).
The full-text version of this article contains a data supplement.