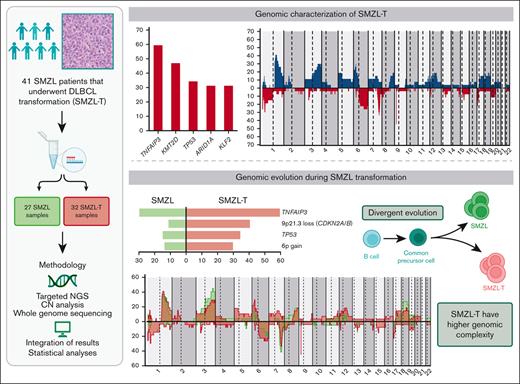
Key Points
SMZL transformation is associated with genomic complexity and distinct genomic alterations (TNFAIP3, CDKN2A/B, TP53, and 6p+).
KLF2 mutations and complex karyotypes in transformed SMZL confer a shorter survival.
Abstract
The genetic mechanisms associated with splenic marginal zone lymphoma (SMZL) transformation are not well defined. We studied 41 patients with SMZL that eventually underwent large B-cell lymphoma transformation. Tumor material was obtained either only at diagnosis (9 patients), at diagnosis and transformation (18 patients), and only at transformation (14 patients). Samples were categorized in 2 groups: (1) at diagnosis (SMZL, n = 27 samples), and (2) at transformation (SMZL-T, n = 32 samples). Using copy number arrays and a next-generation sequencing custom panel, we identified that the main genomic alterations in SMZL-T involved TNFAIP3, KMT2D, TP53, ARID1A, KLF2, 1q gains, and losses of 9p21.3 (CDKN2A/B) and 7q31-q32. Compared with SMZL, SMZL-T had higher genomic complexity, and higher incidence of TNFAIP3 and TP53 alterations, 9p21.3 (CDKN2A/B) losses, and 6p gains. SMZL and SMZL-T clones arose by divergent evolution from a common altered precursor cell that acquired different genetic alterations in virtually all evaluable cases (92%, 12 of 13 cases). Using whole-genome sequencing of diagnostic and transformation samples in 1 patient, we observed that the SMZL-T sample carried more genomic aberrations than the diagnostic sample, identified a translocation t(14;19)(q32;q13) present in both samples, and detected a focal B2M deletion due to chromothripsis acquired at transformation. Survival analysis showed that KLF2 mutations, complex karyotype, and International Prognostic Index score at transformation were predictive of a shorter survival from transformation (P = .001; P = .042; and P = .007; respectively). In summary, SMZL-T are characterized by higher genomic complexity than SMZL, and characteristic genomic alterations that could represent key players in the transformation event.
Introduction
Splenic marginal zone lymphoma (SMZL) is an infrequent low-grade B-cell lymphoma that involves the spleen, bone marrow, and peripheral blood, and accounts for <2% of all lymphoid neoplasms.1-3 Compared with other B-cell lymphomas, SMZL is characterized by few recurrent chromosomal abnormalities, including deletions of 7q31-q32 (30% to 40%) and 6q (8% to 24%), and gains of chromosomes 3/3q (20% to 30%) and 18/18q (8% to 25%).4-8 The most frequent mutations in SMZL include KLF2 (20% to 30%), NOTCH2 (10% to 25%), TP53 (15%), KMT2D (10% to 15%), and TNFAIP3 (7% to 15%).9-13 Recently, Bonfiglio et al14 reported a large comprehensive genomic and transcriptomic characterization of 303 SMZLs at diagnosis, dividing SMZL into 2 main genetic clusters: (1) DMT cluster characterized by alterations in DNA-damage response and mitogen-activated protein kinase, and Toll-like receptor pathways (∼30% of SMZL); and (2) NNK cluster, characterized by alterations on nuclear factor κB (NF-κB), NOTCH2, and KLF2 (∼60% of SMZL), and associated with an inferior survival.14
Despite the indolent clinical course of SMZL, ∼70% of the patients require treatment because of progressive disease, and between 10% to 15% of patients eventually transform to an aggressive lymphoma, generally diffuse large B-cell lymphoma (DLBCL) with dismal prognosis.15-19 The molecular mechanisms involved in the transformation to high-grade lymphomas have been elucidated in other low-grade lymphoid neoplasms20-22 but are not well understood in SMZL. In line with this, only a few clinico-biological parameters have been associated with higher risk of histological transformation, including elevated lactate dehydrogenase, >4 nodal sites involved at diagnosis, or complex karyotype.15,23-25 The recently described C1/BN2 or NOTCH2 molecular DLBCL clusters are defined by alterations in NOTCH2 and genes of the NF-κB pathway, and have been postulated to be of extrafollicular/marginal zone origin,26-29 with genetic features mostly resembling marginal zone lymphoma.
Given that the information on transformed SMZL (SMZL-T) is very limited, we have investigated the SMZL-T genomic landscape using targeted next-generation sequencing (NGS) and copy number (CN) analysis, with the aim of establishing the underlying mechanisms and clonal dynamics of this aggressive transformation.
Methods
Patients
In total, 41 patients diagnosed with SMZL that underwent transformation (SMZL-T) were studied (Table 1). The diagnostic criteria were based on the International Extranodal Lymphoma Study Group and World Health Organization guidelines.1 The pathology and/or flow cytometry data were reviewed upon inclusion in the study. The criterion for considering a case as transformed was histopathological, and was based on the presence of sheets of large cells. In the transformed cases with peripheral blood involvement the presence of large cells was evaluated by immunophenotype and/or cytomorphology. The cases were from the Hospital Clinic de Barcelona, other institutions of the Spanish Lymphoma Group (GELTAMO), and the Royal Marsden National Health Service (NHS) Foundation Trust, London, United Kingdom. Informed consent was obtained in accordance with the institutional review boards of the respective institutions. The study was conducted in accordance with the Declaration of Helsinki. Tumor material was obtained at diagnosis from 9 patients who subsequently transformed but material at transformation was not available for molecular studies; in 18 patients, tumor material was obtained at diagnosis and transformation; and in 14 patients, tumor material was obtained only at transformation; corresponding to a total of 59 samples (supplemental Table 1). We categorized the samples: (1) at diagnosis (SMZL), 27 samples; and (2) at transformation (SMZL-T), 32 samples. Using a QIAmp DNA/RNA Mini Kit (Qiagen, Germany) and AllPrep DNA/RNA FFPE Kit (Qiagen), DNA was extracted from 11 fresh-frozen lymphoma tissues, 6 involved peripheral blood samples, and 42 formalin-fixed paraffin-embedded tissues, respectively. IGHV-IGHD-IGHJ rearrangements were analyzed using IGHV leader primers or consensus primers for IGHV FR1 and/or FR3 regions (supplemental Table 1).30,31 To assess the clonality of the transformed samples, the FR1 region of the IGHV was amplified using BIOMED-2 multiplex polymerase chain reaction protocol. Cytogenetic and fluorescence in situ hybridization (FISH) analyses were performed as previously described32 (supplemental Methods).
Clinico-biological features of patients with SMZL-T at transformation time point.
| Characteristic . | Total (%) . |
|---|---|
| Age >60 y | 24/36 (66.7) |
| Male/female | 14/27 (34.1/65.8) |
| B symptoms | 21/31 (67.7) |
| ECOG performance status ≥2 | 8/32 (25) |
| Ann Arbor stage III–IV | 28/32 (87.5) |
| Bulky disease (>7 cm) | 6/28 (21.4) |
| Hemoglobin < 100 g/L | 10/32 (31.3) |
| Platelets < 100 × 109/L | 5/32 (15.6) |
| Lactate dehydrogenase > UNL | 21/28 (75) |
| B2-microglobulin > UNL | 21/24 (87.5) |
| High–intermediate or high-risk IPI | 20/32 (62.5) |
| Complex karyotype | |
| Diagnosis | 6/12 (50) |
| Transformation | 7/11 (63.6) |
| del7q | |
| Diagnosis | 3/12 (25) |
| Transformation | 3/11 (27.3) |
| FISH at transformation | |
| BCL2 rearrangement | 0/15 (0) |
| BCL6 rearrangement | 2/20 (10) |
| MYC rearrangement | 1/23 (4.3) |
| Histological transformation | |
| At diagnosis | 5/36 (13.9) |
| During follow-up | 31/36 (86.1) |
| Median time to transformation (range), y | 2.42 (0-17) |
| Median time to treatment (range), y | 0.21 (0-11.7) |
| Characteristic . | Total (%) . |
|---|---|
| Age >60 y | 24/36 (66.7) |
| Male/female | 14/27 (34.1/65.8) |
| B symptoms | 21/31 (67.7) |
| ECOG performance status ≥2 | 8/32 (25) |
| Ann Arbor stage III–IV | 28/32 (87.5) |
| Bulky disease (>7 cm) | 6/28 (21.4) |
| Hemoglobin < 100 g/L | 10/32 (31.3) |
| Platelets < 100 × 109/L | 5/32 (15.6) |
| Lactate dehydrogenase > UNL | 21/28 (75) |
| B2-microglobulin > UNL | 21/24 (87.5) |
| High–intermediate or high-risk IPI | 20/32 (62.5) |
| Complex karyotype | |
| Diagnosis | 6/12 (50) |
| Transformation | 7/11 (63.6) |
| del7q | |
| Diagnosis | 3/12 (25) |
| Transformation | 3/11 (27.3) |
| FISH at transformation | |
| BCL2 rearrangement | 0/15 (0) |
| BCL6 rearrangement | 2/20 (10) |
| MYC rearrangement | 1/23 (4.3) |
| Histological transformation | |
| At diagnosis | 5/36 (13.9) |
| During follow-up | 31/36 (86.1) |
| Median time to transformation (range), y | 2.42 (0-17) |
| Median time to treatment (range), y | 0.21 (0-11.7) |
ECOG, Eastern Cooperative Oncology Group; UNL, upper normal limit.
CN analysis and NGS
Copy number alterations (CNAs) were assessed in 49 of 59 samples, corresponding to 22 SMZL at diagnosis and 27 SMZL-T. Oncoscan CNV FFPE assay (ThermoFisher Scientific, Massachusetts, USA) (46 samples) and CytoScan HD assay (3 samples) (Thermo Fisher Scientific) were used based on DNA source. Nexus version 9.0 Discovery Edition software (Biodiscovery, El Segundo, CA) was used for identification and visualization of CNAs using the human genome assembly GRCh37/hg19. Gains and losses of >100kb and copy neutral loss of heterozygosity (CN-LOH) terminal and of >10 Mb were considered. Driver CNAs were determined by GISTIC algorithm (2.0.23).33 Chromothripsis was defined when ≥7 switches between ≥2 CN states were detected on individual chromosomes.34
Single nucleotide variants (SNVs) and insertions/deletions (indels) were assessed in all 59 samples using a targeted NGS panel capturing 37 genes associated with SMZL pathogenesis (supplemental Table 2). Libraries were generated from 150 ng of DNA using molecular-barcoded adapters (ThruPLEX Tag-seq Kit; Takara) coupled with a custom hybridization capture-based method (SureSelectXT Target Enrichment System Capture strategy, Agilent Technologies) and sequenced in a MiSeq instrument (Illumina, 2 × 150 base pairs). NGS data were analyzed using an in-house bioinformatics pipeline previously described (supplemental Methods).35,36 We obtained a mean coverage of 489×, with 88% of the targeted regions of at least 100× (supplemental Table 3; supplemental Figure 1). The panel includes 7 CN regions recurrently altered in SMZL: 3q26.1, 7q32.1-q32.2, 8q24.21, 9p21.3, 12q21.1, 17p13.1, and 18q21.33-q22.1 (supplemental Table 4). Information of these 7 regions was used in the 10 samples lacking SNP-array. CNVkit tool kit37 was used to infer CNA after segmentation into discrete regions. The thresholds (defined without ploidy correction) were a log2 ratio of >0.2 for gains and of <0.2 for losses.
Whole-genome sequencing (WGS)
WGS was performed in 1 patient (SMZL055). Library preparation was performed using TruSeq DNA PCR-Free Kit (Illumina) for germline DNA and transformed samples, and the TruSeq DNA Nano protocol (Illumina) for the diagnostic sample. Libraries were sequenced on a NovaSeq6000 (2 × 151 base pairs) instrument (Illumina). Details of the bioinformatic analysis are provided in supplemental Methods. The mean coverage obtained was 69.46× for SMZL, 70.55× for SMZL-T, and 41.95× for germline sample.
Statistical analysis
Comparison of the frequency of each alteration between diagnosis and transformation was performed using a mixed-effects logistic model, which accounts for the partially paired structure of the data. A fully Bayesian approach was used to estimate this model, which was also used to test the cooccurrence or mutual exclusivity between alterations. Comparison of the total number of CNAs, gains, losses, or mutations between diagnosis and transformation were assessed using mixed-effects negative binomial models, implemented in the glmer.nb function of the lme4 R package. Fisher exact test was used to compare the mutation frequencies at diagnosis between 2 different SMZL series. P values were adjusted using the Benjamini-Hochberg method. Survival from transformation (SFT) was calculated from the time of transformation to the last visit or to the death of the patient. Association between SFT and binary or continuous variables was measured with the log-rank test or Cox regression, respectively. SFT curves were estimated with the Kaplan-Meier method. Details of all statistical analyses are provided in supplemental Methods.
Results
Baseline features
Clinical data of 26 of 41 (63%) cases were previously published.15 Of the study participants, 27 patients were female and 14 male (Table 1; supplemental Table 1). The median age at diagnosis and at transformation was 61 years (range, 41-82 years) and 66 years (range, 43-89 years), respectively. The median time to transformation was 2.4 years (range, 0-17 years). Complex karyotypes were found in 6 of 12 (50%) patients at diagnosis and 7 of 11 (63.6%) patients at transformation. The site of the transformation was nodal in 17 of the 32 cases (53.1%), extranodal in 6 (18.8%) (soft tissues, lachrymal gland and labial vestibule), the spleen in 4 (12.5%), the peripheral blood in 3 (9.4%), and the bone marrow in 2 (6.2%) patients. In the peripheral blood and the bone marrow the diagnosis of transformation was based on the presence of numerous large B cells.
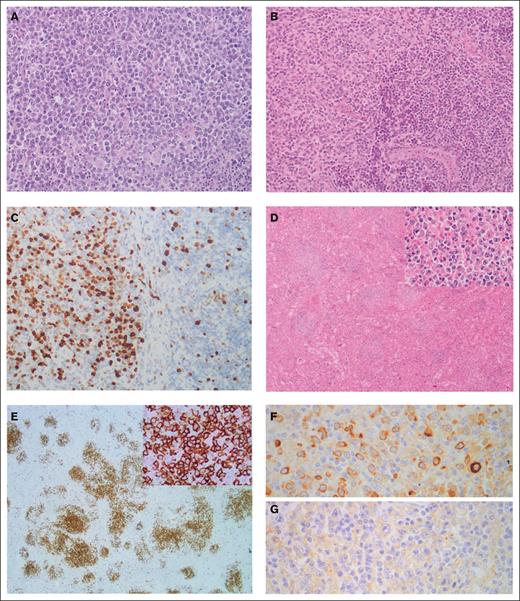
Histological review confirmed transformation to DLBCL, except for 1 case (SMZL017T), in which the neoplastic cells had a blastoid appearance, raising the diagnosis of high-grade B-cell lymphoma, not otherwise specified. This tumor expressed cyclin D1 by immunohistochemistry without t(11;14)(q13;q32) translocation. Consistent with the established diagnostic histopathological criteria, sheets of large cells were observed in all transformed cases. In 11 cases, a coexistence of the low-grade component with the large-cell lymphoma in the transformed biopsy was observed. In 5 patients, transformation was already present at diagnosis. The median Ki67 of SMZL-T was 75% (range 50% to 90%) (Figure 1). FISH studies at transformation showed rearrangements of BCL6 in 10% of cases (2/20), MYC in 4.3% (1/23), and no BCL2 rearrangements in the 15 tumors tested.
Morphological and immunohistochemical features of SMZL-T. (A) Case SMZL058T showing a lymph node with a diffuse proliferation composed of large lymphocytes with a centroblastic appearance (hematoxylin and eosin stain, original magnification ×20). (B-C) Detail of case SMZL07T showing the interphase between a transformed area (left) and an area with remnant marginal splenic B-cell lymphoma (right). In panel B, hematoxylin and eosin stain (original magnification ×20); and in panel C, immunohistochemical staining for Ki67 showing the different proliferative index between both areas (original magnification ×20). (D-G). Case SMZL012T showing the spleen infiltrated by a lymphoid proliferation arranged in a nodular growth pattern, original magnification ×4 (D), constituted of large, atypical cells (inset, original magnification ×40). (E) These cells were positive for CD20 (original magnification ×2; inset, original magnification ×20), (F) had Kappa light chain restriction (original magnification ×40), and (G) were negative for Lambda light chain (original magnification ×40).
Morphological and immunohistochemical features of SMZL-T. (A) Case SMZL058T showing a lymph node with a diffuse proliferation composed of large lymphocytes with a centroblastic appearance (hematoxylin and eosin stain, original magnification ×20). (B-C) Detail of case SMZL07T showing the interphase between a transformed area (left) and an area with remnant marginal splenic B-cell lymphoma (right). In panel B, hematoxylin and eosin stain (original magnification ×20); and in panel C, immunohistochemical staining for Ki67 showing the different proliferative index between both areas (original magnification ×20). (D-G). Case SMZL012T showing the spleen infiltrated by a lymphoid proliferation arranged in a nodular growth pattern, original magnification ×4 (D), constituted of large, atypical cells (inset, original magnification ×40). (E) These cells were positive for CD20 (original magnification ×2; inset, original magnification ×20), (F) had Kappa light chain restriction (original magnification ×40), and (G) were negative for Lambda light chain (original magnification ×40).
Genetic alterations in SMZL-T samples
CNAs in SMZL-T
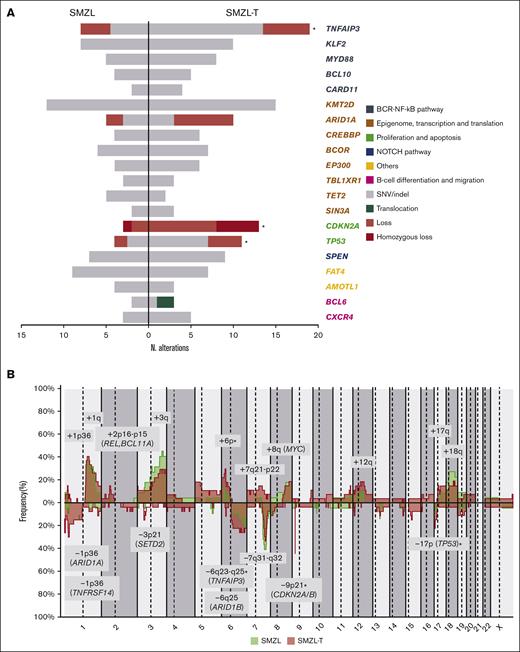
We detected CNA in 24 of 27 (89%) tumors, with a median of 8 CNAs per case (range, 0-28 CNAs), 4 gains (range, 0-20 gains), and 4 losses (range, 0-15 losses). In addition, we identified a total of 32 CN-LOH with a median of 1 CN-LOH per case (range, 0-5 CN-LOH) (supplemental Table 5). The most frequent (n ≥ 14%) alterations were gains of 1p36.12, 1q, 2p16.1-p15 (REL and BCL11A), 3q, 6p, 7q21.11-q22.3, 8q (MYC), 12q, 17q and 18q (BCL2); losses of 1p36.32 (TNFRSF14), 1p36.11 (ARID1A), 3p21.31 (SETD2), 6q23.3-q25.2 (TNFAIP3), 6q25.3 (ARID1B), 7q31-q32, 9p21.3 (CDKN2A/B), and 17p13 (TP53); and CN-LOH of 9p24.3-p21.3 (CDKN2A/B) (Figure 2A). In the 5 cases lacking CN array, we found the following alterations by NGS analyses: 7q32.1-q32.2 losses and 18q21.33-q22.1 gains in 2 cases; and 3q26.1 gain, 9p21.3 loss, 12q21.1 gain, and 17p13.1 loss in 1 case each. Using GISTIC we detected 8 driver CNAs (Q-value < 0.05): gains of 1q, 3q, and 18q (BCL2) and losses in 1p36.11 (ARID1A), 3p21.31 (SETD2), 7q31-q32, 9p21.3 (CDKN2A/B), and 13q14.13-q14.3 (RB1, DLEU1/2) (supplemental Table 6). Furthermore, we observed acquired chromothripsis at transformation in 2 patients, both with biallelic TP53 inactivation (supplemental Figure 2).
Genetic landscape of SMZL-T. (A) CN profile of 27 cases of SMZL-T. On the x-axis, the chromosomes are represented horizontally from 1 to X (chromosome Y is excluded); on the y-axis, the percentages of patients with CNAs are shown, with gains depicted in blue and losses in red. Regions with an incidence of ≥4 cases and potential target genes are indicated. (B) Oncoprint displaying the recurrent alterations (n ≥ 3) found in 32 SMZL-T cases. Each column corresponds to an individual SMZL-T sample. All alterations are displayed by decreasing frequency. In the upper panel, SNVs and indels are shown in gray, deletions in red, and BCL6 translocation in green. In the lower panel, gains (blue), losses (red), and CN-LOH (yellow) of large and focal regions (potential target genes are indicated) are shown.
Genetic landscape of SMZL-T. (A) CN profile of 27 cases of SMZL-T. On the x-axis, the chromosomes are represented horizontally from 1 to X (chromosome Y is excluded); on the y-axis, the percentages of patients with CNAs are shown, with gains depicted in blue and losses in red. Regions with an incidence of ≥4 cases and potential target genes are indicated. (B) Oncoprint displaying the recurrent alterations (n ≥ 3) found in 32 SMZL-T cases. Each column corresponds to an individual SMZL-T sample. All alterations are displayed by decreasing frequency. In the upper panel, SNVs and indels are shown in gray, deletions in red, and BCL6 translocation in green. In the lower panel, gains (blue), losses (red), and CN-LOH (yellow) of large and focal regions (potential target genes are indicated) are shown.
Gene mutations and integrative analysis in SMZL-T
We detected a total of 172 SNVs/indels distributed among 30 of 37 genes analyzed, with a median of 4 mutations per case (range, 1-17 mutations) (supplemental Table 7). We integrated the CNAs, SNVs, and indels and observed that the most frequently altered genes were TNFAIP3 (59.4%), KMT2D (46.9%), TP53 (34.4%), ARID1A (31.3%), and KLF2 (31.3%). The most frequent CNAs were losses of 9p21.3 (CDKN2A/B) (40.6%) and 7q31-q32 (34.4%), and gains of 1q (40.7%) (Figure 2B). Although we identified a low frequency of truncating mutations in NOTCH2 (11.1%) and NOTCH1 (3.7%) genes, this signaling pathway was altered by mutations in other downstream genes, such as SPEN (28.1%) (supplemental Table 7). In addition, we identified homozygous deletions of CDKN2A/B in 5 patients, and biallelic inactivation (deletion and mutation) of TP53 and TNFAIP3 in 5 and 3 patients, respectively.
Although we used a small targeted sequencing panel that does not encompass the full spectrum of mutations required, we applied the LymphGen algorithm to assign the 32 SMZL-T samples to DLBCL molecular subtypes in order to see whether common pathway alterations were present. Of these samples, 56.2% (18 of 32) could be classified: BN2 (11 cases), EZB (2 cases), MCD (2 cases), N1 (1 case), A53 (1 case), and BN2/MCD (1 case) (supplemental Table 1). The fact that the majority of SMZL-T samples were assigned to the BN2 cluster substantiates the postulated extrafollicular/marginal zone origin of this genetic DLBCL subgroup.27
We investigated the cooccurrence and mutual exclusivity of the genetic alterations present in the 41 patients of the study, considering SMZL and SMZL-T samples (supplemental Figure 3). We found coocurrence between TP53 and ARID1A alterations (P value = .001), and MYD88 mutations with 8p23-p22 loss (P value = .004) (supplemental Figure 4).
Genomic evolution during SMZL transformation
Genetic alterations at diagnosis and transformation
We performed CNA (n = 22) and NGS (n = 27) in diagnostic SMZL samples and found that the most frequent alterations (n = 8) were 1q and 3q gains; 7q losses; and KMT2D, FAT4, and KLF2 mutations (supplemental Figure 5). In order to investigate the underlying genomic alterations associated with transformation, we compared the global frequency of genomic alterations at diagnosis with the global frequency observed at transformation, as well as the specific frequencies of those alterations present in at least 5 samples. We observed a higher number of CNAs, gains, and losses at transformation (8 vs 6, P < .001; 4 vs 3, P = .004; and 4 vs 2, P = .001; respectively) (supplemental Figure 6). Losses and mutations of TNFAIP3 and TP53, losses of 9p21.3 (CDKN2A/B), and gains of 6p were also significantly enriched at transformation (59.4% vs 29.6%, P < .001; 34.4% vs 14.8%, P = .04; 40.6% vs 11.1%, P = .001; and 29.6% vs 13.6%, P = .05; respectively). Moreover, we observed an enrichment of deletions at the 1p36.32 (TNFRSF14) region, and CN-LOH of 9p24.3-p21.3 at transformation (25.9% vs 4.5%, P = .06 and 18.5% vs 0%, P = .06; respectively). (Figure 3; supplemental Figures 7-8; supplemental Table 8). TP53 alterations were detected in 11 SMZL-T corresponding to 6 of the 18 paired samples (SMZL/SMZL-T), 4 of these samples had the alteration already at diagnosis (SMZL015D, SMZL022D, SMZL045D, and SMZL052D), with 2 of these the SMZL-T having acquired an alteration of the second TP53 allele, either by mutation or deletion (SMZL015T and SMZL022T); and in 2 other cases the TP53 alteration was acquired at SMZL-T (SMZL007T and SMZL09T). From the 14 samples with DNA samples taken only at transformation, 5 showed TP53 alterations (SMZL0T12, SMZL0T18, SMZL026T, SMZL046T, and SMZL058T) (supplemental Figure 9).
Genomic evolution patterns in SMZL-T. (A) Comparison of frequently altered genes in SMZL (left) and SMZL-T (right). Genes are clustered by pathways (in different colors). Only genes with at least 3 alterations in 1 of the groups are depicted. Asterisks indicate the alterations significantly enriched in 1 of the groups. (B) Comparison of CN profiles of 22 patients with SMZL at diagnosis (green) and 27 patients with SMZL-T at transformation (pink). CN gains are depicted in the upper part of plot, and losses at the bottom. CN regions with recurrence of n ≥ 4 are represented.
Genomic evolution patterns in SMZL-T. (A) Comparison of frequently altered genes in SMZL (left) and SMZL-T (right). Genes are clustered by pathways (in different colors). Only genes with at least 3 alterations in 1 of the groups are depicted. Asterisks indicate the alterations significantly enriched in 1 of the groups. (B) Comparison of CN profiles of 22 patients with SMZL at diagnosis (green) and 27 patients with SMZL-T at transformation (pink). CN gains are depicted in the upper part of plot, and losses at the bottom. CN regions with recurrence of n ≥ 4 are represented.
We applied the genetic cluster classification described by Bonfiglio14 in the diagnostic SMZL, and 66.7% (18 of 27) could be classified: 11 as NNK-SMZL and 7 as DMT-SMZL (supplemental Table 1).
To explore whether certain genetic alterations present at diagnosis could predispose transformation, we compared the genomic alterations of a large published series of 303 SMZL14 cases at diagnosis with the genomic aberrations detected in the diagnostic samples of 27 SMZL that transformed during the follow-up in this study. In our cases, we observed a higher incidence in 83% (19 of 23) of the genes described by Bonfiglio et al14 (supplemental Figure 10; supplemental Table 9).
Clonal evolution
To gain a deeper insight in the evolution of SMZL transformation, we focused on the 13 patients with paired samples (supplemental Table 1) analyzed by both NGS and CNAs assessment. All patients harbored at least 1 lesion shared by SMZL and SMZL-T. We detected the presence of an enriched ancestral common precursor cell in all cases, with a median of 41.6% shared alterations (range, 11% to 100%). This finding supports the clonal relationship between SMZL and SMZL-T (Figure 4A). The transformed tumors had a median of 4 aberrations shared with the diagnostic SMZL (range, 1-12 aberrations). In addition, we observed additional alterations that were unique to the diagnostic sample, suggesting a divergent evolution in 12 of 13 (92%) sample pairs (Figure 4B-C; supplemental Figure 11). Only 1 patient (SMZL045) had a linear evolution acquiring novel alterations (6p gain, 11p12 loss, and 13q12 loss) in the transformed sample, maintaining all the aberrations present at diagnosis (Figure 4D).
Different evolution patterns in SMZL-T. Total number of shared and unique alterations identified in patients with paired diagnostic/transformed samples. (A) Bar plot showing the overall shared aberrations in each case between diagnosis and transformed samples. (B) Unique genomic aberrations identified in each case, at diagnosis (left) and at transformation (right). SNVs, indels, gains, losses, and CN-LOH were considered. The asterisks indicate the cases with no CN array data. Models of divergent (C) and linear (D) evolution patterns during SMZL transformation. Upper panels: simplified models; lower panels: an example of each type, case SMZL059 as an example for divergent evolution (C) and case SMZL045, the only case with linear evolution (D). Green and pink cell aggregates depict the SMZL and SMZL-T clones, respectively, common mutated precursor cell (CPC) in orange, and normal B cell is represented in blue.
Different evolution patterns in SMZL-T. Total number of shared and unique alterations identified in patients with paired diagnostic/transformed samples. (A) Bar plot showing the overall shared aberrations in each case between diagnosis and transformed samples. (B) Unique genomic aberrations identified in each case, at diagnosis (left) and at transformation (right). SNVs, indels, gains, losses, and CN-LOH were considered. The asterisks indicate the cases with no CN array data. Models of divergent (C) and linear (D) evolution patterns during SMZL transformation. Upper panels: simplified models; lower panels: an example of each type, case SMZL059 as an example for divergent evolution (C) and case SMZL045, the only case with linear evolution (D). Green and pink cell aggregates depict the SMZL and SMZL-T clones, respectively, common mutated precursor cell (CPC) in orange, and normal B cell is represented in blue.
WGS
In SMZL055 we had available frozen material from the spleen and the lymph node at diagnosis and transformation, respectively (supplemental Figure 12). The WGS of the SMZL-T carried more genomic aberrations than the diagnostic sample (11917 vs 9818 total mutations; 80 vs 68 coding mutations; 9 vs 8 CNAs; 29 vs 18 structural variants (Figure 5A; supplemental Tables 10-12). Consistent with the NGS and CNA analysis, the WGS confirmed a branching evolution pattern (Figure 5B).
Whole genome landscape of SMZL055 at diagnosis and transformation. (A) Circos plots displaying structural variants and CNAs at diagnosis (left) and transformation (right). (B) Representation of the number of mutations shared by (gray), or specific to, the SMZL (green) and SMZL-T (pink) samples. (C) Representation of the reciprocal translocation t(14;19)(q32;q13). (D) Chromosome 15 of the transformation sample with a chromothripsis pattern inactivating B2M gene, acquired at transformation.
Whole genome landscape of SMZL055 at diagnosis and transformation. (A) Circos plots displaying structural variants and CNAs at diagnosis (left) and transformation (right). (B) Representation of the number of mutations shared by (gray), or specific to, the SMZL (green) and SMZL-T (pink) samples. (C) Representation of the reciprocal translocation t(14;19)(q32;q13). (D) Chromosome 15 of the transformation sample with a chromothripsis pattern inactivating B2M gene, acquired at transformation.
A translocation t(14;19)(q32;q13) present at diagnosis and transformation was identified (Figure 5C). The breakpoints were located at the switch region IGHG2 and downstream of BCL3. We validated this rearrangement using a BCL3 breakapart FISH probe. Although the SMZL-T acquired 4477 specific mutations, we could not identify any additional potential driver gene mutation among them. However, we found that the SMZL-T acquired a deletion of B2M due to a chromothripsis event in chromosome 15 (Figure 5D). In addition, an unbalanced t(1;11) affecting RERE::FCHSD2 genes, a gain of 11q, and a CN-LOH of 6p were acquired at transformation.
Clinical analysis
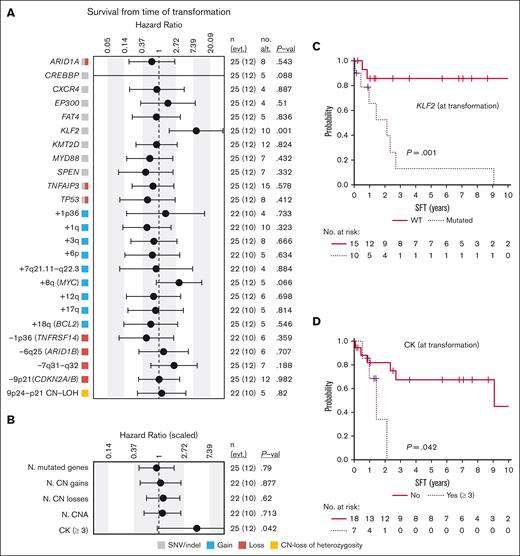
After the diagnosis of histological transformation in the 32 patients with available data, 28 received immunochemotherapy (R-CHOP regimen in 24), followed by autologous stem-cell transplantation in 3 cases. Splenectomy was performed in 3 patients with no further therapy, and the remaining patient died before any therapy could be given. Twenty patients (65%) achieved complete response, 1 partial response, and 10 were refractory to the treatment. Of the patients in complete response, 8 eventually relapsed. Fifty percent (18/36) of the patients eventually died. The median SFT was 6.55 years, and the 5-year SFT was 53.2% (95% confidence interval, 37.1-76.4). The causes of death were: death with progressive lymphoma or during therapy (n = 14), death in the setting of infectious episodes (n = 6), and death from causes unrelated to the lymphoma when in complete response (n = 4; multifocal leukoencephalopaty, interstitial pneumonia, sepsis, and secondary acute myeloid leukemia; 1 each). The main variables predicting shorter SFT were KLF2 mutations (P = .001), complex karyotype (P = .042), and a high-risk international prognostic index score (IPI) (P = .007); there was a trend for MYC gains (P = .066) (Figure 6; supplemental Figure 13).
Survival from time of transformation. (A) Impact of genetic alterations or (B) the cumulative number of (N.) of genetic changes present at transformation on SFT. The impact is quantified with the hazard ratio and its 95% confidence interval. Continuous variables were scaled. The gray, red, blue, and yellow boxes indicate the type of genetic alterations found in each alteration (SNVs/indel, loss, gain, and CN-LOH, respectively). The right columns show: the number of cases (n) and events (evt.), the number of altered cases (no. alt.), and the P value (P-val) of the log-rank test (for dichotomous variables) or of the Cox regression (for continuous variables). Only alterations with at least 4 altered cases at transformation are shown. Complex karyotype (CK) is dichotomized and defined by the presence of 3 or more aberrations. (C-D) Kaplan-Meier curves of SFT based on (C) the presence of KLF2 mutations or (D) complex karyotype at transformation.
Survival from time of transformation. (A) Impact of genetic alterations or (B) the cumulative number of (N.) of genetic changes present at transformation on SFT. The impact is quantified with the hazard ratio and its 95% confidence interval. Continuous variables were scaled. The gray, red, blue, and yellow boxes indicate the type of genetic alterations found in each alteration (SNVs/indel, loss, gain, and CN-LOH, respectively). The right columns show: the number of cases (n) and events (evt.), the number of altered cases (no. alt.), and the P value (P-val) of the log-rank test (for dichotomous variables) or of the Cox regression (for continuous variables). Only alterations with at least 4 altered cases at transformation are shown. Complex karyotype (CK) is dichotomized and defined by the presence of 3 or more aberrations. (C-D) Kaplan-Meier curves of SFT based on (C) the presence of KLF2 mutations or (D) complex karyotype at transformation.
Discussion
We performed a comprehensive genetic characterization of SMZL-T, the molecular pathogenesis of which has, to date, been poorly understood. For the first time, we identified that SMZL-T are characterized by a distinct profile of driver CNAs, including gains of 1q, 3q, and 18q (BCL2) and losses in 1p36 (ARID1A), 3p21 (SETD2), 7q31-q32, 9p21.3 (CDKN2A/B), and 13q14.13-q14.3 (RB1 and DLEU1/2) that are potentially relevant in the transformation event. This profile of alterations is similar to that of de novo DLBCL except for 7q loss, a highly specific alteration of SMZL, rarely found in other small B-cell neoplasms. We reveal that SMZL-T are, on genetic grounds, significantly more complex compared with SMZL, with twice as many alterations. This parallels the findings from other studies documenting a higher genomic complexity in small B-cell neoplasms histologically transformed into high-grade lymphomas, such as follicular lymphoma (FL) and chronic lymphocytic leukemia (CLL).20-22 In line with the high complexity, 3 SMZL-T had chromothripsis, a phenomenon found in several B-cell lymphomas and myelomas,34,38-40 associated with shorter survival and TP53 deficiency.
Two altered regions, 9p21 deletion and 6p gain, were significantly enriched in SMZL-T. To our knowledge, this is the first report of 9p21 deletions in SMZL-T, with the focal deletions delimit a minimal region containing CDKN2A and CDKN2B tumor suppressor genes. This was the most frequent deletion in SMZL-T (40.6%), and was virtually absent in SMZL at diagnostic in our and other published SMZL series.4,6,7,12,41 Moreover, in 6 cases we could demonstrate the acquisition of 9p21 deletion upon transformation. CDKN2A/B deletions have been reported in FL, CLL, and mantle cell lymphoma (MCL), associated with aggressive course, increased risk of transformation,20,22,42,43 and de novo DLBCL.44,45 Of note, a poor overall survival and histological transformation have been described in a subgroup of patients with SMZL that harbored higher promoter methylation, being CDKN2A/B 1 of the regions highly methylated. However, this study only included 5 cases with histological transformation.46 The 6p gains, also enriched in SMZL-T samples, were consistent with their high prevalence in DLBCL,47 especially in the DLBCL C2 cluster,26 in which 6p gains represent late events. Another region highly enriched in SMZL-T was 1p36 deletion, including TNFRSF14 gene. 1p36 loss, CN-LOH, and mutation of TNFRSF14 have been documented in FL, associated with a worse prognosis.48 Although we detected a higher frequency of 1p36 loss in SMZL-T, we were unable to ascertain the incidence of TNFRSF14 mutations, because this gene was not included in our NGS panel. Nevertheless, Bonfiglio et al14 reported a 3% frequency of TNFRSF14 mutations in a large SMZL series, and thus, it seems to be an infrequent event.
The most frequently altered genes in SMZL-T were TNFAIP3 (59.4%), KMT2D (46.9%), KLF2 (31.25%), and TP53 (34.4%). TNFAIP3 (A20) encodes a protein that is a negative regulator of the NF-κB signaling pathway, and is altered by mutations/deletions. TNFAIP3 mutations have previously been found in 7% to 15% of SMZL.9,10,12,14,49 Interestingly, truncating loss-of-function mutations of TNFAIP3 have been described in 32% (6/19) of SMZL that underwent transformation,10 and are enriched in the SMZL NNK cluster, which is characterized by aberrations on NF-κB, NOTCH2, and KLF2, and associated with inferior survival.14,KMT2D was reported in ∼11% to 15% of SMZL cases,10-12,14,49 and is prevalent in other lymphomas such as FL, DLBCL, and MCL.26,38,50 The frequency of KMT2D alterations in our SMZL-T cases is higher than that reported for SMZL. The third most frequently mutated gene in SMZL-T is KLF2, a gene that is involved in marginal zone B-cell homing.9 Loss-of-function KLF2 mutations have been reported in ∼12% to 42% of SMZL,9,10,14,51,52 and are associated with 7q deletion, NOTCH2, TNFAIP3, and ARID1A mutations and IGHV1-02∗02 family usage.9,10,TP53 is the fourth most frequently altered gene in our SMZL-T samples. In previous SMZL studies it had been reported to be mutated or deleted in ∼12% to 16% of cases.9,10,12,14 Of interest, in a study with a small number of patients with SMZL who subsequently transformed, 4 cases with a TP53 mutation at diagnosis further acquired a 17p deletion, inactivating the remaining TP53 allele.10 The frequency of TP53 inactivation in our SMZL-T samples is very high (34.4%) and included cases with TP53 alteration at diagnosis, cases with inactivation of the second allele at transformation, and cases with wild type at diagnosis and acquiring inactivation of TP53 in the transformed sample, highlighting the relevant role of this gene in the transformation process. Other frequently altered genes in SMZL-T, previously described in SMZL, are ARID1A (31.25%), SPEN (28.1%), NOTCH2 (11.1%), and NOTCH1 (3.7%). Of note, we observed a lower frequency of NOTCH2 mutations compared with previous studies; however, we detected a high prevalence of SPEN mutations, a gene involved in NOTCH signaling, which might be complementary. Overall, we show that SMZL-T have preferential deregulation of the cell cycle (CDKN2A/B, TP53, and TNFRSF14), DNA damage response (TP53, and ARID1A), and the NF-κB pathway (TNFAIP3), suggesting that a deregulation of these pathways could represent key features in the development of SMZL-T.
The activating MYD88L265P is a well-known recurrent mutation in lymphoplasmacytic lymphoma but is also found in CLL and other lymphomas, including SMZL.9,10,12,39,53,54 MYD88L265P mutations in SMZL have been described as alternative to other drivers, such as TP53 and NOTCH2, and are associated with favorable overall survival.10 In our SMZL-T cohort, all cases with MYD88L265P had concomitant alterations in: TNFAIP3 (4 cases), TP53 (3), CDKN2A/B (3), and, in 1 case, also concomitant KLF2, KMT2D, and NOTCH2 alterations. Globally, the mechanism of transformation of SMZL with MYD88L265P mutation seems to be similar to cases without this alteration.
We documented a clonal relationship in all 18 SMZL/SMZL-T sample pairs. Furthermore, in all except 1 case, the SMZL-T clone arose from an altered common precursor through the acquisition of independent genetic events (divergent evolution), an evolution pattern described for most FL transformed to DLBCL.20,55-57
To date, WGS has been documented in only 6 SMZL,11 none of which were SMZL-T. In the patient evaluated in this study, we observed higher complexity in the transformed sample, detected a divergent evolution pattern, and identified relevant genomic alterations acquired at transformation (ie, B2M). In this context, B2M genomic aberrations have been documented to be specifically acquired at transformation in 3 FL that transformed to DLBCL.20
An interesting finding, not previously documented, is the prognostic impact of KLF2 mutations in SMZL-T. KLF2 mutation is considered to be an early event in SMZL, and has been associated with a short median time to first treatment.10 We have shown that KLF2 is the only mutation associated with shorter survival from the time of transformation. Of note, a high prevalence of KLF2 mutations (21.7%) has been described in the BN2 DLBCL molecular subgroup compared with the other subgroups.27 Mutations in other genes that are reportedly associated with histological transformation, shorter overall survival, and event-free survival in SMZL include TNFAIP3 and TP53.9,10 Our findings further support these studies but with a much higher incidence of mutations of both genes in SMZL-T (34.4% TP53 and 59.4% TNFAIP3). We have not established an unfavorable prognostic of NOTCH2 mutations, whose impact in small series of SMZL cases in the literature has been controversial.10-12,58
In conclusion, our study has identified potential molecular players responsible for the transformation in the largest series of SMZL-T reported to date. Genomic alterations affecting the NF-κB signaling pathway (TNFAIP3), cell cycle control, and DNA damage responses (CDKN2A/B, TP53) are acquired distinctively at transformation, and KLF2 mutations, complex karyotypes, and high IPI in transformed SMZL are associated with short survival from transformation.
Acknowledgments
The authors thank the Hematopathology Collection registered at the Biobank of Hospital Clínic-IDIBAPS for sample procurement. We want to particularly acknowledge patients and Biobank HUB-ICO-IDIBELL (PT20/00171) integrated in the Spanish Biobank Network and Xarxa Banc de Tumors de Catalunya (XBTC) for their collaboration. The authors are indebted to the IDIBAPS Genomics core facility and are grateful to Miriam Prieto and Silvia Martín for their technical and logistic assistance, and Jose Alamo for immunophenotype and cytomorphology analysis. Some figures or parts have been created with BioRender.com.
This study was supported by Fundación Asociación Española Contra el Cancer (AECC)/Centro de Investigación Biomédica en Red de Cáncer (CIBERONC): PROYE18020BEA (S.B.), fondo de Investigaciones Sanitarias, Instituto de Salud Carlos III “Cofinanciado por la Unión Europea” and Fondos FEDER: European Regional Development Fund “Una manera de hacer Europa”: PI17/01061 (S.B.), PI19/00887 (A.L.-G. and E.G.), INT20/00050 (A.L.-G.), MaratÓ TV3-Cancer/201904-30 (S.B.), Generalitat de Catalunya Suport Grups de Recerca AGAUR (2021-SGR-01293 [S.B.] and 2021-SGR-01172 [E.C.]), and Ministerio de Ciencia e Innovación (PID2021-123054OB-I00 [E.C.]). C.L. is supported by postdoctoral Beatriu de Pinós grant, Secretaria d’Universitats i Recerca del Departament d’Empresa i Coneixement de la Generalitat de Catalunya and by Marie Sklodowska-Curie COFUND program from H2020 (2018-BP-00055). E.C. is an Academia Researcher of the "Institució Catalana de Recerca i Estudis Avançats" of the Generalitat de Catalunya. This work was mainly developed at the Centre Esther Koplowitz (CEK), Barcelona, Spain.
Authorship
Contribution: M.G., C.L., A.N., and G.F. analyzed and interpreted data and wrote the manuscript; F.N. performed bioinformatic analysis; G.C. performed statistical and clinical analyses; G.F. and E.C. reviewed the hematoxylin and eosin stains and immunostains; A.N. and G.B.-M. performed sample preparation, and provided and centralized clinical data; M.A., M.J.B., M.B., M.L.-G., D. Colomer, E.D.-D., L.E., P.F., E.G., O.R, A.R.-D., L.V.F., A.W., F.C., A.L.-G., and E.M. provided samples and/or clinical data; D. Costa provided cytogenetic data; A.E. contributed to panel design; M.G., C.L., A.N., G.F., F.N., G.C., A.L.-G, E.M., and S.B. analyzed and interpreted data; S.B. and E.M. designed the study, supervised the research, interpreted data, and wrote the manuscript; and all authors read, commented on, and approved the manuscript.
Conflict-of-interest disclosure: F.N. has received honoraria from Janssen and AbbVie for speaking at educational activities. M.J.B. is currently an employee of Swedish Orphan Biovitrum. E.C. has been a consultant for Takeda, NanoString, AbbVie, and Illumina; has received research support from AstraZeneca; received honoraria from Janssen, EUSPharma, and Roche for speaking at educational activities; and is an inventor on a Lymphoma and Leukemia Molecular Profiling Project patent ‘Method for subtyping lymphoma subtypes by means of expression profiling’ (PCT/US2014/64161). A.L.-G. served on the advisory board of Roche, Celgene, Novartis, and Gilead/Kite, and received grants from Celgene and Gilead/Kite. The remaining authors declare no competing financial interests.
The current affiliation for M.J.B. is Swedish Orphan Biovitrum, Portugal.
Correspondence: Sílvia Beà, Molecular Pathology of Lymphoid Neoplasms, Fundació Clínic per a la Recerca Biomèdica - Instituto de Investigaciones Biomédicas August Pi i Sunyer, Rosselló, 153, 08036 Barcelona, Spain; e-mail: sbea@clinic.cat.
References
Author notes
∗M.G., C.L., A.N., and G.F. contributed equally to this study.
#A.L.-G., E.M., and S.B. jointly supervised this study.
Presented in abstract form at the 27th Congress of the European Hematology Association in June 2022.
Genomic data set reported in this article, including whole-genome sequencing, next-generation sequencing and copy number alteration arrays, have been deposited at the European Genome-phenome Archive (accession number: EGAS00001006389; link will be available at the moment of publication).
Data are available upon request from the corresponding author, Silvea Beà (sbea@clinic.cat).
The full-text version of this article contains a data supplement.