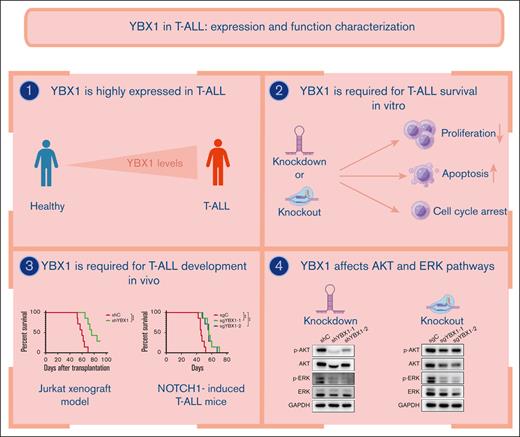
Key Points
YBX1 is required for survival in T-ALL by augmenting leukemia cell viability and cell cycle progression and inhibiting cell apoptosis.
YBX1 depletion impedes leukemogenesis in human T-ALL xenograft and NOTCH1-induced T-ALL model.
Abstract
Y-box-binding protein 1 (YBX1), a member of the RNA-binding protein family, is a critical regulator of cell survival in various solid tumors and acute myeloid leukemia. However, the function of YBX1 in T-cell acute lymphoblastic leukemia (T-ALL) remains elusive. Here, we found that YBX1 was upregulated in patients with T-ALL, T-ALL cell lines, and NOTCH1-induced T-ALL mice. Furthermore, depletion of YBX1 dramatically reduced cell proliferation, induced cell apoptosis, and induced G0/G1 phase arrest in vitro. Moreover, YBX1 depletion significantly decreased the leukemia burden in the human T-ALL xenograft and NOTCH1-induced T-ALL mice model in vivo. Mechanistically, downregulation of YBX1 markedly inhibited the expression of total AKT serine/threonine kinase (AKT), p-AKT, total extracellular signal-regulated kinase (ERK), and p-ERK in T-ALL cells. Taken together, our results uncovered a critical role of YBX1 in the leukemogenesis of T-ALL, which may have great potential as a biomarker and therapeutic target in T-ALL.
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) is a hematologic malignancy characterized by uncontrolled accumulation of malignant precursor T cells. Although many treatment strategies, including intensive combination chemotherapy regimens and allogeneic hematopoietic cell transplantation, are currently available, T-ALL remains an important cause of morbidity in both children and adults.1,2 Resistance to multidrug therapy and relapse remain significant clinical problems.3-6 Numerous genes associated with survival are thought to be the main cause of drug resistance and relapse.7-10 Identifying the genes involved in leukemogenesis is of great value in the development of targeted therapies.
Y–box binding protein 1 (YBX1), a member of the cold-shock protein family, contains 3 recognized domains: an amino-terminal region, a highly conserved central cold-shock domain, and a C-terminal domain.11 YBX1 specifically binds to DNA and RNA to regulate many biological processes, including DNA replication, transcription, repair, chromatin remodeling, messenger RNA (mRNA) translation, and pre-mRNA splicing.12-17 YBX1 recognizes the consensus sequence 5′-CTGATTGG-3′ to regulate transcriptional activation of genes implicated in cell proliferation, differentiation, migration, epithelial-mesenchymal transition, and chemoresistance.11,13,14,17 YBX1 has been identified as an oncogene that regulates tumorigenesis, recurrence, and metastasis.18,19 Aberrant high expression of YBX1 has been widely reported in multiple solid tumors.12,17-22 However, the expression and function of YBX1 in T-ALL remain elusive.
In this study, we investigated the expression, function, and potential underlying mechanism of YBX1 in T-ALL cells. Our results demonstrated that YBX1 was aberrantly elevated at both the mRNA and protein levels in T-ALL compared with that in healthy control. We then analyzed cell proliferation, apoptosis, and the cell cycle in YBX1-silenced cells using 2 short hairpin RNA (shRNAs) and found that the loss of YBX1 inhibited cell proliferation and induced cell apoptosis and cell cycle arrest. In addition, the biological function of YBX1 in T-ALL was confirmed in YBX1 knockout Jurkat cells using 2 single guide RNAs (sgRNAs). Furthermore, YBX1 depletion impeded leukemogenesis in the Jurkat xenograft model and in NOTCH1-induced mice. Mechanistically, we found that YBX1 regulates leukemogenesis through the AKT and extracellular signal-regulated kinase (ERK) signaling pathways.
Materials and methods
Patients and cell culture
Bone marrow samples from patients with T-ALL and healthy donors were obtained with informed consent from the First Affiliated Hospital of the University of Science and Technology of China. Jurkat, Molt4, CCRF-CEM and HEK293T were purchased from the Cell Resource Center, Peking Union Medical College (the headquarter of the National Science & Technology Infrastructure, National Biomedical Cell-Line Resource). Leukemia cell lines were cultured in complete RPMI-1640 supplemented with 10% fetal bovine serum (FBS; Gibco), and HEK293T cells were maintained in Dulbecco’s modified Eagle medium with 10% FBS (Gibco). All the cell lines were incubated at 37°C with 5% CO2.
RNA extraction and qRT-PCR
To synthesize complementary DNA, we used TaKaRa’s PrimeScript RT Reagent Kit following the manufacturer’s instructions after RNA extraction, using an RNAiso Plus Kit (TaKaRa, Japan). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using a 7500 RT-PCR System (Applied Biosystems). Glyceraldehyde-3-phosphate dehydrogenase or β-actin was used as the housekeeping gene, and relative mRNA expression was determined as the fold change using the comparative Ct method. Three or six independent experiments were conducted to collect all the data. Primer sequences are shown in supplemental Table 1.
Western blot analysis
Cells were lysed in radio immunoprecipitation assay lysis buffer (RIPA) buffer with a protease inhibitor cocktail (Selleck), and proteins were separated via sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Polyvinylidene fluoride (PVDF) membranes were incubated with primary antibodies at 4°C overnight after blocking for 30 minutes with milk. Membranes were incubated with chemiluminescent substrate (MIKX, China) after incubation with secondary antibodies for 2 hours at room temperature. Chemiluminescent signal was detected using a Chemiluminescent Imaging System (Tanon 5200, Shanghai, China).
Primary antibodies used are as follows: AKT (1:1000; 9272, Cell Signaling Technology), p-AKT (1:1000; 4060, Cell Signaling Technology), ERK (1:1000; 4370, Cell Signaling Technology), p-ERK (1:1000; 4695, Cell Signaling Technology), YBX1 (1:1000; 20339-1-AP, Proteintech), and glyceraldehyde-3-phosphate dehydrogenase (1:50000; 60004-1-Ig, Proteintech).
Lentiviral vector construction and transduction
To generate YBX1 knockdown cell lines, 2 independent shRNAs were designed using bioinformatics tools from MISSION shRNA. The target sequences of shYBX1 are listed in supplemental Table 1. The shRNA fragments were subcloned into the pLKO.1–green fluorescent protein (GFP) lentiviral vector following the manufacturer’s instructions. Using the viral packaging constructs pMD2.G and pSPAX2, lentiviruses were produced in HEK293T cells using polyethyleneimine (PEI). T-ALL cell lines were transfected with the prepared shRNA lentivirus using 8 μg/mL polybrene. At 48 hours after infection, GFP+ cells were sorted using a FACS Aria III (BD Biosciences, USA).
To generate YBX1 knockout cell lines, 2 independent sgRNAs were designed using online CRISPR design tool by Zhang Lab (http://crispr.mit.edu/). The sequences of sgRNA are listed in supplemental Table 1. The 2 fragments were subcloned into the LentiCRISPR-v2 lentiviral vector following the manufacturer’s instructions. Lentiviruses were produced from transfected HEK293T cells and used for infection experiment as described previously. At 48 hours after transfection, 2 μg/mL puromycin was added to the medium for 48 hours to select positive cells.
MTS assay
Cell viability was measured using the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)- 2H-tetrazolium (MTS) assay after depletion of YBX1 or treatment with the YBX1 inhibitor SU056. The cells were plated in 96-well plates at a density of 1 × 104 cells per well. 10 μL MTS (Promega) was added to each well at different time points (0, 24, 48, and 72 hours). The plate was subsequently incubated for 2 hours at 37°C. A Synergy H4 Hybrid Microplate Reader was used to measure the absorbance value in each well at 490 nm.
Flow cytometry analysis
Approximately 1 × 106 cells were harvested on day 3 after transfection or on days 1 and 2 with SU056 treatment to detect cell apoptosis and cell cycle via flow cytometry. For apoptosis assessment, cells were washed with phosphate-buffered saline (PBS) and resuspended in 100 μL binding buffer. The cells were then stained with annexin V–APC and propidium iodide (PI) per the manufacturer’s instructions (Biolegend). For cell cycle analysis, the cells were fixed with 75% ethanol overnight. Cells were washed once with cold PBS, treated with RNase A (10 mg/mL) for 15 minutes at room temperature, and stained with 50 μg/mL PI for 10 minutes in the dark. Apoptosis and the cell cycle were determined via flow cytometry (Beckman, USA) after staining.
Mice
C57BL/6J and NCG mice were purchased from GemPharmatech (Nanjing) and were housed in a sterile facility. The Shenzhen University Institutional Animal Care and Use Committee approved the study protocol and this work was conducted in accordance with animal ethical standards.
Human T-ALL xenograft
Jurkat xenografts were carried out as previously described.23 Briefly, 4.5 × 106 Jurkat cells infected with YBX1 sgRNA were transplanted intravenously into the 6- or 8-week-old female NCG mice. As the mice in the control group became moribund, 4 mice from each group were euthanized. Bone marrow was collected by flushing the mice with PBS, stained using human CD45 antibody, and analyzed via flow cytometry.
Generation of the murine NOTCH1-induced T-ALL model
The shRNAs targeting mouse YBX1 were designed using bioinformatics tools from MISSION shRNA. The target sequences of shYBX1 are shown in supplemental Table 1. The shRNA fragments were subcloned into the pLKO.1-puromaycin lentiviral vector per the manufacturer’s instructions. Lentivirus packaging was performed in HEK293T cells. The intracellular domain of human NOTCH1 (ICN1) was subcloned into the retroviral vector MSCV-IRES-GFP, which was kindly provided by Jianxiang Wang from the Chinese Academy of Medical Sciences & Peking Union Medical College, China. Retrovirus packaging was performed in HEK293T cells using PEI containing the packaging plasmid eco. CD117+ cells (c-Kit+) were enriched with a hematopoietic stem/progenitor cell enrichment kit from the bone marrow extracted from 6 or 8 week old C57BL/6J mice. c-Kit+ cells were cultured in Iscove modified Dulbecco medium (Hyclone) supplemented with 10% FBS (Gibco), 10 ng/mL recombinant murine interleukin-3 (PeproTech), 10 ng/mL recombinant murine interleukin-6 (PeproTech), and 50 ng/mL recombinant murine SCF (PeproTech). The c-Kit+ cells were first infected with MSCV-ICN1-IRES-GFP retroviruses in the presence of 8 μg/mL polybrene on day 1. The infected cells were infected with the prepared PLKO.1-puromysin lentivirus on day 2. At 48 hours after transfection, 4 μg/mL puromycin was added to the medium to select positive cells for the next 2 days. GFP+ cells were sorted using FACS Aria III (BD Biosciences) and then transplanted into lethally irradiated (9 Gy) C57BL/6J mice via tail vein injection. Leukemia progression was monitored based on the percentage of GFP+ cells in peripheral blood and analyzed via flow cytometry. Control mice became moribund. Three mice from each group were euthanized to analyze leukemia burden.
RIP
RNA immunoprecipitation (RIP) was performed as previously described, with some modifications.23 Briefly, HEK293T cells expressing flag-YBX1 were harvested and lysed in RIP lysis buffer. The cleared lysate was incubated with anti-immunoglobulin G (negative control) or anti-flag antibody conjugated to protein A/G magnetic beads overnight at 4°C. Beads were resuspended in 80 μL PBS after washing with RIP buffer 5 times, followed by DNase I and Proteinase K digestion. Total RNA was extracted from both the input and immunoprecipitated RNA and analyzed via RT-PCR.
RNA stability assays
HEK293T cells with or without YBX1 knockdown were treated with actinomycin D at a final concentration of 5 μg/mL and collected at the indicated time points. The mRNAs were analyzed via qRT-PCR.
RNA sequencing assay
Total RNA from Jurkat cells with shC or shYBX1 was isolated using total RNA isolation solution (TRIzol). Complementary DNA library construction, sequencing, and transcriptome data analysis were performed by Gene Denovo Co Ltd (Guangzhou, China).
Statistical analysis
All data are presented as the mean ± standard deviation. Statistical analyses were performed using GraphPad Prism 7.0 software or SPSS software. Survival analyses were conducted using the Kaplan-Meier method, and log-rank tests were used to evaluate the differences. Statistical significance was calculated using a 2-tailed Student t test. P < .05 was considered significant. Asterisks indicate significant differences (∗P < .05; ∗∗P < .01; ∗∗∗P < .001).
Results
YBX1 is highly expressed in T-ALL
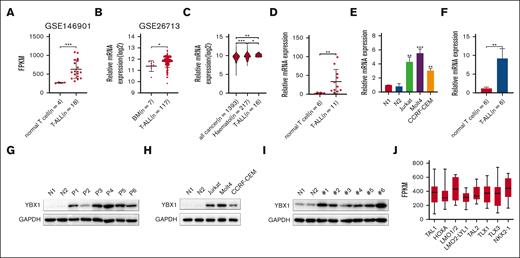
To investigate the expression of YBX1 in T-ALL, we first analyzed the gene expression profiles of YBX1 from publicly available data sets. The data from the Gene Expression Omnibus data sets (GSE146901 and GSE26713) revealed higher mRNA expression of YBX1 in patients with T-ALL than that in T cells or bone marrow from healthy donors (Figure 1A-B). By analyzing the Cancer Cell Line Encyclopedia, we also observed that YBX1 was overexpressed in T-ALL cell lines compared with that in all other cancer types and hematologic malignancies (Figure 1C). We further detected the mRNA expression of various T-ALL samples via qRT-PCR. Elevated mRNA expression level of YBX1 was detected in samples from patient with T-ALL (Figure 1D) and T-ALL cell lines (Figure 1E). Moreover, a higher mRNA expression level of YBX1 was observed in samples from NOTCH1-induced T-ALL mice than in healthy control cells (Figure 1F). These results indicated that YBX1 mRNA expression was significantly upregulated in T-ALL cells. To confirm the protein expression of YBX1 in leukemia, western blot was used to detect the expression of YBX1 protein in T-ALL cells. Consistent with the results of mRNA expression analysis, higher protein expression of YBX1 was detected in primary human T-ALL (Figure 1G) than in healthy donors. Similar results were observed in T-ALL cell lines (Figure 1H) and NOTCH1-induced T-ALL mice (Figure 1I). A total of 264 primary T-ALL samples were analyzed to determine YBX1 expression in different subtypes of T-ALL based on unique gene expression signatures that reflect different stages of arrest.24 Interestingly, YBX1 mRNA levels varied, with the highest expression in the LMO1/2 and NKX2-1 subtypes (Figure 1J). Taken together, these data demonstrated that YBX1 is highly expressed in T-ALL at both the mRNA and protein levels.
The expression of YBX1 is elevated in T-ALL. (A) The mRNA levels of YBX1 were analyzed in healthy T cells and primary T-ALL cells using microarray data set GSE146901. (B) YBX1 mRNA levels in healthy bone marrow (BM) and T-ALL BMs from microarray data set GSE26713 were analyzed. (C) mRNA expression analysis of YBX1 in T-ALL compared with all cancer types and other hematologic cancers in the Cancer Cell Line Encyclopedia (CCLE) database. (D) The mRNA expression of YBX1 in T cells from healthy donors and patients with T-ALL was determined via qRT-PCR. (E) Relative YBX1 mRNA levels in T cells derived from healthy donors, Jurkat, Molt4, and CCRF-CEM cells. (F) YBX1 mRNA expression was detected in healthy T cells from wild-type mice and leukemia cells from NOTCH1-driven T-ALL mice. (G-I) Western blot analysis of YBX1 expression in healthy T cells and leukemia cells from patients with T-ALL (G), various patient-derived leukemia cell lines (H), and NOTCH1-induced T-ALL mice (I). Glyceraldehyde-3-phosphate dehydrogenase was used as an internal control. (J) Analysis of YBX1 expression in 264 primary T-ALL samples classified into 8 subtypes. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. N, normal T cells.
The expression of YBX1 is elevated in T-ALL. (A) The mRNA levels of YBX1 were analyzed in healthy T cells and primary T-ALL cells using microarray data set GSE146901. (B) YBX1 mRNA levels in healthy bone marrow (BM) and T-ALL BMs from microarray data set GSE26713 were analyzed. (C) mRNA expression analysis of YBX1 in T-ALL compared with all cancer types and other hematologic cancers in the Cancer Cell Line Encyclopedia (CCLE) database. (D) The mRNA expression of YBX1 in T cells from healthy donors and patients with T-ALL was determined via qRT-PCR. (E) Relative YBX1 mRNA levels in T cells derived from healthy donors, Jurkat, Molt4, and CCRF-CEM cells. (F) YBX1 mRNA expression was detected in healthy T cells from wild-type mice and leukemia cells from NOTCH1-driven T-ALL mice. (G-I) Western blot analysis of YBX1 expression in healthy T cells and leukemia cells from patients with T-ALL (G), various patient-derived leukemia cell lines (H), and NOTCH1-induced T-ALL mice (I). Glyceraldehyde-3-phosphate dehydrogenase was used as an internal control. (J) Analysis of YBX1 expression in 264 primary T-ALL samples classified into 8 subtypes. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. N, normal T cells.
YBX1 silencing reduces cell viability in T-ALL
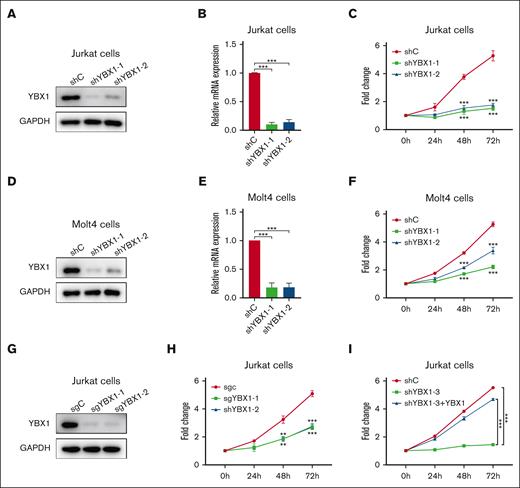
To determine the role of YBX1 in T-ALL cell viability, YBX1 was silenced in Molt4 and Jurkat cells using 2 shRNAs. As shown in Figure 2, the protein and mRNA expression of YBX1 was significantly reduced in Jurkat and Molt4 cells transduced with shRNA targeting YBX1 (Figure 2A-B, 2D-E). Compared with the control cells, T-ALL cells with YBX1 depletion by shRNAs showed a large decrease in cell viability (Figure 2C,F). Similar results were observed in Jurkat cells in which YBX1 gene was knocked out using sgRNA (Figure 2G-H). To rule out the possibility of off-target depletion, we restored YBX1 expression in YBX1 knockdown cells (supplemental Figure 1). As expected, restoration of YBX1 completely rescued the effect on the viability caused by YBX1 knockdown in Jurkat cells (Figure 2I), indicating that the reduced cell viability was not caused by an off-target shRNA effect. These results demonstrated that YBX1 may play a critical role in maintaining cell viability in T-ALL.
YBX1 inhibition compromises T-ALL cell viability. (A,D) The protein expression of YBX1 was detected via western blot in Jurkat (A) and Molt4 cells (D) with YBX1 knockdown. (B,E) qRT-PCR analysis of YBX1 mRNA expression in Jurkat (B) and Molt4 cells (E). (C,F) Cell viability was determined using MTS assay in Jurkat (C) and Molt4 cells (F). (G) Western blot analysis of YBX1 in Jurkat cells with YBX1 knockout (sgYBX1-1 or sgYBX1-2). (H-I) Cell viability was assessed via MTS after transduction with the indicated lentiviruses in Jurkat cells. ∗∗P<.01; ∗∗∗P<.001.
YBX1 inhibition compromises T-ALL cell viability. (A,D) The protein expression of YBX1 was detected via western blot in Jurkat (A) and Molt4 cells (D) with YBX1 knockdown. (B,E) qRT-PCR analysis of YBX1 mRNA expression in Jurkat (B) and Molt4 cells (E). (C,F) Cell viability was determined using MTS assay in Jurkat (C) and Molt4 cells (F). (G) Western blot analysis of YBX1 in Jurkat cells with YBX1 knockout (sgYBX1-1 or sgYBX1-2). (H-I) Cell viability was assessed via MTS after transduction with the indicated lentiviruses in Jurkat cells. ∗∗P<.01; ∗∗∗P<.001.
YBX1 depletion induces apoptosis in T-ALL cells
Next, we examined whether YBX1 affects the apoptosis of T-ALL cells. Wright staining was performed to observe morphological characteristics of Jurkat and Molt4 cells following YBX1 silencing with 2 shRNAs. A typical apoptotic morphology, including vacuolar degeneration and nuclear fragmentation, was observed when YBX1 was knocked down (Figure 3A). We also detected cell apoptosis via flow cytometry using annexin V/PI staining. Flow cytometry analysis showed that silencing of YBX1 caused a significant increase in apoptosis (Figure 3B-C; supplemental Figure 2A-B). Moreover, CRISPR/Cas9-mediated knockout of YBX1 in Jurkat cells significantly induced apoptosis, consistent with the results of shRNA-mediated YBX1 knockdown (Figure 3D; supplemental Figure 2C). We further determined whether the exogenous expression of YBX1 could rescue cell apoptosis caused by YBX1 knockdown. Overexpression of YBX1 significantly reduced cell apoptosis mediated by YBX1 depletion (Figure 3E). We also observed depletion of the expression of YBX1 in HEK293T cells. Silencing YBX1 in HEK293T cells had little effect on cell apoptosis (supplemental Figure 2D-E), suggesting that depletion of YBX1 in HEK293T cells was not toxic. These results suggest that suppression of YBX1 by either shRNAs or sgRNAs induces cell apoptosis in T-ALL cell lines in vitro. We further assessed the effects of YBX1 knockout on the expression profiles of several apoptotic signaling-related molecules. Lower mRNA expression levels of Myc and higher mRNA expression levels of Bad and Bim were detected in Jurkat cells with YBX1 knockout than in the control cells (Figure 3F). These results suggested that YBX1 may be involved in the regulation of the apoptotic signaling pathway in T-ALL cells.
YBX1 depletion induces apoptosis and cell cycle arrest at the G0/G1 phase in Molt4 and Jurkat cells. (A) Representative images of Wright–Giemsa staining for the morphological analysis of Jurkat and Molt4 cells. (B-C) Cell apoptosis was examined in Jurkat (B) and Molt4 cells (C) with YBX1 knockdown by flow cytometry using annexin V/PI staining. Quantification of cell apoptosis in Jurkat (B) and Molt4 cells (C). (D) Jurkat cells were infected via lentiviruses expressing YBX1 sgRNA (sgYBX1-1 or sgYBX1-2). Quantification of apoptosis in Jurkat cells with YBX1 knockout. (E) Percentages of apoptotic leukemia cells after transduction on day 3. (F) qRT-PCR analysis of mRNA expression of cell apoptosis-related genes. (G-J) Cell cycle was analyzed using flow cytometry with PI staining. (K) Relative mRNA expression of cell cycle-related genes. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001.
YBX1 depletion induces apoptosis and cell cycle arrest at the G0/G1 phase in Molt4 and Jurkat cells. (A) Representative images of Wright–Giemsa staining for the morphological analysis of Jurkat and Molt4 cells. (B-C) Cell apoptosis was examined in Jurkat (B) and Molt4 cells (C) with YBX1 knockdown by flow cytometry using annexin V/PI staining. Quantification of cell apoptosis in Jurkat (B) and Molt4 cells (C). (D) Jurkat cells were infected via lentiviruses expressing YBX1 sgRNA (sgYBX1-1 or sgYBX1-2). Quantification of apoptosis in Jurkat cells with YBX1 knockout. (E) Percentages of apoptotic leukemia cells after transduction on day 3. (F) qRT-PCR analysis of mRNA expression of cell apoptosis-related genes. (G-J) Cell cycle was analyzed using flow cytometry with PI staining. (K) Relative mRNA expression of cell cycle-related genes. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001.
YBX1 inhibition induces cell cycle arrest at G0/G1 phase
To test the effect of YBX1 depletion on the cell cycle of T-ALL cells, a cell cycle assay was performed via flow cytometry with PI staining. Cell cycle analysis showed that knockdown of YBX1 induced cell cycle arrest at the G0/G1 phase in Jurkat cells (Figure 3G; supplemental Figure 3A) and Molt4 cells (Figure 3H; supplemental Figure 3B). Cell cycle arrest was also observed in Jurkat cells with YBX1 knockout (Figure 3I; supplemental Figure 3C). Restoration of YBX1 effectively rescued cell cycle arrest caused by YBX1 knockdown (Figure 3J). Furthermore, we examined the expression of several cell cycle-related genes via qRT-PCR. The mRNA expression of p18, p19, p21, p27, and p53 was significantly upregulated in Jurkat cells carrying sgYBX1 compared with that in control cells carrying sgC (Figure 3K). These results indicated that YBX1 could modulate the cell cycle of T-ALL cells.
YBX1 knockout impedes T-cell leukemogenesis in the Jurkat xenograft
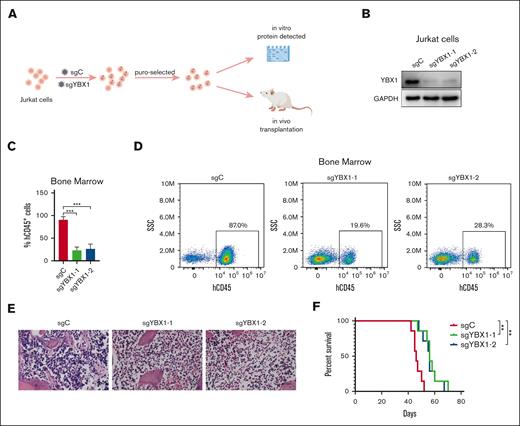
To evaluate the role of YBX1 in T-cell leukemogenesis in vivo, we established a human xenograft model using Jurkat cells (Figure 4A). Jurkat cells were infected via lentiviruses encoding sgC or sghYBX1, with puromycin to select stable clones. The knockout of YBX1 was confirmed via western blot before transplantation (Figure 4B). Puromycin-selected Jurkat cells (4.5 × 106 cells) were engrafted intravenously into the NCG mice. The engraftment of Jurkat cells in NCG mice was determined by quantifying the percentage of hCD45+cells in the bone marrow by flow cytometry. Knockout of YBX1 suppressed CD45+ leukemia cell dissemination in the bone marrow 42 days after transplantation (Figure 4C-D). Consistently, hematoxylin and eosin staining confirmed that the loss of YBX1 significantly reduced the leukemia burden in the bone marrow (Figure 4E). Furthermore, Kaplan-Meier survival analysis revealed that YBX1 knockdown significantly prolonged the survival of mice (Figure 4F). These results clearly demonstrated that YBX1 may be involved in T-ALL leukemogenesis process.
YBX1 knockout decreases leukemia burden in the Jurkat xenograft model. (A) Graphical illustration of the Jurkat xenograft model. Jurkat cells were infected with lentiviruses carrying control (sgC) or YBX1 sgRNA (sgYBX1-1 or sgYBX1-2). The protein level of YBX1 was detected via western blot on day 5 after transfection and before transplantation. Approximately 4.5 × 106 cells were injected into NCG mice intravenously, followed by the assessment of leukemia cell dissemination. (B) Protein analysis via western blot. (C) Flow cytometry analysis showing the percentage of hCD45+ cells in the BM on day 42 after transplantation. (D) Representative flow plot showing the frequency of hCD45+ leukemia cells in the BM. (E) Representative hematoxylin and eosin staining images of the BM from each group. (F) Survival curves are displayed using Kaplan-Meier plots (n = 7; long-rank test). ∗∗P < .01; ∗∗∗P < .001. The pattern diagram was drawn using Figdraw.
YBX1 knockout decreases leukemia burden in the Jurkat xenograft model. (A) Graphical illustration of the Jurkat xenograft model. Jurkat cells were infected with lentiviruses carrying control (sgC) or YBX1 sgRNA (sgYBX1-1 or sgYBX1-2). The protein level of YBX1 was detected via western blot on day 5 after transfection and before transplantation. Approximately 4.5 × 106 cells were injected into NCG mice intravenously, followed by the assessment of leukemia cell dissemination. (B) Protein analysis via western blot. (C) Flow cytometry analysis showing the percentage of hCD45+ cells in the BM on day 42 after transplantation. (D) Representative flow plot showing the frequency of hCD45+ leukemia cells in the BM. (E) Representative hematoxylin and eosin staining images of the BM from each group. (F) Survival curves are displayed using Kaplan-Meier plots (n = 7; long-rank test). ∗∗P < .01; ∗∗∗P < .001. The pattern diagram was drawn using Figdraw.
YBX1 is required for NOTCH1-induced T-ALL development
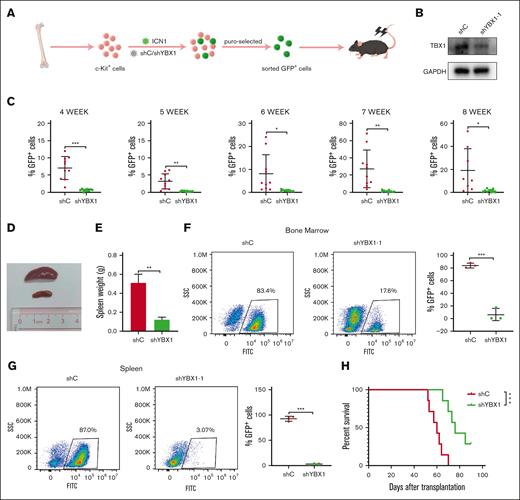
The NOTCH1 gene is a major oncogene in T-ALL. Aberrant activation of NOTCH signaling is crucial for the pathogenesis of T-ALL. Next, we assessed the function of YBX1 in NOTCH1-induced T-ALL. c-Kit+ cells from 6 or 8 week old C57BL/6 mice were infected with ICN1/shC or ICN1/shYBX1. GFP+ cells were sorted using FACS Aria III from puromycin-selected cells (Figure 5A). Before transplantation, protein level was detected via western blot to evaluate the efficiency of shRNA interference. As shown in Figure 5B, YBX1 protein was significantly decreased compared with those cells transduced with the control shRNA. Approximately 2 × 105 sorted cells were transplanted into lethally irradiated (9.0 Gy) C57BL/6 mice via tail vein injection. During the latency period of leukemia development, GFP+ leukemia cells in peripheral blood were monitored via flow cytometry at different time points. Analysis of GFP+ cells from peripheral blood showed that silencing of YBX1 significantly decreased the percentage of GFP+ leukemia in vivo (Figure 5C). A significant decrease in spleen size and weight was observed in ICN1/shYBX1 mice compared with that in ICN1/shC mice at week 8 after transplantation (Figure 5D-E). Moreover, the knockdown of YBX1 resulted in a lower percentage of GFP+ cells in the bone marrow and spleen (Figure 5F-G). In line with these results, mice from the ICN1/shYBX1 group displayed markedly longer latency, which correlated with efficient YBX1 depletion (supplemental Figure 4), indicating that the depletion of YBX1 significantly delayed the development of T-ALL. These results suggest that YBX1 may play a vital role in T-cell leukemogenesis in vivo.
YBX1 knockdown impedes leukemogenesis in NOTCH1-induced T-ALL model. (A) Graphical illustration of NOTCH1-induced T-ALL model. C-Kit+ stem and progenitor cells isolated from the BM of 6- or 8-week-old C57/BL6 mice were infected with a retrovirus carrying ICN1 and lentivirus expressing shC or shYBX1. Approximately 2.0 × 105 GFP+ transduced cells were intravenously injected into lethally irradiated mice. The percentage of GFP+ cells in peripheral blood was quantified using flow cytometry on the subsequent days. (B) YBX1 was analyzed before transplantation. (C) Percentages of GFP+ leukemia cells in the peripheral blood were measured via flow cytometry at 4, 5, 6, 7, and 8 weeks after transplantation. (D) Representative spleen images for each group. (E) Spleen weights of the mice in each group were measured. (F-G) GFP+ leukemia cells in the BM (F) and spleen (G) were quantified using flow cytometry on day 53. (H) Survival curves were plotted using the Kaplan-Meier curves (n = 7; long-rank test). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. The pattern diagram was drawn using Figdraw.
YBX1 knockdown impedes leukemogenesis in NOTCH1-induced T-ALL model. (A) Graphical illustration of NOTCH1-induced T-ALL model. C-Kit+ stem and progenitor cells isolated from the BM of 6- or 8-week-old C57/BL6 mice were infected with a retrovirus carrying ICN1 and lentivirus expressing shC or shYBX1. Approximately 2.0 × 105 GFP+ transduced cells were intravenously injected into lethally irradiated mice. The percentage of GFP+ cells in peripheral blood was quantified using flow cytometry on the subsequent days. (B) YBX1 was analyzed before transplantation. (C) Percentages of GFP+ leukemia cells in the peripheral blood were measured via flow cytometry at 4, 5, 6, 7, and 8 weeks after transplantation. (D) Representative spleen images for each group. (E) Spleen weights of the mice in each group were measured. (F-G) GFP+ leukemia cells in the BM (F) and spleen (G) were quantified using flow cytometry on day 53. (H) Survival curves were plotted using the Kaplan-Meier curves (n = 7; long-rank test). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. The pattern diagram was drawn using Figdraw.
YBX1 affects the AKT and ERK signaling pathways
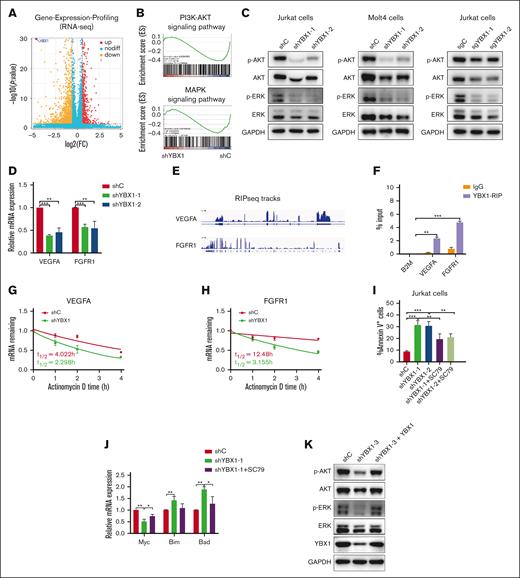
To better understand the molecular mechanism by which YBX1 regulates T-ALL leukemogenesis, we performed RNA sequencing (RNA-seq) analysis 3 days after YBX1 knockdown in Jurkat cells. The results of the RNA-seq showed that 622 genes were upregulated and 1328 genes were downregulated in Jurkat cells with YBX1 knockdown (Figure 6A). Interestingly, gene set enrichment analysis showed an enrichment on phosphoinositide 3 kinase (PI3K)/AKT pathway and mitogen-activated protein kinase (MAPK)pathway (Figure 6B). Furthermore, we investigated the effect of YBX1 on the expression of AKT and ERK signaling-related molecules via western blot. As shown in Figure 6C, the protein expression of total AKT, p-AKT, total ERK, and p-ERK was dramatically downregulated in YBX1-silenced Molt4 and Jurkat cells compared with that in cells expressing shC or sgC (Figure 6C; supplemental Figure 5A-C), suggesting that YBX1 may be involved in the regulation of expression of the AKT and ERK signaling molecules. To further investigate how YBX1 regulates the AKT and ERK signaling pathways, differential gene expression analysis showed that VEGFA and FGFR1, which have been reported to regulate both signaling pathways,25,26 were significantly downregulated by YBX1 deletion. To validate the RNA-seq data, we performed RT-PCR. The RT-PCR results were consistent with the RNA-seq results, which demonstrated that the mRNA levels of vascular endothelial growth factor A (VEGFA) and fibroblast growth factor receptor 1 (FGFR1) were greatly downregulated in YBX1-silenced Jurkat cells (Figure 6D). Considering that YBX1 regulates gene expression by binding and stabilizing mRNAs has been previously demonstrated.23 Therefore, we aimed to assess whether these 2 mRNAs are the binding targets for YBX1. To evaluate this hypothesis, we analyzed a previously published RIP sequencing data set23 and used RIP followed by qPCR in HEK293T cells. The results helped confirm that VEGFA and FGFR1 are the targets of YBX1 binding (Figure 6E-F). Functionally, we found that YBX1 knockdown significantly reduced the half-life of VEGFA and FGFR1 mRNA (Figure 6G-H). Thus, these results indicate that YBX1 regulates the AKT and ERK signaling pathways, in part by regulating the mRNA stability of VEGFA and FGFR1. Considering that depletion of YBX1 induced a decrease in the total protein of AKT, we used SC79, a specific AKT activator that enhances AKT phosphorylation to reactivate the AKT signaling pathway (supplemental Figure 5D). Therefore, shYBX1-induced apoptosis was partially rescued by SC79 (Figure 6I). Moreover, SC79 treatment partially restored the decreased mRNA of Myc and the increased mRNA of some apoptosis-related genes (Bim and Bad) and cell cycle-related genes (p53, p21, and p27) caused by YBX1 knockdown (Figure 6J; supplemental Figure 5E). Furthermore, the decreased protein expression of total AKT, p-AKT, total ERK, and p-ERK induced by YBX1 knockdown was totally restored by YBX1 overexpression (Figure 6K). Overall, YBX1 regulates the AKT and ERK signaling pathways in T-ALL.
YBX1 regulates the AKT and ERK signaling pathways in T-ALL cells. (A) Volcano plot of differentially expressed genes in RNA-seq at day 3 after knockdown of YBX1 in Jurkat cells. (B) Enrichment of genes in the PI3K/AKT and MAPK pathways using gene set enrichment analysis. (C) Whole cell lysates collected from shC and shYBX1 groups 3 days after infection were analyzed. Western blot was performed to detect the protein expression of p-AKT, total AKT, p-ERK, and total ERK in Jurkat and Molt4 cells. (D) qRT-PCR analysis of mRNA expression of VEGFA and FGFR1. (E) RIP sequencing integrative genomics viewer (IGV) tracks of selected differential gene expression (DEGs) in GSE159153. (F) YBX1 RIP-qPCR analysis showing YBX1 binding to VEGFA and FGFR1 mRNA in HEK293T cells. (G-H) The mRNA half-life of VEGFA (G) and FGFR1 (H) in HEK293T cells. (I-J) Jurkat cells were pretreated with SC79 (1 μg/mL) for 24 hours 2 days after transduction with lentiviruses expressing shRNAs. The percentage of apoptosis was detected via flow cytometry (I), and the mRNA expression of apoptosis-related genes was detected via qRT-PCR (J). (K) Protein expression of p-AKT, total AKT, p-ERK, and total ERK in Jurkat cells was detected via western blot. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001.
YBX1 regulates the AKT and ERK signaling pathways in T-ALL cells. (A) Volcano plot of differentially expressed genes in RNA-seq at day 3 after knockdown of YBX1 in Jurkat cells. (B) Enrichment of genes in the PI3K/AKT and MAPK pathways using gene set enrichment analysis. (C) Whole cell lysates collected from shC and shYBX1 groups 3 days after infection were analyzed. Western blot was performed to detect the protein expression of p-AKT, total AKT, p-ERK, and total ERK in Jurkat and Molt4 cells. (D) qRT-PCR analysis of mRNA expression of VEGFA and FGFR1. (E) RIP sequencing integrative genomics viewer (IGV) tracks of selected differential gene expression (DEGs) in GSE159153. (F) YBX1 RIP-qPCR analysis showing YBX1 binding to VEGFA and FGFR1 mRNA in HEK293T cells. (G-H) The mRNA half-life of VEGFA (G) and FGFR1 (H) in HEK293T cells. (I-J) Jurkat cells were pretreated with SC79 (1 μg/mL) for 24 hours 2 days after transduction with lentiviruses expressing shRNAs. The percentage of apoptosis was detected via flow cytometry (I), and the mRNA expression of apoptosis-related genes was detected via qRT-PCR (J). (K) Protein expression of p-AKT, total AKT, p-ERK, and total ERK in Jurkat cells was detected via western blot. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001.
YBX1 inhibitor SU056 exerts antileukemia activity in T-ALL in vitro and in vivo
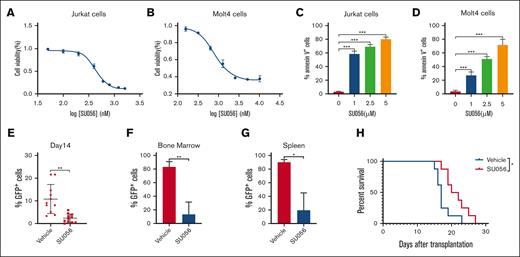
Given the important role of YBX1 in regulating leukemogenesis, we evaluated the therapeutic potential of SU056, an inhibitor of YBX1, in T-ALL. Firstly, we analyzed the effect of SU056 on the proliferation and apoptosis of T-ALL cells. Jurkat and Molt4 cells were treated with SU056 at different concentrations for 24 and 48 hours. Treatment with SU056 significantly suppressed cell proliferation and induced cell apoptosis at 24 and 48 hours in a dose-dependent manner (supplemental Figure 6A-D; Figure 7A-D). We, then, investigated the in vivo antitumor effect of SU056 in a NOTCH1-driven murine T-ALL model. Mice that received transplantation with NOTCH1-induced T-ALL leukemia cells were treated for 15 consecutive days with vehicle or SU056 (20 mg/kg, intraperitoneally). Notably, SU056 treatment significantly decreased the tumor burden, as assessed via fluorescence-activated cell sorting detection of leukemic GFP+ cells in the peripheral blood (Figure 7E), bone marrow (Figure 7F), and spleen (Figure 7G). Moreover, SU056 treatment prolonged the survival of T-ALL mice (Figure 7H). These results indicate that the YBX1 inhibitor SU056 might be a potential candidate for a novel therapeutic drug for T-ALL.
SU056 treatments demonstrate antileukemia efficacy both in vitro and in vivo. (A-B) Cell viability was determined using MTS assay in Jurkat (A) and Molt4 (B) cells treated with SU056 at various concentrations for 48 hours. (C-D) Quantification of cell apoptosis in Jurkat (C) and Molt4 (D) cells treated with SU056 for 48 hours. (E) Percentages of GFP+ leukemia cells in peripheral blood were measured via flow cytometry on day 14. (F-G) GFP+ leukemia cells in the BM (F) and spleen (G) were quantified via flow cytometry on day 16. (H) Survival curves were plotted using the Kaplan-Meier curves (n = 8; long-rank test). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001.
SU056 treatments demonstrate antileukemia efficacy both in vitro and in vivo. (A-B) Cell viability was determined using MTS assay in Jurkat (A) and Molt4 (B) cells treated with SU056 at various concentrations for 48 hours. (C-D) Quantification of cell apoptosis in Jurkat (C) and Molt4 (D) cells treated with SU056 for 48 hours. (E) Percentages of GFP+ leukemia cells in peripheral blood were measured via flow cytometry on day 14. (F-G) GFP+ leukemia cells in the BM (F) and spleen (G) were quantified via flow cytometry on day 16. (H) Survival curves were plotted using the Kaplan-Meier curves (n = 8; long-rank test). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001.
Discussion
YBX1 is a well-known multifunctional oncoprotein and has been identified as a prognostic clinical biomarker. The overexpression of YBX1 is associated with poor prognoses in various cancers, such as breast cancer, prostate cancer, hepatocellular cancer, colorectal cancer, lung cancer, multiple myeloma, osteosarcoma, synovial sarcoma, and ovarian cancer.12,17-22,27-29 YBX1 is also involved in leukemogenesis in hematologic malignancies. It has been reported that YBX1 is highly expressed in acute myeloid leukemia.23,30 YBX1 could promote leukemogenesis by amplifying the translation of Myc or interacting with IGF2BPs and stabilizing m6A-tagged RNA.23,30 YBX1 was found to be required for the survival of human myeloid leukemia cells. Recently, Zhao et al showed that heterogeneous nuclear ribonucleoprotein D and YBX1 are required for T-cell lymphoma–associated lncRNA1 (TCLlnc1)-mediated regulation of the transforming growth factor–β signaling pathway as well as T–lymphoma cell proliferation and migration in peripheral T-cell lymphoma.31 However, the role of YBX1 in T-ALL remains unknown. In this study, we analyzed the mRNA expression using a public database and further assessed the expression of YBX1 mRNA and protein in patients with T-ALL, lymphoid cell lines, and NOTCH1-induced T-ALL mice. Elevated YBX1 expression was observed in patients with T-ALL. Our results indicated that YBX1 may be a potential molecular marker of T-ALL. Downregulation of YBX1 significantly decreased cell viability, induced cell apoptosis, and induced G0/G1 phase arrest in T-ALL cells. Importantly, depletion of YBX1 impeded T-cell leukemogenesis in a human T-ALL xenograft and NOTCH1-induced T-ALL mouse model in vivo. Taken together, our results revealed that YBX1 may play a crucial role in the pathogenesis of T-ALL. However, the clinical association between high YBX1 expression and poor prognosis remains poorly defined. Further studies to investigate this should be performed in the future.
YBX1 has been reported to be implicated in the regulation of multiple signaling pathways. It can modulate the expression of a large number of genes with a variety of functions, such as important components of growth factor–mediated pathways (EGFR, HER2/ErbB2, and HDGF),32-34 oncogenic factors (Myc and HOXC8),24,35,36 cell cycle- and apoptosis-related genes (cyclin D1, p16, p21, Bcl-2, and BAX),23,37-40 and chemoresistance-related genes (MDR1, ABCB1, MVP/LRP, and CD44).41-44 Perner et al reported that YBX1 could modulate translational output by recruiting relevant mRNA to polysomal chains in acute myeloid leukemia.30 Feng et al also showed that YBX1 depletion could promote mRNA decay of Myc and Bcl-2 in an m6A-dependent manner in myeloid leukemia.23 In T-ALL cells, we observed that knockout of YBX1 was accompanied by downregulation of the cell survival-related gene Myc; upregulation of cell cycle-related genes p18, p19, p21, p27, and p53; and cell apoptosis-related genes Bad and Bim. YBX1 depletion also decreased the protein expression of total AKT, p-AKT, total ERK, and p-ERK in T-ALL cells. Differential gene expression analysis revealed that silencing of YBX1 significantly downregulated the mRNA expression of VEGFA and FGFR1, which have been reported to regulate the AKT and ERK signaling pathways.25,26 Furthermore, we showed that FGFR1 and VEGFA were the binding targets of YBX1. Knockdown of YBX1 markedly decreased VEGFA and FGFR1 mRNA half-life in T-ALL cells. Our results provide new insight into the mechanism underlying YBX1-mediated T-ALL leukemogenesis.
Given that YBX1 is dispensable for normal hematopoiesis, YBX1 may have great potential as a biomarker and therapeutic target in T-ALL. Blocking YBX1-mediated signaling is an alternative way to develop new drugs for T-ALL treatment. Recently, an inhibitor of YBX1 activity (SU056) was reported to inhibit ovarian cancer cell survival.22 Our results demonstrated that treatment with SU056 impaired T-ALL cell survival in vitro and in vivo, indicating that targeting YBX1 is effective for the treatment of T-ALL. It will be interesting to evaluate the therapeutic potential of YBX1 inhibitors in patients with T-ALL in the future.
Acknowledgments
This research was funded by the National Natural Science Foundation of China (31970739) and Anhui Natural Science Foundation (2108085QH322).
Authorship
Contribution: H.L. and D.Z. designed and performed the experiments; Q.F., S.W., X. Zhang, and X.C. contributed to the hematoxylin and eosin assays; X. Zhu, N.A., Y.C., and L.Z. interpreted the data and contributed to manuscript preparation; H.L. wrote the manuscript; and D.L. and N.Z. conceived, directed, and funded the project; Z.W. provided suggestions for how to answer the comments of this manuscript from reviewers.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Na Zhao, Department of Hematology, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Lujiang Rd No 17, Hefei 230001, China; e-mail: zhaonamed@126.com; and Desheng Lu; e-mail: delu@szu.edu.cn.
References
Author notes
∗H.L., D.Z., D.L., and N.Z. contributed equally to this work.
The raw RNA sequence data reported in this article have been deposited in the Genome Sequence Archive in National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA-Human, HRA004741) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa-human.
Data are available on request from the corresponding authors, Na Zhao (zhaonamed@126.com) and Desheng Lu (delu@szu.edu.cn).
The full-text version of this article contains a data supplement.