Key Points
In a phase 3 clinical trial of FL, AICDA-mediated intraclonal heterogeneity was not prognostic.
TP53 mutated populations are common, subclonal, and their absence confers sensitivity to a radiation-containing regimen.
Abstract
Although TP53 is commonly mutated in transformed follicular lymphoma, mutations are reported in <5% of pretreatment follicular lymphoma (FL) specimens. We assayed archival follicular B-cell non-Hodgkin lymphoma specimens from a completed clinical trial, Southwest Oncology Group S0016, a phase 3 randomized intergroup trial of CHOP (cyclophosphamide, hydroxydaunorubicin, oncovin, and prednisone) chemotherapy plus R-CHOP (rituximab-CHOP) compared with CHOP chemotherapy plus 131-iodine tositumomab (radioimmunotherapy [RIT]-CHOP). Subclonal TP53 mutations (median allele frequency 0.02) were found in 25% of diagnostic FL specimens and in 27% of a separate validation cohort. In the R-CHOP arm, pathogenic TP53 mutations were not associated with progression-free survival (PFS) (10-year PFS 43% vs 44%). In contrast, among patients with no detectable pathogenic TP53 mutation, RIT-CHOP was associated with a longer PFS than with R-CHOP (10-year PFS 67% vs 44%; hazard ratio = 0.49; P = .008). No relationship was detected between PFS and the extent of activation-induced cytidine deaminase (AICDA)–mediated heterogeneity. In summary, subclonal TP53 mutations are common in FL and are a distinct phenomenon from AICDA-mediated genetic heterogeneity. The absence of a detectable subclonal mutation in TP53 defined a population that particularly benefited from RIT.
Introduction
Follicular lymphoma (FL) is a mature B-cell neoplasm whose cell-of-origin, the germinal center B cell, expresses activation-induced cytidine deaminase (AICDA) which physiologically targets the immunoglobulin genes. Aberrant targeting of AICDA to other genes creates not only innumerable passenger mutations but may also create a smaller number of additional driver mutations that lead to progression.1-7 This hypothesis suggests that greater apolipoprotein B messenger RNA–editing enzyme, catalytic polypeptide–mediated genetic heterogeneity will be associated with a greater likelihood of tumor evolution. Previous studies of FL have yielded a range of conclusions regarding relationships among AICDA expression, AICDA-mediated mutations, tumor heterogeneity, and outcomes.5,8-13
Although TP53 is the most commonly mutated gene in human cancers, mutations are reported in <5% of pretreatment FL specimens.3,10,14 In contrast, TP53 is reported to be mutated in ∼20% of transformed and/or relapsed FL specimens. This disparity suggests that TP53-mutated subpopulations, which are too small for detection, might be present at the time of diagnosis in tumors destined to relapse. There is precedence for the clinical significance of subclonal, TP53-mutated populations; for example, in chronic lymphocytic leukemia, the median variant allele frequency (VAF) of TP53 mutations is 0.02 at the time of detection, and any detectable subclonal TP53 mutation, regardless of VAF, is associated with shorter progression-free survival (PFS) after immunochemotherapy.15
To investigate the impact of genetic heterogeneity, we developed a deep-sequencing assay to quantify subclonal mutations in archival formalin-fixed paraffin-embedded (FFPE) specimens and applied this assay to a National Clinical Trials Network phase 3 trial in FL with mature results, SWOG (formerly Southwest Oncology Group) S0016 (www.clinicaltrials.gov; accessed April 2021). In this study, we used SWOG 0016 to test if 2 forms of heterogeneity, which are driven by AICDA and marked by dysfunctional TP53, are associated with outcomes in the setting of chemotherapy combined either with immunotherapy or with radioimmunotherapy (RIT).16,17
Methods
SWOG and Alliance (formerly Cancer and Leukemia Group B) compared 2 immunochemotherapy regimens R-CHOP (rituximab-cyclophosphamide, doxorubicin, vincristine, and prednisone) vs CHOP followed by consolidation with iodine I-131 tositumomab RIT-CHOP) in a phase 3 randomized intergroup protocol (SWOG S0016), enrolling 554 patients with untreated, advanced-stage FL (bulky stage II, III, or IV; any grade) between 1 March 2001 and 15 September 2008.16 FFPE diagnostic specimens were submitted for central banking but were not required for enrollment.
An independent validation cohort consisted of 75 FFPE B-cell non-Hodgkin lymphoma specimens from 37 patients diagnosed with FL, enrolled in 1 of several SWOG clinical trials in B-cell non-Hodgkin lymphoma (S0016, S0433, S0801, and S0433) and treated at the University of Rochester. Of these, 30 patients had a pretreatment FL specimen.
Eighty-five specimens (11 nodular lymphocyte predominant Hodgkin lymphoma [nLPHL], 38 mantle cell lymphomas [MCL], 36 marginal zone lymphoma [MZL] specimens) served as methodologic controls. These specimens were processed identically to the FL specimens and analyzed using the same pipeline.
The polymerase chain reaction (PCR) strategy in this study includes 2 features to allow high sensitivity while suppressing false discovery owing to a range of sources (refer to supplement for rationale, methods, and data). First, genomic template concentration is as high as achievable in order to detect low frequency sequence variants and to bias early rounds of PCR to use guide DNA (gDNA) as template. Second, we use an internal control sequence (eukaryotic ultraconserved region [eUCR]), to provide specimen- and sequence-specific thresholds for false discovery to account for PCR amplification-related errors and formalin-induced sequence alterations. We validated the reproducibility of the methods and showed that the results are not meaningfully affected by repair of formalin-associated DNA lesions and are independent of PCR-cycle number (refer to supplemental Results and Methods).
Each specimen fulfilled the following criteria: (1) sufficient DNA for 4 multiplexed PCR-reactions each with 250 ng gDNA to allow amplification of the TP53 (all exons), the 5′ UTR of BCL2 (uBCL2, a region of the gene with particularly high numbers of passenger mutations that is reliably amplifed)18 and the clonal IGHV gene and (2) appropriate size distribution of DNA products after library preparation (∼300 base pairs)
To obtain a false discovery rate of 1% for single nucleotide variants (SNV) with a VAF = 0.002, each multiplex reaction included an amplicon of eUCR41 to provide specimen-, reaction-, and sequence-specific metrics of DNA-damage and sequencing noise. Ultradeep, amplicon-based sequencing (∼10 000×) (Ilumina 300 nt paired end) were merged, mapped to reference, and SNV were called using LoFreq based on individualized error thresholds (derived from the eUCR41 amplicon; refer to supplemental Methods).
TP53 sequence variants were classified as pathogenic when both the Seshat and International Agency for Research on Cancer databases concurred that the variant was likely associated with cancer.
PFS was defined as the time from date of registration to the date of first observation of progressive disease or death owing to any cause. Patients last known to be alive and progression-free were censored at the date of last contact. Cox regression model was used to estimate hazard ratio (HR) and 95% confidence interval (CI).
For clarity, VAF are expressed in decimal format (range 0-1, in which 0.05 corresponds to 5%) and fraction of specimens affected are expressed in percentage format.
Results
Diagnostic biopsies were available for 230 of 571 enrollees in SWOG S0016. DNA and sequence data from 147 samples (TP53 147, BCL2 146, and IGHV 120) fulfilled the quality metrics (refer to "Methods” and supplement for CONSORT [Consolidated Standards of Reporting Trials] diagram). For patients with (147) and without (389) completed sequencing, the median duration of follow-up (both 14.7 years), the number of patients who experienced progression of disease or died (both 58%), and other patient characteristics (demographics, histology, stage, FLIPI, treatment arm, and PFS) were comparable (with the exception of serum β-2 microglobulin) (supplemental Table 1).
Twenty-five percent of FL specimens from patients with untreated advanced-stage disease have a TP53 mutation
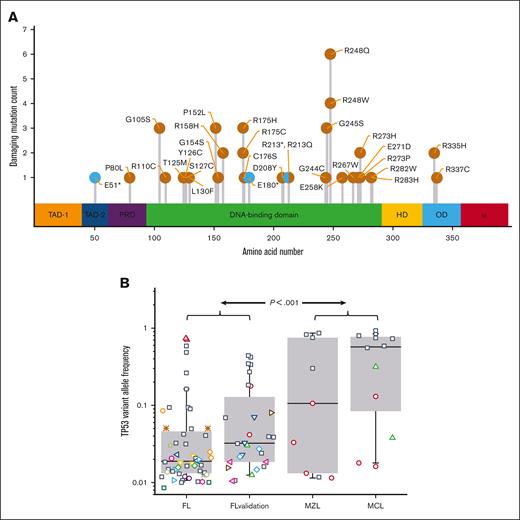
The 147 FFPE specimens from S0016 (80 RIT-CHOP; 67 R-CHOP) had 51 mutations within TP53; the variants in 37 patients (25%) created missense or nonsense mutations that met established criteria for pathogenicity (Figure 1A; supplemental Table 2). With the exception of sex, clinical characteristics at presentation of patients with and without TP53 mutations were indistinguishable (males were overrepresented in the nonmutated group; supplemental Table 3).
TP53 mutations are common in S0016 FL specimens and have a low VAF. (A) The pathogenic mutations detected in S0016 include canonical hotspots for TP53 mutations. The location and numbers of mutations at each site are indicated as missense (orange) or nonsense mutations (cyan). The colored regions indicate assigned functionality of each protein domain (from left, transactivation 1, transactivation 2, proline-rich domain, DNA binding, hinge domain, oligomerization domain, and alpha domain). (B) The median VAF of pathogenic mutations in FL is low. In the 37 of 147 FL specimens with a pathogenic TP53 mutation, the median VAF for these mutations is 0.02 (FL). In a validation set, 28 mutations were detected in 19 specimens (FL validation) with a median VAF of 0.03. Additional control samples of MZL and mantle cell lymphoma MCL (36 and 38 specimens) have a substantially higher median VAF. Boxes contain second and third quartiles, and whiskers 10 to 90 percentiles. Specimens with a single detectable mutation are noted with black squares. For specimens with >1 TP53 mutation, all the mutations in that specimen share a unique symbol.
TP53 mutations are common in S0016 FL specimens and have a low VAF. (A) The pathogenic mutations detected in S0016 include canonical hotspots for TP53 mutations. The location and numbers of mutations at each site are indicated as missense (orange) or nonsense mutations (cyan). The colored regions indicate assigned functionality of each protein domain (from left, transactivation 1, transactivation 2, proline-rich domain, DNA binding, hinge domain, oligomerization domain, and alpha domain). (B) The median VAF of pathogenic mutations in FL is low. In the 37 of 147 FL specimens with a pathogenic TP53 mutation, the median VAF for these mutations is 0.02 (FL). In a validation set, 28 mutations were detected in 19 specimens (FL validation) with a median VAF of 0.03. Additional control samples of MZL and mantle cell lymphoma MCL (36 and 38 specimens) have a substantially higher median VAF. Boxes contain second and third quartiles, and whiskers 10 to 90 percentiles. Specimens with a single detectable mutation are noted with black squares. For specimens with >1 TP53 mutation, all the mutations in that specimen share a unique symbol.
Pathogenic TP53 mutations in FL are typically subclonal and often multiple
In the initial data set, the VAFs for pathogenic TP53 mutations ranged from 0.009 to 0.74 (median 0.02) (Figure 1B). Twenty-six samples had a single mutation whereas 11 had >1 (8 with 2 and 3 with 3 mutations). In the validation set of 75 samples from a total of 37 patients, 14 samples (19%) had 28 pathogenic TP53 mutation with a median VAF of 0.03 (range 0.01-0.44). To assess if the low VAFs of TP53 mutations in FL specimens might be because of nonspecific artifacts associated with PCR or the analytical pipeline, we assayed 85 samples from 3 other B-cell lymphomas, MCL (38), in which TP53 mutations have been reported to be predominantly clonal,19 MZL (36), and nLPHL (11). No mutations were detected in the 11 nLPHL specimens. In contrast to FL, TP53 mutations in MCL and MZL had a median VAF of 0.32, indicating that the abundance of low VAF mutations in the FL specimens is not due to processing artifacts (Figure 1B).
Subclonal TP53 mutation status affects progression only in patients treated with RIT-CHOP
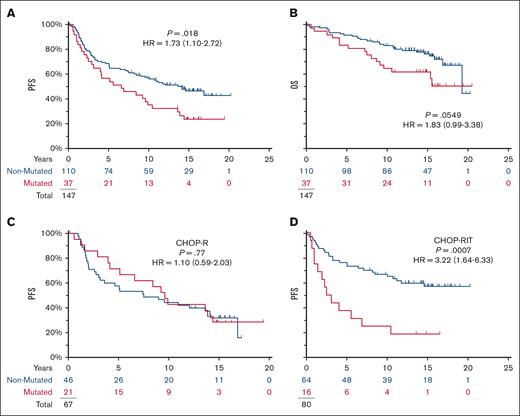
Although almost all the mutations were subclonal, pathogenic TP53 mutations were associated with a shortened PFS (HR = 1.73; P = .0183; 95% CI for HR, 1.10-2.72) (Figure 2A). In contrast, no impact was seen on PFS in the CHOP-R arm (10-year PFS 43% vs 44%) (Figure 2C); the effect of mutation status on the PFS seen in the full-study population (Figure 2A) is explained by the separation of the PFS curves for the RIT-CHOP arm (10-year PFS 25% vs 67%; HR = 3.22; P = .0007) (Figure 2D).
TP53 mutations are prognostic in S0016 and specifically affect the prognosis on the RIT-CHOP arm but not on the R-CHOP arm. Of 147 patients, 72 showed disease progression and 44 died with a median of 15 years follow-up among those last known alive. (A,B) Mutations in TP53 were associated with shortened PFS in S0016 but not OS. (C,D) TP53 mutations affected PFS on RIT-CHOP arm but not on the R-CHOP arms (blue, no mutation detected; red, mutation detected).
TP53 mutations are prognostic in S0016 and specifically affect the prognosis on the RIT-CHOP arm but not on the R-CHOP arm. Of 147 patients, 72 showed disease progression and 44 died with a median of 15 years follow-up among those last known alive. (A,B) Mutations in TP53 were associated with shortened PFS in S0016 but not OS. (C,D) TP53 mutations affected PFS on RIT-CHOP arm but not on the R-CHOP arms (blue, no mutation detected; red, mutation detected).
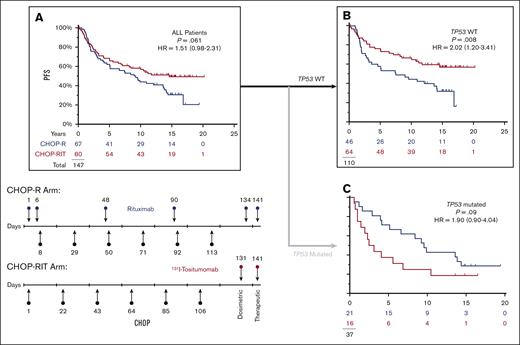
RIT-CHOP prolongs PFS particularly in patients lacking TP53 mutations
In this subset of patients with available specimens, the impact of the therapies is almost superimposable on the entire population as last reported17 (Figure 3A). Considering only the 110 patients with no evidence of a TP53 mutation, RIT-CHOP was associated with a longer PFS than R-CHOP (10-year PFS 67% vs 44%; HR = 0.49; P = .008; 95% CI, 0.29-0.83) (Figure 3B). In the corresponding subset (the 37 patients with a pathogenic TP53 mutation), PFS with RIT-CHOP was not significantly different from that obtained with R-CHOP (10-year PFS 25% vs 43%; HR = 1.90; P = .09; 95% CI, 0.90-4.04) (Figure 3C) (refer to supplemental Data for overall survival).
In patients with NO detectable TP53 mutation, CHOP-RIT is associated with longer PFS. (A) There is a trend to longer PFS with CHOP-RIT similar to that previously published for the entire study.17 (B) Removing the 37 patients with TP53 mutations, the PFS was significantly longer on the RIT-CHOP arm compared with the R-CHOP arm. (C) In the subset of 37 patients with a TP53 mutation, there is an opposite trend with shortened PFS on the RIT-CHOP arm. (Red, RIT-CHOP; Blue R-CHOP).
In patients with NO detectable TP53 mutation, CHOP-RIT is associated with longer PFS. (A) There is a trend to longer PFS with CHOP-RIT similar to that previously published for the entire study.17 (B) Removing the 37 patients with TP53 mutations, the PFS was significantly longer on the RIT-CHOP arm compared with the R-CHOP arm. (C) In the subset of 37 patients with a TP53 mutation, there is an opposite trend with shortened PFS on the RIT-CHOP arm. (Red, RIT-CHOP; Blue R-CHOP).
Treatment and time do not consistently affect the VAFs for TP53 mutations
In the serial samples assembled in the validation cohort, the VAFs of pathogenic TP53 mutations did not increase with time or therapy (Figure 4). Of the 21 pathogenic TP53 mutations observed at any time point, various patterns were seen including persistence, emergence, emergence with subsequent persistence, loss, and emergence with subsequent loss. Only 2 patients (patients 2 and 35) showed stable VAFs. Of the 11 TP53 mutations with VAF <0.1, only 1 was observed at 2 distinct time points (patient 38 C141Y). One specimen showed a complete loss of a relatively abundant TP53 mutation (patient 1; mutation R110C) whereas in another a relatively high VAF TP53 mutation emerged (patient 25; mutation C176R). Two patients (38 and 42) showed transformation to diffuse large B-cell lymphoma; neither case showed acquisition of a high VAF TP53 mutation. Regardless of the details of the time courses, a consistent increase in the VAF of pathogenic TP53 mutation was not observed in FL treated with CHOP-based regimens.
TP53 mutations are evanescent and their VAF does not overtly increase with relapse. Out of 22 patients with serial specimens, 11 patients had at least 1 specimen with a pathogenic mutation. The VAFs for these mutations are shown in Figure 1B FL validation. To estimate changes in tumor cell content, we determined the sum of VAFs for mutations in a single amplicon of uBCL2 (solid black lines with orange blocks); this provides an estimate of the minimum tumor content. The TP53 mutations are depicted on the thick gray lines (red blocks mark the VAF; blue blocks denote that the mutation was not detected at that time point [for TP53] or only germ line sequence for uBCL2).
TP53 mutations are evanescent and their VAF does not overtly increase with relapse. Out of 22 patients with serial specimens, 11 patients had at least 1 specimen with a pathogenic mutation. The VAFs for these mutations are shown in Figure 1B FL validation. To estimate changes in tumor cell content, we determined the sum of VAFs for mutations in a single amplicon of uBCL2 (solid black lines with orange blocks); this provides an estimate of the minimum tumor content. The TP53 mutations are depicted on the thick gray lines (red blocks mark the VAF; blue blocks denote that the mutation was not detected at that time point [for TP53] or only germ line sequence for uBCL2).
Intraclonal tumor genetic heterogeneity does not explain the impact of low abundance TP53 mutations on the clinical course of FL
The detection of low abundance mutations in any gene might mark tumors that have an increased intratumoral genetic heterogeneity. We sought to test the alternative hypothesis that intraclonal heterogeneity itself, rather than the presence of a low abundance mutation specifically in TP53, would correlate with outcome or predict the presence of a detectable pathogenic mutation in TP53.
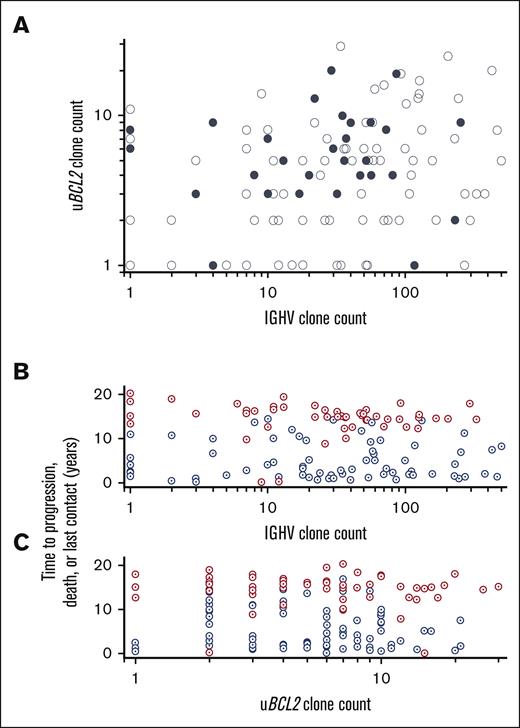
AICDA-mediated sequence variants in IGHV and uBCL2 were used to assess intraclonal genetic heterogeneity in 147 specimens (120 IGHV and 146 uBCL2) also assessed for TP53 variants. Abundant AICDA-mediated mutations were present such that the clonal IGHV was highly mutated (<0.98 germ line; 98/105 patients; median, 0.89; quartiles 1-3, 0.87-0.92) as was uBCL2 (136/146 patients; 828 SNVs, median of 5 per specimen; range, 1-32). Mutations in uBCL2 were strongly skewed to the AICDA motif (P < .001) and sequences with VAFs implying that clonal dominance were common (median, 0.28; quartiles 2-3, 0.14-0.40) (supplemental Table 4). Similar to our previous work, the numbers of subclones based on IGHV (range, 1-501; median, 30) and uBCL2 (range, 1-32; median, 5) were highly variable; furthermore, in the 115 samples with at least 1 subclone marked by uBCL2 or IGHV sequence variants, the numbers of subclones were independent of each other (Figure 5A).18 The detection of a pathogenic mutation in TP53 was not associated with the number of subclones detected in either uBCL2 or IGHV sequences (Figure 5A).
The extent of AICDA-mediated diversity as marked by uBCL2 and IGVH varies widely and is unrelated to the presence of a TP53 mutation. (A) The number of distinct sequences for uBCL2 is the maximum number seen for a single amplicon within that region (refer to Methods). The number of distinct IGVH sequences is the number of distinct variants detected in the clonal IGVH sequence (refer to Methods). Patients with no variants are scored as 1. No relationship is apparent among the number of distinct uBCL2 sequences, the number of distinct IGVH sequences, and the status of TP53 (filled denotes detectable pathogenic lesion; open denotes no detectable lesion). (B-C) No relationship is apparent between the number of variant uBCL2 or IGVH sequences and the time to progression or progression at any time. The number of variant IGVH and uBCL2 sequences was not different between the patients who showed disease progression (blue) or who did not show disease progression (red). Furthermore, there is no suggestion that the number of sequence variants detected differed between those who showed disease progression within 2 years and those who showed disease progression later.
The extent of AICDA-mediated diversity as marked by uBCL2 and IGVH varies widely and is unrelated to the presence of a TP53 mutation. (A) The number of distinct sequences for uBCL2 is the maximum number seen for a single amplicon within that region (refer to Methods). The number of distinct IGVH sequences is the number of distinct variants detected in the clonal IGVH sequence (refer to Methods). Patients with no variants are scored as 1. No relationship is apparent among the number of distinct uBCL2 sequences, the number of distinct IGVH sequences, and the status of TP53 (filled denotes detectable pathogenic lesion; open denotes no detectable lesion). (B-C) No relationship is apparent between the number of variant uBCL2 or IGVH sequences and the time to progression or progression at any time. The number of variant IGVH and uBCL2 sequences was not different between the patients who showed disease progression (blue) or who did not show disease progression (red). Furthermore, there is no suggestion that the number of sequence variants detected differed between those who showed disease progression within 2 years and those who showed disease progression later.
Although the data using subclones marked by AICDA-mediated mutations in uBCL2 and IGHV show that diagnostic FL specimens are composed of varying numbers of subclones, neither the number of these mutations nor the number of subclones denoted by these mutations were prognostic (Figure 5B; supplemental File 4). Four metrics were assessed: the number of mutations present in the most mutated sequence and the number of distinct sequences detected. Cox regression showed no relationship between PFS and the maximum number of mutations in uBCL2 (HR, 1; 95% CI, 0.94-1.03) or the subclone count based on uBCL2 variants (HR, 1; 95% CI, 0.95-1.03). Furthermore, receiver operator curves did not suggest a cut point for any of these measures that would predict disease progression at any time (areas under the curve are indistinguishable from 0.5; supplemental File 5). No relationship was observed between progression and the percent identity IGHV to germ line for the most frequent IGHV sequence in each case (HR, 0.85; 95% CI, 0.50-1.45; relative to 10% increase in percent identity) and the number of subclones defined by IGHV sequences (HR, 1.02; 95% CI, 0.99-1.04; relative to 10 counts increase in subclone). Similar results were observed when whether the subclone count was considered as a continuous variable or stratified as quartiles. Furthermore, none of these measures was associated with the likelihood of progression within 24 months, a time point used as a prognostic indicator in clinical practice.20
Discussion
These data suggest that TP53 mutations are a predictive biomarker for RIT in FL. Specifically, in patients without any detectable TP53 mutation, RIT-CHOP had a substantially improved PFS compared with R-CHOP (67% vs 44%, 10-year PFS). Although tositumomab is no longer available, our results may be paradigmatic with relevance to other settings in which radiation is used.
Characteristics of subclonal TP53 mutations in FL
Although TP53 mutations have been described as infrequent in low grade FL, our ultradeep sequencing shows that 25% of advanced–stage FL specimens had at least 1 subpopulation with a mutation. Although the TP53 mutations observed in FL are at the same hotspots as in other lymphoid malignancies, the incidence, VAFs, and time course of TP53 mutations in FL appear distinctive. First, in contrast to TP53 mutations in MCL and MZL, the VAF is almost uniformly low in FL. Second, multiple pathogenic TP53 mutations per case were more frequent in FL specimens than in MZL or MCL, consistent with a recent study of 26 cases of MCL that reported that 1 of 9 TP53 mutated specimens had >1 mutation.19 Third, although TP53 mutant populations are reported to expand after therapy in chronic lymphocytic leukemia,21 we did not identify a single instance in which the VAF of a pathogenic TP53 mutation showed an interval increase.
The relationship between TP53 mutation status and outcome depended on therapy
Several publications have suggested that although infrequent, high VAF TP53 mutations are associated with a poor prognosis in FL and are more common in grade 3 and transformed FL.3,10,14 Our salient finding is that the lack of detectable low VAF TP53 mutations is associated with improved PFS for RIT-CHOP compared with that for R-CHOP (Figure 3B).
In contrast to patients treated on the RIT-CHOP arm, TP53 mutation status did not affect PFS for patients treated with R-CHOP (Figure 2C). Similarly, Pastore et al did not identify TP53-mutations as prognostic for patients treated with R-CHOP (although the methods may not have detected subclonal mutations).22 The lack of a discernable effect of TP53 mutations in the R-CHOP arm may reflect that overexpression of BCL2, a hallmark feature of FL, creates a high barrier for TP53-dependent apoptosis. A major mechanism of TP53 function is induction of BAX, which in turn inhibits BCL2; even in the presence of fully functional TP53, CHOP-induced activation of BAX may be insufficient to overcome BCL2-mediated blockade of apoptosis in FL.23
Our observation suggesting that TP53 mediates response to radiotherapy is consistent with previous reports. More than a quarter century ago, Lowe et al24 demonstrated that external beam radiation rapidly shrinks the murine thymus because of apoptosis of lymphocytes, whereas the thymic T cells of TP53 knock-out mice are resistant. Knoops et al showed that the efficacy of low-dose external beam radiation in FL depends on TP53.25 Therefore, functional TP53 is likely the basis for the efficacy of RIT and can be considered as a predictive biomarker in this regard.
AICDA-mediated genetic heterogeneity does not correspond to clinical outcome
Although intraclonal genetic heterogeneity in FL is well-documented, the association of diversity and subclonal populations with outcome has not been extensively investigated. Early studies showed extensive AICDA-mediated diversification of IGHV sequences.26 Furthermore, sequencing studies of serial specimens show that the dominant subclones vary over time.3,27 Regardless, there are fewer data addressing whether the number of subclones or the type of intraclonal heterogeneity affect outcome in FL.11 Although a large study has detected an association between intraclonal heterogeneity and outcome in a range of solid tumors, similar studies have not, to our knowledge, been conducted in lymphoma.28 To provide a metric of AICDA-mediated intraclonal heterogeneity, we focus on the loci most highly targeted by AICDA, uBCL2, and IGHV, in which the density of mutations is at least an order of magnitude greater than in other well-characterized AICDA targets (eg, PIM1, MYC, and BCL6).18
Neither the number of mutations in uBCL2 and IGHV nor the number of subclones marked by these mutations correlated with the presence of low VAF TP53 mutations. Therefore, detection of low VAF TP53 mutations does not appear to be a surrogate marker for greater intraclonal genetic heterogeneity. These data suggest a paradox; although AICDA-mediated mutations are reported to be more abundant in progressed or transformed FL, we find that the level of AICDA-mediated mutation in diagnostic specimens is not related to the likelihood of subsequent progression.3 Two models could resolve this paradox. AICDA-mediated mutagenesis is episodic in lymphoma cell lines in which after many cell cycles with no detectable AICDA-mediated mutagenesis, transient bursts of mutagenesis occurred in individual cells.29 In a mouse model of FL, lymphomagenesis occurred through multiple cycles of B cells through germinal centers, each cycle including episodic activation of AICDA and an opportunity to acquire additional driver mutations.30 Therefore, AICDA-mediated mutagenesis may be a driver of transformation, but because it occurs intermittently, it cannot serve as a prognostic marker. Alternatively, others have suggested that AICDA-mediated epigenetic heterogeneity rather than genetic heterogeneity drives progression.31 In either case, our data refine hypotheses regarding the relationship between AICDA and progression by discrediting the hypothesis that AICDA-mediated genetic heterogeneity can serve as a prognostic marker in FL. Given the dominant role of APOBECs (apolipoprotein B messenger RNA–editing enzyme, catalytic polypeptides, the gene fanliy that includes AICDA) in sculpting the genome in a wide range of malignancies,32 our observation may have implications beyond FL.33
Limitations
We cannot formally exclude that clonal hematopoiesis instead of tumor-intrinsic somatic mutations was the source of the low-level TP53-mutations seen in 25% of the patients. Were the TP53 mutations due to infiltrating myeloid cells, one might suggest that the rapid progression seen in TP53-mutated subjects treated with RIT-CHOP was due to the emergence of a myelodysplasia. However, progression in this study was defined by lymphoma-specific findings. Furthermore, Bennett et al34 explicitly addressed this issue in S0016 that no increase in marrow pathology was detected in the RIT-CHOP arm, suggesting that the shortened PFS and overall survival associated with these low VAF TP53 mutations is not due to an effect of radiation on the marrow.
Although our methods address TP53 point mutations, the approach does not address alternative oncogenic mechanism, particularly deletion. Qu et al35 studied these same samples from S0016 and reported that high VAF loss of TP53 (at least 20%-30% of alleles lost) is associated with shortened PFS. Assessing whether such copy number losses are present at VAFs similar to 0.02 (2%) in FFPE specimens which would require technologies that are currently speculative.
We could not locate another phase 3 trial collection with which to independently test whether subclonal TP53 mutations represent a predictive marker for regimens including radiation. Our observation in this study depends on the decades-long stewardship of specimens and data, and the lack of a validation cohort highlights a critical limitation in trial programs. We hope that this missed opportunity stimulates funders and trialists to enhance support for the collection of trial-associated specimens.
Summary
Low VAF TP53 mutations are predictive of therapeutic inefficacy for patients with FL treated with CHOP-RIT. The relative efficacy of RIT-CHOP compared with R-CHOP is apparent in the 75% of patients who lack a detectable TP53 mutation. These observations call for enhanced precision medicine approaches through the use of deep sequencing in FL, as clinically relevant cohorts are defined not only by driver mutations but also by the mutations not found with a highly sensitive assay.
Acknowledgments
The authors recognize the late Oliver Press for his contributions to SWOG and study S0016. John Spence and Phil Rock provided essential computational and technical support. C. Ross (University of Michigan) supported efforts to locate additional specimens.
This work was supported by National Institutes of Health/National Cancer Institute grants U10CA180888, U10CA180819, U10CA180821, R21CA198072 (W.R.B.), and in part by GlaxoSmithKline.
Authorship
Contribution: W.R.B. designed and performed research, analyzed data, and wrote the manuscript; H.L. and M.L.L. designed research and analyzed data; D.A. designed and performed research, analyzed data, and wrote the manuscript; J.M.S. designed and performed research, and analyzed data; C.M.S. and L.M.R. performed research; S.M.S., M.S., J.P.L., and M.S.K. conducted the trial; and J.W.F. conducted the trial and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: W. Richard Burack, Department of Medicine, University of Rochester, Pathology and Laboratory Medicine, 601 Elmwood Ave, Box 626, Rochester, NY 14642; e-mail: richard_burack@urmc.rochester.edu.
References
Author notes
Data are available on request from the corresponding author, W. Richard Burack (richard_burack@urmc.rochester.edu).
The full-text version of this article contains a data supplement.

![TP53 mutations are evanescent and their VAF does not overtly increase with relapse. Out of 22 patients with serial specimens, 11 patients had at least 1 specimen with a pathogenic mutation. The VAFs for these mutations are shown in Figure 1B FL validation. To estimate changes in tumor cell content, we determined the sum of VAFs for mutations in a single amplicon of uBCL2 (solid black lines with orange blocks); this provides an estimate of the minimum tumor content. The TP53 mutations are depicted on the thick gray lines (red blocks mark the VAF; blue blocks denote that the mutation was not detected at that time point [for TP53] or only germ line sequence for uBCL2).](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/7/17/10.1182_bloodadvances.2022009467/2/m_blooda_adv-2022-009467-gr4.jpeg?Expires=1769105362&Signature=kO-1aqq52GhDwScpc0PyYsyfzEzqmhU6B2MNAmm-8tRD2BbV~0vOmHOEhgFMfTBP3YL4WXZK4kU0X2C2UUG414Nlj-pdc~ZP258ul~LPv0yu9dmHNbc7JmlTEspHlh~-ti788DGfVLBT~M1M~ckaZ4EGBtTB6HMKoW44ijRkP6LLCMhMXwfmInA~RHWn4K0j7t84Hzib1McHcAAMA2duL~toAlmPB~rflGK1Qv8hRca75dgYBFqb4HIs4qRrtkE99A0oxtLJsoAmSo63k4pZ7oqCuxovv7FLowRzh4PhgPukyxjppHEshf4CS6DPZ2wKkO-piZbxaPCVQugif0fI~Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)