Key Points
mcfDNA is increased during neutropenia after allo-HCT.
Studying the taxa-of-origin of this circulating DNA may provide novel insights into post-HCT biology.
Abstract
We used a next-generation sequencing platform to characterize microbial cell–free DNA (mcfDNA) in plasma samples from patients undergoing allogeneic hematopoietic stem cell transplantation (allo-HCT). In this observational study, we sought to characterize plasma mcfDNA in order to explore its potential association with the immunologic complications of transplantation. We compared serially collected patient samples with plasma collected from healthy control subjects. We observed changes in total mcfDNA burden in the plasma after transplantation, which was most striking during the early posttransplant neutropenic phase. This elevation could be attributed to a number of specific bacterial taxa, including Veillonella, Bacteroides, and Prevotella (genus level). For an additional cohort of patients, we compared the data of mcfDNA from plasma with 16s-ribosomal RNA sequencing data from stool samples collected at matched time points. In a number of patients, we confirmed that mcfDNA derived from specific microbial taxa (eg, Enterococcus) could also be observed in the matched stool sample. Quantification of mcfDNA may generate novel insights into mechanisms by which the intestinal microbiome influences systemic cell populations and, thus, has been associated with outcomes for patients with cancer.
Introduction
Hematopoietic stem cell transplantation (allo-HCT) is a curative-intent therapy for patients with hematological malignancies. Allo-HCT involves preconditioning with chemotherapy (and/or radiation) and the infusion of a stem cell graft from a carefully selected donor. Conditioning regimens are typically of sufficient toxicity to damage the mucosa of the intestinal tract. This damage peaks during the period of posttransplant neutropenia, which occurs because of ablation of host hematopoiesis and the lag-time in the engraftment of donor cells. In addition to restoring the hematopoietic compartment, the graft contributes a curative graft-versus-leukemia effect. This desirable immunologic effect is challenging to separate from graft-versus-host disease (GVHD), a systemic inflammatory syndrome predominantly affecting the skin, liver, and gastrointestinal tract. Mouse models have facilitated the detailed study of GVHD pathogenesis, and the translocation of bacterial products (eg, the bacterial cell-wall product lipopolysaccharide) is thought to be a critical component of GVHD initiation.1
The bacterial communities residing in the intestinal tract have been of interest in the field of allo-HCT for many years. Recent advances in sequencing technology have facilitated deep exploration of the intestinal microbiome, and we now know that allo-HCT is accompanied by marked changes in the diversity and composition of intestinal microbial communities.2 It is also clear that there are a number of specific microbial taxa in the gut that are associated with the complications of transplantation, including infection, GVHD, relapse and poor immune recovery.3-8 Translocation of viable intestinal bacteria into the systemic circulation is thought to be a key mechanism for bloodstream infections that occur in the setting of mucosal barrier injury.9,10 In addition to being a reservoir of potential pathogens, the intestinal microbiome is a rich source of potential immunomodulators (eg, bacterial antigens and metabolites). Given the associations between the microbiome and transplant complications, many of which do not have a gut-associated pathogenesis, it is reasonable to speculate that these immunomodulators may act in the periphery as well as within the gut lumen. Using highly sensitive sequencing technology it is now possible to measure circulating microbial cell–free DNA (mcfDNA), which may allow us to identify specific microbiome-derived molecules that interact with immune cells in the periphery.
Methods
Patient selection and clinical data
We included all patients treated at Memorial Sloan Kettering Cancer Center who received an unmodified peripheral blood stem cell allograft and who had biobanked samples collected during their neutropenic nadir and at recovery (demographics shown in Table 1; GVHD described in supplemental Table 1). For 18 additional patients, we could expand the analysis and include a closely matched stool and an additional pretransplant blood sample. The demographics of the subcohort of patients with stool samples are described in supplemental Table 2, and the time points for sample collection in supplemental Table 3.
Patient characteristics
| Characteristic . | N (%) . |
|---|---|
| n | 70 (100) |
| age (mean [SD]) | 56.80 (13.99) |
| Gender | |
| Female | 28 (40.0) |
| Male | 42 (60.0) |
| Disease | |
| Leukemia | 33 (47.1) |
| Myelodysplastic syndrome | 13 (18.6) |
| Myeloproliferative disorder | 4 (5.8) |
| Non-Hodgkin lymphoma | 15 (21.4) |
| Other | 5 (7) |
| Conditioning intensity | |
| Myeloablative | 23 (32.9) |
| Reduced intensity | 42 (60.0) |
| Nonablative | 5 (7.1) |
| Conditioning regimen∗ | |
| Fludarabine/melphalan | 36 (51.4) |
| Busulfan/fludarabine | 14 (20.0) |
| Fludarabine/melphalan/thiotepa | 5 (7.1) |
| Other (containing TBI) | 12 (16.8) |
| Other (TBI-free) | 3 (4.2) |
| Degree of HLA match | |
| Matched unrelated | 42 (60.0) |
| Matched related | 16 (22.8) |
| Mismatched/haplo-related | 7 (10) |
| Mismatched unrelated | 5 (7.1) |
| Acute GVHD (before day 100) | |
| No | 30 (42.9) |
| Yes | 40 (57.1) |
| Characteristic . | N (%) . |
|---|---|
| n | 70 (100) |
| age (mean [SD]) | 56.80 (13.99) |
| Gender | |
| Female | 28 (40.0) |
| Male | 42 (60.0) |
| Disease | |
| Leukemia | 33 (47.1) |
| Myelodysplastic syndrome | 13 (18.6) |
| Myeloproliferative disorder | 4 (5.8) |
| Non-Hodgkin lymphoma | 15 (21.4) |
| Other | 5 (7) |
| Conditioning intensity | |
| Myeloablative | 23 (32.9) |
| Reduced intensity | 42 (60.0) |
| Nonablative | 5 (7.1) |
| Conditioning regimen∗ | |
| Fludarabine/melphalan | 36 (51.4) |
| Busulfan/fludarabine | 14 (20.0) |
| Fludarabine/melphalan/thiotepa | 5 (7.1) |
| Other (containing TBI) | 12 (16.8) |
| Other (TBI-free) | 3 (4.2) |
| Degree of HLA match | |
| Matched unrelated | 42 (60.0) |
| Matched related | 16 (22.8) |
| Mismatched/haplo-related | 7 (10) |
| Mismatched unrelated | 5 (7.1) |
| Acute GVHD (before day 100) | |
| No | 30 (42.9) |
| Yes | 40 (57.1) |
Haplo, haploidentical; SD, standard deviation; TBI, total body irradiation.
No patients received antithymocyte globulin as in vivo T-cell depletion.
Samples from healthy volunteers
Plasma samples were collected from healthy donors from 5 geographically diverse areas of the United States (age, 18-65 years, screened for common health conditions, including infectious diseases, through a questionnaire and standard blood donor screening assays; Serologix and StemExpress). All samples (from patients and healthy volunteers) were obtained with written informed consent under institutional review board–approved protocols.
Stool and plasma sample sequencing
The complete cfDNA sequencing was performed using plasma samples as previously described and as outlined in the supplemental Methods.11,12 Sequencing of the 16s ribosomal RNA (rRNA) gene from V4 to V5 variable region was performed using stool samples processed as previously described2-4 and also outlined in supplemental Methods.
Statistical analysis
Statistical tests were performed using GraphPad Prism. For Figure 1B, paired Wilcoxon testing was used. For other unpaired data, Mann-Whitney nonparametric testing was used.
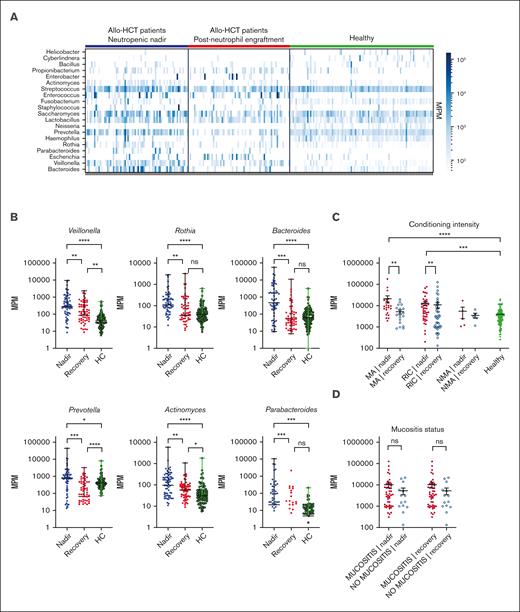
Total MPM and a number of specific taxa are elevated in samples from patients after transplantation. (A) Heatmap of bacterial mcfDNA mapped to genus-level bacterial taxonomy in patient plasma samples (n = 71 patients with 142 samples; 99 healthy control subjects with 99 unique samples). Each column is a plasma sample; each row is a genus. For this analysis, taxa shown are those present in >10 samples with >200 MPM and aggregated at the genus level. (B) Quantification of top differentially abundant taxa (genus). Each point represents a blood sample. (C) Correlation of total MPM with conditioning intensity. (D) Correlation of total MPM with the presence of oropharyngeal mucositis (no comparisons were statistically significant). Nonparametric (Wilcoxon) testing was used throughout (∗∗∗∗P <.0001; ∗∗∗P 0.0001-0.001; ∗∗P = .001-0.01; ∗P = .05-0.01). HC, healthy control; ns, not significant.
Total MPM and a number of specific taxa are elevated in samples from patients after transplantation. (A) Heatmap of bacterial mcfDNA mapped to genus-level bacterial taxonomy in patient plasma samples (n = 71 patients with 142 samples; 99 healthy control subjects with 99 unique samples). Each column is a plasma sample; each row is a genus. For this analysis, taxa shown are those present in >10 samples with >200 MPM and aggregated at the genus level. (B) Quantification of top differentially abundant taxa (genus). Each point represents a blood sample. (C) Correlation of total MPM with conditioning intensity. (D) Correlation of total MPM with the presence of oropharyngeal mucositis (no comparisons were statistically significant). Nonparametric (Wilcoxon) testing was used throughout (∗∗∗∗P <.0001; ∗∗∗P 0.0001-0.001; ∗∗P = .001-0.01; ∗P = .05-0.01). HC, healthy control; ns, not significant.
Results
We assembled a cohort of allo-HCT recipients in order to measure the circulating mcfDNA during the neutropenic period and neutrophil engraftment. We studied 142 unique blood samples from 71 patients (Table 1) and analyzed samples from 99 healthy volunteers. The sequencing assay used in this study (the commercially available Karius Test) facilitates the quantification of microbial DNA molecules (molecules per microliter [MPM]). In clinical practice, the Karius Test is used as an adjunct diagnostic for patients in whom infection is clinically suspected, but standard evaluations fail to identify a causative organism.13,14 Patients with a microbiologically confirmed bloodstream infection were excluded from this cohort, because we specifically sought to explore the mcfDNA profiles that correlated with noninfectious transplant outcomes.
We identified a number of bacterial mcfDNA signatures characteristic of the neutropenic state in patients, in comparison with the same patients after neutrophil recovery and with that of the independent healthy control cohort (Figure 1A). When we quantified the absolute number of molecules attributable to each genus, we found the highest relative increases in the numbers of Parabacteroides (8.4-fold), Bacteroides (7.9-fold), and Prevotella (7.2-fold) during the neutropenic phase compared with that during the postrecovery time point (Figure 1B). Smaller increases were seen in the numbers of Veillonella (3.2-fold), Actinomyces (3.0-fold), and Rothia (1.9-fold; Figure 1B). This may suggest that bacteria, or DNA, from these genera can preferentially translocate into the circulation during periods of impaired barrier function without corresponding to a clinically or microbiologically confirmed infection. None of these taxa are considered common pathogens, but they have been correlated with clinical outcomes in prior studies of the intestinal microbiome after allo-HCT.4,6,15 High relative abundance of the Prevotella genus has been reported in a number of studies on patients with autoimmune conditions and associated with low abundance of immunoregulatory bacteria.16 In contrast, the Bacteroides genus includes a number of species that are thought to induce regulatory T cells, a cell population linked with tolerance after allo-HCT.17,18Veillonella is a member of the core oral microbiota and has been previously linked to a higher risk of GVHD-related mortality.6
To deepen our understanding of the clinical factors linked with mcfDNA burden after transplant, we analyzed patients according to the intensity of the conditioning regimen and examined the quantitative differences in mcfDNA at both neutropenic nadir and after recovery. Patients who receive myeloablative (MA) chemotherapy experienced the most severe transplant-related mucosal damage. In the patients who received reduced-intensity conditioning (RIC), regimens resulted in less toxicity; in the non-MA group, conditioning-related toxicity was minimal. In recipients of MA conditioning and RIC transplants, there was a statistically significant increase in the mcfDNA concentration (MPM) during neutropenia compared with that in samples from these same patients after engraftment and compared with that in samples from healthy controls. No statistically significant difference was seen between the mcfDNA burden during the neutropenic phase for the patients who received MA conditioning and RIC transplants (Figure 1C). In the small number of recipients of non-MA transplant (n = 5), we did not observe any increase in mcfDNA above that in healthy controls. Thus, focusing on the MA conditioning and RIC recipients, we next questioned whether mcfDNA burden was associated with clinically evident oral mucositis during the neutropenic period after transplant. Given that many of the taxa observed in the samples are typically considered oral flora, we hypothesized that severe oral mucositis (requiring opiate analgesia) would be associated with higher MPM of mcfDNA. In contrast to our expectations, we did not observe a higher MPM of mcfDNA in patients with more severe symptomatic mucosal barrier damage (Figure 1D). This might have been due to our small cohort size, or the difference in timing of sample collection (ie, not all samples were collected during the peak of clinical mucositis). Furthermore, we did not see a relationship between mcfDNA and those with exposure to antibiotics , those with diarrhea, or with specific exposure to total body irradiation or melphalan (supplemental Figures 1 and 2).
To explore the contribution of the lower gastrointestinal tract microbiota to mcfDNA, we identified a subset of patients from whom stool and blood samples had been collected at similar time points (supplemental Tables 2 and 3). We analyzed plasma samples to quantify bacterial mcfDNA in circulation and stool samples to establish the relative abundance of genus-specific rRNA (using 16s rRNA amplicon–based sequencing). We defined “shared MPM” as bacterial mcfDNA in blood that mapped to genus-level sequences that were also present in stool. As shown in Figure 2A, the number of taxa measured in plasma that can be mapped to microbial taxa in the intestinal tract increased significantly during neutropenic nadir and remained elevated into the recovery phase (nadir: 25.2-fold above the pretransplant baseline; neutrophil recovery: 11.12-fold).
Plasma mcfDNA molecules can be mapped to taxa found in matched stool samples. (A) Quantified MPM of taxa that were identified using both plasma mcfDNA sequencing and 16s rRNA sequencing of matching stool samples (n = 18 unique patients). Each point is a blood sample; the vertical axis plots the aggregated MPM in plasma, at the genus level, that were also observed in the paired fecal sample. (B) A single patient time course, in a recipient of a reduced-intensity (fludarabine/melphalan) allograft, who developed acute GVHD on day 21. In addition to the chemotherapy and antibiotic therapy shown in the top panel, the patient received acyclovir as antiviral prophylaxis and micafungin (until day 7), followed by voriconazole, consistent with institutional antifungal prophylaxis practice. Red squares represent blood sampling points. IV, intravenous; PO, per os.
Plasma mcfDNA molecules can be mapped to taxa found in matched stool samples. (A) Quantified MPM of taxa that were identified using both plasma mcfDNA sequencing and 16s rRNA sequencing of matching stool samples (n = 18 unique patients). Each point is a blood sample; the vertical axis plots the aggregated MPM in plasma, at the genus level, that were also observed in the paired fecal sample. (B) A single patient time course, in a recipient of a reduced-intensity (fludarabine/melphalan) allograft, who developed acute GVHD on day 21. In addition to the chemotherapy and antibiotic therapy shown in the top panel, the patient received acyclovir as antiviral prophylaxis and micafungin (until day 7), followed by voriconazole, consistent with institutional antifungal prophylaxis practice. Red squares represent blood sampling points. IV, intravenous; PO, per os.
With a total cohort size of 18 patients, further analysis of clinical contributors to high mcfDNA burden is challenging. However, we were intrigued to note that there was significant heterogeneity in the postneutropenic nadir mcfDNA burden, with that in some patients returning to zero or baseline and in others continuing to harbor mcfDNA concentrations comparable with those seen during neutropenic nadir (Figure 2A; supplemental Figure 3). As patient numbers are insufficient for robust conclusions, we selected 1 patient in whom a high bacterial mcfDNA burden was maintained into the recovery phase and explored their clinical course in detail. This patient, subject 2, received a reduced-intensity allograft and developed severe acute GVHD involving the gastrointestinal tract (time course shown in Figure 2B). During neutropenic nadir, we observed a significant increase in the shared microbial taxa, with Streptococcus and Enterococcus present at a high relative abundance in both the stool and plasma. On day 20, the patient was readmitted to the hospital for investigation of gastrointestinal symptoms, and a histopathological diagnosis of gastrointestinal GVHD was made on day 21. As highlighted in Table 2, several bacterial taxa were identified in both the plasma and stool samples. This case study highlights that mcfDNA may have the potential to predict intestinal GVHD before the onset of clinical symptoms.
Comparison of taxa present in the blood and stool of subject 2
| Timepoint . | Genus . | Plasma relative abundance (fraction) . | Stool relative abundance (fraction) . |
|---|---|---|---|
| Before transplantation | Streptococcus | (0.318) | (0.0785) |
| Neutropenic nadir | Streptococcus | (0.0625) | (0.0195) |
| Enterococcus | (0.0176) | (0.423) | |
| GVHD onset | Streptococcus | (0.206) | (0.0410) |
| Lachnoclostridium | (0.0517) | (0.023) | |
| Blautia | (0.0491) | (0.153) | |
| Enterococcus | (0.0487) | (0.346) | |
| Clostridium | (0.0453) | (0.00501) | |
| Actinomyces | (0.0147) | (0.0205) | |
| Lactobacillus | (0.00736) | (0.00874) | |
| Akkermansia | (0.00534) | (0.0168) |
| Timepoint . | Genus . | Plasma relative abundance (fraction) . | Stool relative abundance (fraction) . |
|---|---|---|---|
| Before transplantation | Streptococcus | (0.318) | (0.0785) |
| Neutropenic nadir | Streptococcus | (0.0625) | (0.0195) |
| Enterococcus | (0.0176) | (0.423) | |
| GVHD onset | Streptococcus | (0.206) | (0.0410) |
| Lachnoclostridium | (0.0517) | (0.023) | |
| Blautia | (0.0491) | (0.153) | |
| Enterococcus | (0.0487) | (0.346) | |
| Clostridium | (0.0453) | (0.00501) | |
| Actinomyces | (0.0147) | (0.0205) | |
| Lactobacillus | (0.00736) | (0.00874) | |
| Akkermansia | (0.00534) | (0.0168) |
Discussion
In summary, in this study we report that plasma mcfDNA is measurable, undergoes dynamic change dictated by the phase of transplantation, and appreciably differs from that seen in healthy people. Therefore, mcfDNA represents a potentially useful source of information regarding the composition of the intestinal microbiome, and a functional measure of the integrity of the gut barrier. Further prospective studies of mcfDNA in the population of patients receiving allo-HCT will facilitate a deeper understanding of mcfDNA as a potential biomarker and may uncover new biological interactions between the microbiome and the human host.
Acknowledgments
M.R.M.v.d.B. was supported by the following NCI awards: MSKCC Cancer Center Core Grants P30-CA008748, R01-CA228358, R01-CA228308, and P01-CA023766; National Heart, Lung, and Blood Institute award R01-HL125571 and R01-HL123340; NIA National Institute on Aging award Project 2 of P01-AG052359; National Institute of Allergy and Infectious Diseases award U01 AI124275; Tri-Institutional Stem Cell Initiative award 2016-013; The Lymphoma Foundation; The Susan and Peter Solomon Divisional Genomics Program; and the Parker Institute for Cancer Immunotherapy at Memorial Sloan Kettering Cancer Center. J.U.P. reports funding from National Heart, Lung, and Blood Institute National Institutes of Health Award K08HL143189. K.A.M. acknowledges funding from the Parker Institute for Cancer Immunotherapy, DKMS, The American Australian Association, The Haematology Society of Australia and New Zealand, the Royal Australasian College of Physicians, and the American Society of Hematology.
Authorship
Contribution: L.B. designed and performed computational analyses; J.A.-Z. performed computational analyses; O.C.S. and A.E.W. contributed to writing the manuscript and advised on data analysis strategies; Z.A.K., P.G., and J.S. collected clinical data and oversaw clinical sampling; S.B. contributed to study design and the manuscript; M.-A.P., Y.T., and M.R.M.v.d.B. contributed to data analysis and study design; J.U.P. contributed to study design and edited the manuscript; and K.A.M. designed the study, analyzed data, prepared figures, and wrote the manuscript.
Conflict-of-interest disclosure: M.-A.P. reports honoraria from AbbVie, Astellas, Bristol Myers Squibb, Celgene, Equilium, Incyte, Karyopharm, Kite/Gilead, Merck, Miltenyi Biotec, MorphoSys, Novartis, Nektar Therapeutics, Omeros, Takeda, and VectivBio AG, and Vor Biopharma; serves on data safety monitoring boards of Cidara Therapeutics, Medigene, Sellas Life Sciences, and Servier and the scientific advisory board of NexImmune; has ownership interests in NexImmune and Omeros; has received research support for clinical trials from Incyte, Kite/Gilead, Miltenyi Biotec, and Novartis; and serves in a volunteer capacity as a member of the Board of Directors of the American Society for Transplantation and Cellular Therapy and Be The Match (National Marrow Donor Program) as well as on the CIBMTR Cellular Immunotherapy Data Resource Executive Committee. M.R.M.v.d.B. has received research support and stock options from Seres Therapeutics and stock options from Notch Therapeutics and Pluto Therapeutics; has received royalties from Wolters Kluwer; has consulted, received honorarium from or participated in advisory boards for Seres Therapeutics, Vor Biopharma, Rheos Medicines, Frazier Healthcare Partners, Nektar Therapeutics, Notch Therapeutics, Ceramedix, Lygenesis, Pluto Therapeutics, GlaxoSmithKline, Da Volterra, Thymofox, Garuda, Novartis (spouse), Synthekine (spouse), BeiGene (spouse), and Kite (spouse); has intellectual property licensing with Seres Therapeutics and Juno Therapeutics; and holds a fiduciary role in the foundation board of DKMS (a nonprofit organization). J.U.P. reports research funding, intellectual property fees, and travel reimbursement from Seres Therapeutics, and consulting fees from DaVolterra, CSL Behring, and from MaaT Pharma; serves on an advisory board of and holds equity in Postbiotics Plus Research; and has filed intellectual property applications related to the microbiome (reference numbers #62/843 849, #62/977 908, and #15/756 845). Memorial Sloan Kettering Cancer Center has financial interests relative to Seres Therapeutics. K.A.M. serves on the advisory board and holds equity in Postbiotics Plus Research, and she has consulted for Incyte. The remaining authors declare no competing financial interests.
Correspondence: Kate A. Markey, Translational Science and Therapeutics Division, Fred Hutchinson Cancer Center, 1100 Fairview Ave N, Mailstop S3-204, Seattle, WA 98109; e-mail: kmarkey@fredhutch.org.
References
Author notes
Sequencing data files of stool samples have been deposited in BioProject (accession numbers are listed in the supplemental Material).
Plasma sequencing of mcfDNA was performed using the Karius Test, a proprietary laboratory-developed assay; therefore, the raw sequencing data are subject to intellectual property restrictions.
The full-text version of this article contains a data supplement.