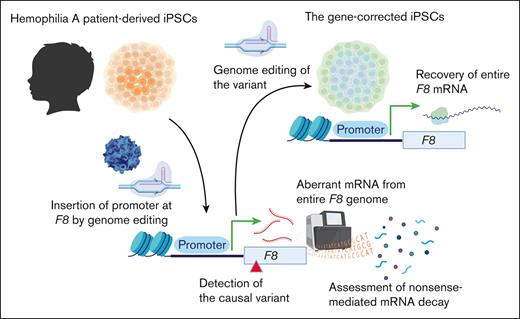
Key Points
Pathogenic deep intron variants in hemophilia A can be identified from patient iPSCs via genome editing.
Our approach is potentially useful to verify the therapeutic effect of personalized genome editing.
Abstract
The importance of genetic diagnosis for patients with hemophilia has been recently demonstrated. However, the pathological variant cannot be identified in some patients. Here, we aimed to identify the pathogenic intronic variant causing hemophilia A using induced pluripotent stem cells (iPSCs) from patients and genome editing. We analyzed siblings with moderate hemophilia A and without abnormalities in the F8 exon. Next-generation sequencing of the entire F8 revealed 23 common intron variants. Variant effect predictor software indicated that the deep intronic variant at c.5220-8563A>G (intron 14) might act as a splicing acceptor. We developed iPSCs from patients and used genome editing to insert the elongation factor 1α promoter to express F8 messenger RNA (mRNA). Then, we confirmed the existence of abnormal F8 mRNA derived from aberrant splicing, resulting in a premature terminal codon as well as a significant reduction in F8 mRNA in iPSCs due to nonsense-mediated RNA decay. Gene repair by genome editing recovered whole F8 mRNA expression. Introduction of the intron variant into human B-domain–deleted F8 complementary DNA suppressed factor VIII (FVIII) activity and produced abnormal FVIII lacking the light chain in HEK293 cells. Furthermore, genome editing of the intron variant restored FVIII production. In summary, we have directly proven that the deep intronic variant in F8 results in aberrant splicing, leading to abnormal mRNA and nonsense-mediated RNA decay. Additionally, genome editing targeting the variant restored F8 mRNA and FVIII production. Our approach could be useful not only for identifying causal variants but also for verifying the therapeutic effect of personalized genome editing.
Introduction
Hemophilia is an inherited bleeding disorder caused by a genetic abnormality of blood coagulation factor VIII (FVIII) or IX. The main treatment for hemophilia has long focused on protein replacement therapy, which replaces the missing coagulation factor in the blood with a coagulation factor preparation. Very recently, gene therapies using adeno-associated virus (AAV) vectors for hemophilia have been approved by the European Medicines Agency and the US Food and Drug Administration.1,2 Current gene therapy is essentially gene replacement therapy, in which a functional coagulation factor gene is delivered to liver cells by a vector.3 On the other hand, genome editing therapies that directly target chromosomal DNA are expected in the near future as a curative treatment for hemophilia and other genetic diseases.4,5 Among them, techniques that specifically normalize a patient's genetic variant, including base editing or prime editing, are considered the ultimate personalized therapy for genetic diseases.6 We succeeded in repairing a single nucleotide variant in human induced pluripotent stem cells (iPSCs) derived from a patient with severe hemophilia B using a base-editing approach.7 It is essential to reliably identify the disease-causing variant to achieve gene repair in individual cases.
In contrast, in some patients with hemophilia, the disease-causing variant cannot be identified by conventional genetic analysis; identification is especially difficult in patients with mild and moderate hemophilia A. The Canadian National Program for Hemophilia Variant Testing reported that the variant could not be identified in 103 of 1177 patients with hemophilia A.8 Recent advances in next-generation sequencing (NGS) have allowed for easy identification of intronic variants,9 and it is well known that deep intronic variations may be etiological in some patients with hemophilia. Although no exonic variant could be found in 10 of 49 Taiwanese patients with mild hemophilia A, a deep intronic variant in F8 intron 18 leading to alternative splicing might be a hot spot causal variant in these cases.10 It was reported that a deletion of 13 bases in intron 13 of F8 occurs in 6% of French mild hemophilia A cases.11 In addition, there are several reports showing that F8 deep intronic variations are assessed as likely pathogenic according to the American College of Medical Genetics and Genomics (ACMG) criteria.12-16 The functional impact of these variations was supported by either ectopic F8 messenger RNA (mRNA) analysis of white blood cells or a minigene assay. However, these analyses were unable to clarify how the actual expression of the entire F8 gene is regulated in patients.
In this study, we analyzed sibling cases of moderate hemophilia A without etiologic variants in exons. Whole F8 analysis by NGS identified several common variants in the intron sequence, and variant effect predictor (VEP) software implicated the possibility that a variant at c.5220-8563A>G (intron 14) acted as a splicing acceptor. The intronic variant has already been identified as a causal variant in the previous literature, because aberrant ectopic F8 transcript fragments have been observed in white blood cells.14 However, this finding was considered inconclusive because aberrant splicing was also observed in a reconstitution experiment with a wild-type minigene.14 Here, we established patient-derived iPSCs and proved the existence of 2 mechanisms for the disease by intronic variation, that is, aberrant splicing and resultant nonsense-mediated RNA decay. Furthermore, genome editing of the variant repaired full-length F8 expression in patient-derived iPSCs and FVIII production in HEK293 cells harboring F8 complementary DNA (cDNA) with the intronic variant.
Materials and methods
Patients
The experiments were approved by the ethical committee of Jichi Medical University (approval number 18-18) and Tokyo Medical University (approval number SH3710), and written informed consent was obtained from the participants.
Sequencing analysis
Genomic DNA and RNA were extracted using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) and RNeasy Mini Kit (Qiagen), respectively. cDNA was synthesized using the PrimeScript real-time polymerase chain reaction (PCR) Kit (Takara Bio, Shiga, Japan).
NGS against the entire F8 genomic locus was amplified in 14 overlapping regions (5-23 kb) by long-range PCR using KOD FX Neo DNA polymerase (Toyobo, Osaka, Japan), as described previously.9 The DNA library was prepared by fragmentation using a Nextera XT DNA sample preparation kit (Illumina, San Diego, CA). The paired-end adapter-ligated fragments of the pooled libraries were sequenced using Illumina MiSeq (Illumina). The obtained sequences were aligned to the GRCh37/hg19 coordinates of an F8 reference sequence (ENSG000000185010) using the Burrows-Wheeler Aligner. The variants were detected using the Genome Analysis Toolkit and annotated by the VariantStudio software program (Illumina).
Long-read RNA-seq analysis (Iso-seq) using PacBio Sequel II (Pacific Bioscience, Menlo Park, CA) was performed, and the data were analyzed by Takara Bio. The analysis data at the F8 genomic region were visualized using integrative genomic viewer software (Broad Institute, Cambridge, MA).
When indicated, DNA and mRNA sequences (cDNA) were confirmed by Sanger sequencing at Eurofins Genomics Japan (Tokyo, Japan). Primer sequences are described in supplemental Table 2.
PCR
PCR was performed using Ex Premier DNA Polymerase (Takara Bio). The PCR products were analyzed by microchip gel electrophoresis (MultiNA; Shimazu, Kyoto, Japan). When indicated, quantitative real-time PCR was performed using Thunderbird Probe quantitative PCR Mix (Toyobo) or Thunderbird SYBR qPCR Mix (Toyobo) on a QuantStudio 12K Flex (Thermo Fisher Scientific, Waltham, MA). Reactions were analyzed in triplicates, and expression levels were normalized to GAPDH mRNA levels. Predesigned primer and probe sets were used for the experiments to determine F8 expression levels (TaqMan Gene Expression Assays, Thermo Fisher Scientific, F8: Hs00252034_m1, GAPDH: Hs02758991_g1). The other primer sequences are described in supplemental Table 2.
Human iPSC generation and culture
iPSCs were generated from peripheral blood mononuclear cells (PBMCs), as previously described.7 PBMCs were isolated using Lymphocyte Separation Solution (Nacalai Tesque, Kyoto, Japan) and cultured in StemFitAK02N (Ajinomoto Healthy Supply, Tokyo, Japan) without supplement C containing 50 ng/mL human interleukin-6 (BioLegend, San Diego, CA), 100 ng/mL human stem cell factor (BioLegend), 10 ng/mL human thrombopoietin (BioLegend), and 100 ng/mL human Fms-related tyrosine kinase ligand (BioLegend) for 3 days. The cells were then transduced with Sendai virus vectors expressing reprogramming genes (CytoTune-iPS 2.0, ID Pharma, Tokyo, Japan). Four days after transduction, the cells were harvested and seeded on plates coated with iMatrix-511 (Matrixome, Osaka, Japan) and cultured in StemFitAK02N with all supplements. The medium was changed every other day until colonies appeared. Established iPSCs were maintained in StemFitAK02N on plates coated with Matrigel or iMatrix-511. To inhibit nonsense-mediated mRNA decay, semiconfluent iPSCs were cultured in culture medium containing 100 μM cycloheximide (FUJIFILM Wako, Osaka, Japan) for 24 hours.
Plasmid construction and AAV vector preparation
The fragment of mini-intron 14 with the patient’s variant (c.5220-8563A>G) was synthesized by Eurofins Genomics Japan (Tokyo, Japan). The wild-type sequence was generated by mutagenesis. The fragment was introduced between exon 14 and exon 15 of pcDNA3 expressing B-domain–deleted (BDD) F8 cDNA17 by the In-Fusion HD Cloning Kit (Takara Bio). The cDNAs of SpCas9 or SpCas9-NG18 were kindly provided by Nishimasu and Nureki (The University of Tokyo) and introduced into the pcDNA3 vector.
To insert the human elongation factor 1α (EF1α) promoter just upstream of the transcriptional start site of F8 by homology-directed repair (HDR), we generated a donor AAV vector (serotype 3B) harboring the EF1α promoter with homology arms. We introduced the EF1α promoter sequence derived from pBApo-EF1α Neo (Takara Bio) with a homology arm obtained by PCR between inverted terminal repeats of the plasmid AAV (pAAV). The AAV3B vector was produced by transfection of AAVpro293 cells (Takara Bio) with the pAAV, pRep/Cap (3B), and pHelper (Takara Bio).19 The AAV vector was purified using the AAVpro Purification Kit (all serotypes; Takara Bio). The quantification of the AAV vector was measured by the quantification of the SV40 polyadenylation signal, as previously described.19
Genome editing of iPSCs
To insert the EF1α promoter just before the transcriptional start site of F8, iPSCs (2 × 104 cells) were seeded in a 24-well plate 1 day before transfection. SpCas9 ribonucleoprotein (RNP; 4 μM) was transfected into the cells using Lipofectamine Stem Transfection Reagent (Thermo Fisher Scientific), and the cells were cultured in StemFitAK02N with 10 μM Y-27632 (Fujifilm Wako) and 5 μM L-755507 (Sigma-Aldrich). The cells were further transduced with the AAV vector (5 × 104 viral genomes per cell) on the following day. To generate the gene-corrected iPSCs, cells were transfected with the P3 Primary Cell 4D-Nucleofector X Kit S (Lonza, Basel, Switzerland). Briefly, 5 × 105 cells were suspended in a nucleofector solution mixed with single-stranded oligodeoxynucleotides (4 μM) and SpCas9 RNPs (4 μM). Cell suspension mixtures were then transferred to cuvettes, and the nucleofector process was initiated using a 4D-Nucleofector Core Unit (Lonza). The cells were then seeded onto iMatrix-511–coated wells and cultured in StemFit-AK02N with 10 μM Y-27632 and 5 μM L-755507. After 2 passages, single-cell–derived colonies were isolated, and genomic DNA was assessed by Sanger sequencing.
Generation of HEK293 cells stably expressing F8 cDNA with mini-intron 14
HEK293 cells were maintained in Dulbecco modified Eagle medium (Sigma-Aldrich) containing 10% fetal bovine serum (Thermo Fisher Scientific), GlutaMAX supplement (Thermo Fisher Scientific), and penicillin-streptomycin solution (Fujifilm Wako). To generate HEK293 cells stably expressing F8 cDNA with mini-intron 14 derived without or with the patient variant (c.5220-8563A>G), HEK293 cells were transfected with the linearized plasmid using Lipofectamine 3000 Reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. After transfection, 400 μg/mL G418 (Nacalai Tesque) was added to the culture medium to obtain stably expressed clones.
Immunohistochemistry
The cells were fixed with 4% paraformaldehyde for 20 minutes, permeabilized with phosphate-buffered saline containing 0.3% triton X-100, and blocked with 2% goat serum for 1 hour. The cells were then incubated with sheep anti-human FVIII polyclonal antibody (50 μg/mL; Haematologic Technologies, Essex Junction, VT) overnight at 4°C. Samples were incubated with a goat anti-sheep immunoglobulin G (IgG) antibody conjugated to Alexa Fluor 594 (2 μg/mL; Thermo Fisher Scientific) for 1 hour at room temperature. Slides were mounted with Vectashield mounting medium containing 4′,6-diamidino-2-phenylindole (Vector Laboratories, Newark, CA) and observed and photographed using a confocal microscope (Leica TCS SP8; Leica Microsystems, Wetzlar, Germany).
FVIII activity and antigen
FVIII activity (FVIII:C) was measured with a 1-stage clotting time assay or chromogenic assay (Revohem FVIII, Sysmex, Kobe, Japan) using an automated coagulation analyzer (Sysmex CA-1600 analyzer; Sysmex). To measure FVIII antigen (FVIII:Ag), enzyme-linked immunosorbent assay was performed using the Factor VIII Paired Antibody Set (Affinity Biologicals, Ancaster, Canada). To detect FVIII containing light chain, an anti-FVIII light chain-specific monoclonal antibody (GMA-8020, Green Mountain Antibodies, Burlington, VT) was used as a detector antibody in an enzyme-linked immunosorbent assay. Antibody binding was detected with an anti-mouse IgG goat antibody conjugated with horseradish peroxidase (SeraCare Life Science, Milford, MA) using ABTS microwell peroxidase substrate (SeraCare Life Science).
Immunoblotting
The medium of the cells was changed to chemically defined serum-free medium (HE 1000; Gmep, Fukuoka, Japan) and cultured for the following 48 hours. The cell supernatant was collected after centrifugation, mixed with sodium dodecyl sulfate sample buffer containing 2-mercaptoethanol, and then boiled for 5 minutes. Proteins were resolved by 7.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and then transferred to a polyvinylidene difluoride membrane. Immunoblotting with the specific antibody was performed, as described previously.20 To detect FVIII protein in the supernatant, we used sheep anti-human FVIII polyclonal antibody (0.8 μg/mL; Haematologic Technologies). The bound antibody was detected with a peroxidase-labeled anti-sheep IgG antibody (0.1 μg/mL, SeraCare Life Science) and visualized with Immobilon Western Chemiluminescent (Merck Millipore, Burlington, MA) and an ImageQuant 800 digital imaging system (Cytiva, Tokyo, Japan).
T7 endonuclease assay
The PCR product was denatured and reannealed using a thermal cycler. Genomic alterations were detected using T7 endonuclease I (Nippon Gene Co, Tokyo, Japan) and analyzed by microchip gel electrophoresis (MultiNA).
Statistical analyses
Statistical analyses were performed using Prism 9 software (Graph-Pad Software, San Diego, CA). All data are presented as the mean ± standard deviation. Statistical significance was calculated using two-tailed Student t test or one-way analysis of variance with post hoc Tukey multiple comparison test. All P values are described in the figures.
Results
Common intronic variants in the moderate hemophilia A siblings
We first examined the F8 sequence in the 2 moderate hemophilia A siblings (FVIII:C, 1∼2%). We could not detect any disease-causing variants in the coding sequence in F8 exons. Entire F8 sequencing by NGS revealed 23 common intronic variants in both patients (supplemental Table 1). VEP software implicated the possibility that a variant at c.5220-8563A>G (intron 14) acted as a splicing acceptor (supplemental Table 1). This variant had a high Δ score of acceptor gain, strongly suggesting the possibility of a new splice acceptor. We carefully investigated the impact of each variant using several in silico prediction tools. Potentially damaging/deleterious variants were identified by the VEP. The prediction of aberrant splicing resulting from intron variants was assessed by SpliceAI (supplemental Table 1), MaxEntScan, and dbscSNV. However, no variants other than c.5220-8563A>G were suggested to explain the phenotype of the patient. Furthermore, all other variants were registered as single nucleotide polymorphisms in the dbSNP database. Therefore, we considered these variants unlikely to be etiologic due to their high allele frequency.
Identification of aberrant F8 mRNA in iPSCs derived from a patient with moderate hemophilia A
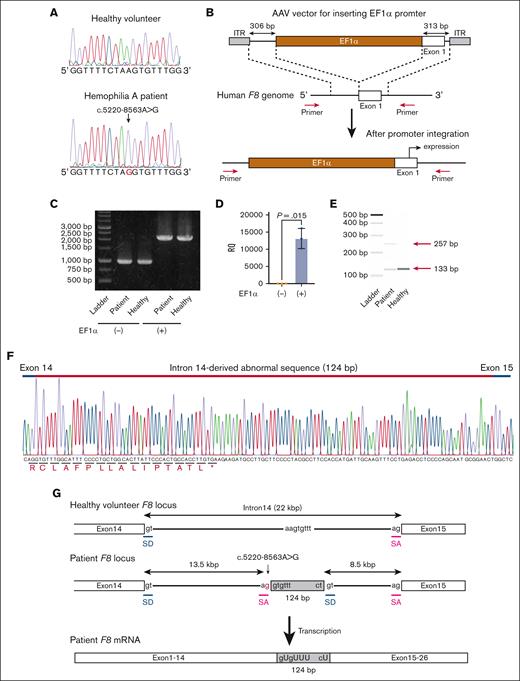
Next, we developed iPSCs from the PBMCs of the patient. We confirmed that the established iPSCs contained the same variant in intron 14 of F8 (Figure 1A). To express the full-length F8 mRNA from undifferentiated iPSCs, we inserted the EF1α promoter just before the transcriptional start site of F8 by genome editing. We created an AAV vector harboring the EF1α promoter with homology arms and transduced the iPSCs with SpCas9 RNP together with the AAV vector (Figure 1B). The promoter was predicted to be inserted into the target site by HDR. The transduced iPSCs were passaged by serial dilution in single-cell culture conditions, and the insertion of the EF1α promoter at F8 in cloned iPSCs was confirmed by PCR (Figure 1B-C; supplemental Figure 1). We confirmed the expression of F8 mRNA in undifferentiated iPSCs derived from healthy volunteers by insertion of the EF1α promoter (Figure 1D). We further amplified the mRNA sequence between exons 14 and 15 and found the existence of abnormal transcripts consisting of 257 bases in iPSCs derived from the patient (Figure 1E). We confirmed the insertion of a 124-base sequence derived from intron 14 (Figure 1F). These data suggested that the additional splice acceptor site caused by the patient variant (c.5220-8563G>A) induced aberrant splicing (Figure 1G).
The expression of aberrant F8 transcripts in patient-derived iPSCs by insertion of the EF1α promoter. (A) DNA sequencing (a part of F8 intron 14) of iPSCs derived from a healthy volunteer and the patient with hemophilia A. The red nucleotide represents the variant. (B-F) Healthy donor and patient-derived iPSCs were treated with the SpCas9 RNP, and the AAV vector harboring the EF1α promoter with homology arms, and then single clones were isolated. (B) Experimental scheme for inserting the EF1α promoter before the transcriptional start site of F8. The red arrows indicate the primers used to confirm the insertion. (C) The insertion of the EF1α promoter was confirmed by PCR in an isolated clone. (D) Relative F8 mRNA expression assessed by quantitative reverse transcription (RT) PCR in healthy volunteer–derived iPSCs without or with the insertion of the EF1α promoter. Values represent the mean ± standard deviation (n = 3). Statistical analysis was performed by two-tailed Student t test. (E-F) Identification of aberrant F8 transcripts between exons 14 and 15 by RT-PCR in patient-derived iPSCs with insertion of the EF1α promoter. (E) The transcript between exons 14 and 15 assessed by microchip gel electrophoresis. The red arrows indicate the size of the amplicons. (F) Sequencing of the aberrant F8 transcript between exons 14 and 15 from patient-derived iPSCs. Additional sequences (124 bp) were inserted between exons 14 and 15. (G) Hypothesis for insertion of the aberrant mRNA sequence by abnormal splicing in the patient. The variant (red) at intron 14 in the patient’s F8 genome may provide an abnormal splice acceptor site, leading to the insertion of 124 bp of the pseudoexon just after the variant. The gray boxes indicate the pseudoexon. ITR, inverted terminal repeat; RQ, relative quantification; SA, splicing acceptor; SD, splice donor.
The expression of aberrant F8 transcripts in patient-derived iPSCs by insertion of the EF1α promoter. (A) DNA sequencing (a part of F8 intron 14) of iPSCs derived from a healthy volunteer and the patient with hemophilia A. The red nucleotide represents the variant. (B-F) Healthy donor and patient-derived iPSCs were treated with the SpCas9 RNP, and the AAV vector harboring the EF1α promoter with homology arms, and then single clones were isolated. (B) Experimental scheme for inserting the EF1α promoter before the transcriptional start site of F8. The red arrows indicate the primers used to confirm the insertion. (C) The insertion of the EF1α promoter was confirmed by PCR in an isolated clone. (D) Relative F8 mRNA expression assessed by quantitative reverse transcription (RT) PCR in healthy volunteer–derived iPSCs without or with the insertion of the EF1α promoter. Values represent the mean ± standard deviation (n = 3). Statistical analysis was performed by two-tailed Student t test. (E-F) Identification of aberrant F8 transcripts between exons 14 and 15 by RT-PCR in patient-derived iPSCs with insertion of the EF1α promoter. (E) The transcript between exons 14 and 15 assessed by microchip gel electrophoresis. The red arrows indicate the size of the amplicons. (F) Sequencing of the aberrant F8 transcript between exons 14 and 15 from patient-derived iPSCs. Additional sequences (124 bp) were inserted between exons 14 and 15. (G) Hypothesis for insertion of the aberrant mRNA sequence by abnormal splicing in the patient. The variant (red) at intron 14 in the patient’s F8 genome may provide an abnormal splice acceptor site, leading to the insertion of 124 bp of the pseudoexon just after the variant. The gray boxes indicate the pseudoexon. ITR, inverted terminal repeat; RQ, relative quantification; SA, splicing acceptor; SD, splice donor.
Restoration of F8 mRNA by the correction of the intronic variant in patient-derived iPSCs
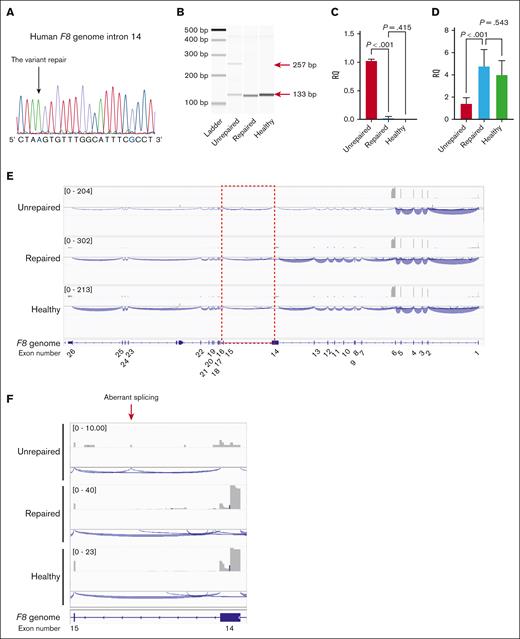
To directly demonstrate whether the variant caused abnormal splicing, we corrected the variant by genome editing. We transduced patient iPSCs expressing F8 mRNA with SpCas9 RNP together with single-stranded oligodeoxynucleotides for repair (supplemental Figure 2). We inserted the mutation at the protospacer adjacent motif (PAM) sequence (G>C) to prevent cleavage after repair (supplemental Figure 2). We selected gene-corrected patient iPSCs (Figure 2A) and confirmed the disappearance of abnormal splicing (Figure 2B-C). Moreover, the decrease in F8 mRNA in the patient-derived iPSCs was restored by gene correction (Figure 2D). The decrease in aberrant F8 transcripts in the patient-derived iPSCs was recovered by pretreatment with the nonsense-mediated mRNA decay inhibitor (supplemental Figure 3). Long-read RNA-seq analysis (Iso-seq) showed the disappearance of aberrant splicing and the recovery of whole F8 mRNA expression in the iPSCs after gene correction (Figure 2E-F).
Restoration of F8 mRNA expression by correction of the intronic variant in patient-derived iPSCs. The patient-derived iPSCs with the insertion of the EF1α promoter before the transcriptional start site of F8 were transduced with SpCas9 (RNP) and an 83-nucleotide single oligonucleotide to correct the variant; then a single clone was isolated. (A) Confirmation of the gene correction by DNA sequencing at the variant in the isolated clone. The color blue indicates the replaced nucleotides derived from the single oligonucleotide. (B) Disappearance of aberrant F8 transcripts between exons 14 and 15 by gene correction was assessed with RT-PCR. The red arrows indicate the size of the amplicons. (C-D) Relative expression of the aberrant transcript (C) and F8 mRNA between exons 25 and 26 (D). Values represent the mean ± standard deviation (n = 3). Statistical analysis was performed using one-way analysis of variance with Tukey post hoc multiple comparison test. (E) Representative results of the long-read RNA-seq of the F8 full-length transcript. (F) Magnified image of intron 14. The gray boxes represent the depth of exon coverage, and the blue lines indicate the splicing junctions of F8 mRNA. The exons in the F8 gene are shown at the bottom. Unrepaired, patient-derived iPSCs with insertion of the EF1α promoter; repaired, gene-corrected patient-derived iPSCs with insertion of the EF1α promoter; healthy, healthy donor-derived iPSCs with insertion of the EF1α promoter.
Restoration of F8 mRNA expression by correction of the intronic variant in patient-derived iPSCs. The patient-derived iPSCs with the insertion of the EF1α promoter before the transcriptional start site of F8 were transduced with SpCas9 (RNP) and an 83-nucleotide single oligonucleotide to correct the variant; then a single clone was isolated. (A) Confirmation of the gene correction by DNA sequencing at the variant in the isolated clone. The color blue indicates the replaced nucleotides derived from the single oligonucleotide. (B) Disappearance of aberrant F8 transcripts between exons 14 and 15 by gene correction was assessed with RT-PCR. The red arrows indicate the size of the amplicons. (C-D) Relative expression of the aberrant transcript (C) and F8 mRNA between exons 25 and 26 (D). Values represent the mean ± standard deviation (n = 3). Statistical analysis was performed using one-way analysis of variance with Tukey post hoc multiple comparison test. (E) Representative results of the long-read RNA-seq of the F8 full-length transcript. (F) Magnified image of intron 14. The gray boxes represent the depth of exon coverage, and the blue lines indicate the splicing junctions of F8 mRNA. The exons in the F8 gene are shown at the bottom. Unrepaired, patient-derived iPSCs with insertion of the EF1α promoter; repaired, gene-corrected patient-derived iPSCs with insertion of the EF1α promoter; healthy, healthy donor-derived iPSCs with insertion of the EF1α promoter.
Inhibition of FVIII production by the introduction of the variant in HEK293 cells
Next, we examined whether the patient’s variant abolished FVIII protein expression. We generated HEK293 cells stably expressing human BDD F8 cDNA with the mini-intron 14 sequence without or with the patient’s variant (c.5220-8563A>G) (Figure 3A). We identified aberrant F8 expression in HEK293 cells stably expressing human BDD F8 cDNA with the mini-intron 14 sequence harboring the patient’s variant (Figure 3B). Contrary to the results from iPSCs (Figure 2D), mRNA expression of F8 was not reduced in HEK293 cells expressing human BDD F8 cDNA with the patient’s variant (Figure 3C), suggesting that the minigene assay does not assess nonsense-mediated RNA decay. Although FVIII protein was similarly expressed within the cells (Figure 3D), FVIII:C and FVIII:Ag in the cell supernatant were significantly reduced in HEK293 cells expressing human BDD F8 cDNA with the patient’s variant (Figure 3E-F). The FVIII:C results for both the 1-stage clotting time assay and the chromogenic assay were comparable (supplemental Figure 4). The supernatant FVIII from HEK293 cells expressing human BDD F8 cDNA with the patient’s variant had a lower specific FVIII:C than those produced from the HEK293 cells expressing wild-type human BDD F8 (Figure 3G). As expected from the results of mRNA sequencing (Figure 1F), FVIII derived from F8 cDNA with the patient’s variant lacked a light chain (Figure 3H; supplemental Figure 5). FVIII:Ag containing a light chain could not be detected in the supernatant in HEK293 cells expressing F8 cDNA with the patient’s variant (supplemental Figure 4), suggesting that the variant acts as a strong splice acceptor.
Inhibition of FVIII production by the introduction of the patient variant in HEK293 cells. HEK293 cells were transfected with the plasmid for the expression of BDD F8 cDNA containing mini-intron 14 derived without or with the patient variant (c.5220-8563A>G). (A) Schematic presentation of F8 cDNA with mini-intron 14. The gray boxes indicate the abnormal exon. (B) Identification of aberrant F8 transcripts between exons 14 and 15 by RT-PCR in HEK293 cells with the variant. The red arrows indicate the size of the amplicons. (C) Relative expression of F8 mRNA between exons 25 and 26. Values represent the mean ± standard deviation (n = 3). Values represent the mean ± standard deviation (n = 3). Statistical analysis was performed by two-tailed Student t test. (D) FVIII expression within the cells was evaluated by immunohistochemical staining and photographed by conformal microscopy (Leica, TCS SP8). Red, FVIII; blue, 4′,6-diamidino-2-phenylindole (DAPI). The scales in merged images represent 10 μm. (E-G) FVIII activity (FVIII:C) (E), FVIII antigen (FVIII:Ag) (F), and the ratio of FVIII:C to FVIII:Ag (G) derived from the cell supernatant. Values are the mean ± standard deviation (n = 3). Statistical analysis was performed using two-tailed Student t test (comparison between 2 groups) or one-way analysis of variance with post hoc Tukey multiple comparison test (comparison between 2 values among 3 groups). (H) Immunoblotting of FVIII protein in the supernatant. The red and blue arrows represent the heavy chain (87.5 kilodalton [kDa]) and the light chain (79 kDa), respectively. Variant, HEK293 cells expressing F8 with the patient’s intronic variant; wild-type, HEK293 cells expressing F8 without the variant; control, parental HEK293 cells.
Inhibition of FVIII production by the introduction of the patient variant in HEK293 cells. HEK293 cells were transfected with the plasmid for the expression of BDD F8 cDNA containing mini-intron 14 derived without or with the patient variant (c.5220-8563A>G). (A) Schematic presentation of F8 cDNA with mini-intron 14. The gray boxes indicate the abnormal exon. (B) Identification of aberrant F8 transcripts between exons 14 and 15 by RT-PCR in HEK293 cells with the variant. The red arrows indicate the size of the amplicons. (C) Relative expression of F8 mRNA between exons 25 and 26. Values represent the mean ± standard deviation (n = 3). Values represent the mean ± standard deviation (n = 3). Statistical analysis was performed by two-tailed Student t test. (D) FVIII expression within the cells was evaluated by immunohistochemical staining and photographed by conformal microscopy (Leica, TCS SP8). Red, FVIII; blue, 4′,6-diamidino-2-phenylindole (DAPI). The scales in merged images represent 10 μm. (E-G) FVIII activity (FVIII:C) (E), FVIII antigen (FVIII:Ag) (F), and the ratio of FVIII:C to FVIII:Ag (G) derived from the cell supernatant. Values are the mean ± standard deviation (n = 3). Statistical analysis was performed using two-tailed Student t test (comparison between 2 groups) or one-way analysis of variance with post hoc Tukey multiple comparison test (comparison between 2 values among 3 groups). (H) Immunoblotting of FVIII protein in the supernatant. The red and blue arrows represent the heavy chain (87.5 kilodalton [kDa]) and the light chain (79 kDa), respectively. Variant, HEK293 cells expressing F8 with the patient’s intronic variant; wild-type, HEK293 cells expressing F8 without the variant; control, parental HEK293 cells.
Restoration of FVIII production by genome editing of the variant in HEK293 cells
We further examined whether gene disruption of the variant would recover FVIII production. Because SpCas9 cleaves between 3 and 4 bases upstream of the PAM sequence, sequence alteration mainly occurs around the cleavage site.5 In addition, because the PAM sequence for SpCas9 (NGG) could not be found around the target site, we applied PAM-flexible SpCas9-NG, which recognized only G as a PAM sequence.18 We designed a 20 nt guide RNA just before the AGA sequence in the minus strand (supplemental Figure 6A). HEK293 cells expressing human BDD F8 cDNA with the patient’s variant were transfected with the plasmid harboring SpCas9 or SpCas9-NG. We confirmed the induction of double-strand breaks (DSBs) at the target site (supplemental Figure 6B-C) and the recovery of FVIII production (supplemental Figure 6D) after genome editing. Genome editing with SpCas9-NG more efficiently induced DSBs and produced FVIII in the supernatant.
Identification of the intron BPS for abnormal splicing caused by the variant
We then tried to identify the intron branch point sequence (BPS) for abnormal splicing. We predicted 4 candidate BPSs before the variant by Splice Site Prediction by Neural Network software (https://www.fruitfly.org/seq_tools/splice.html)21 (supplemental Figure 7A). BPSs just before the variant were removed in the expression plasmid of BDD F8 cDNA containing mini-intron 14 without or with the variant (supplemental Figure 7A). The removal of candidate BPS eliminated abnormal splicing (supplemental Figure 7B-C) and restored FVIII production in HEK293 cells (supplemental Figure 7D).
Classification of the c.5220-8563A>G variant according to the ACMG guidelines
We finally assessed the variant according to the ACMG guidelines with InterVar (https://wintervar.wglab.org/). We presumed that the identified variant (c.5220-8563A>G) could cause splice aberrations based on the in silico analysis (supplemental Table 1). Furthermore, we identified aberrant mRNAs using iPSCs and splice aberrations in the minigene assay (Figures 1-3). We incorporated additional information about the variant (PM2: variant not referenced in gnomAD; PP3: in silico analysis predicted splice impact; and PS3: ectopic RNA analysis supported splice impact) and found that the variation was categorized as likely pathogenic.
Discussion
Genetic diagnosis has become increasingly important in hemophilia in recent years. Genetic abnormalities affect disease phenotypes, such as severity and the development of coagulation factor inhibitors.22-24 In addition, the importance of genetic diagnosis in the identification of female carriers has been advocated because measurements of plasma coagulation factor levels cannot be used for diagnosis.25 The guidelines published by the World Federation of Hemophilia recommend pursuing genetic diagnosis for any patients and their families who desire it.26 The development of NGS technology has enabled whole-genome analysis and the identification of causal variants in numerous diseases.27 However, additional tools are needed to identify which variant causes the disease and to analyze the mechanisms by which the variant causes the disease, especially deep intronic variants.
Prediction by in silico analysis is insufficient for genetic diagnosis for F8 deep intronic variants.12,13 Hence, the effects of these variants should be confirmed by the identification of aberrant F8 transcripts and/or minigene assays. Because FVIII is produced from sinusoidal endothelial cells,28,29 abnormal mRNA could be obtained from liver tissue. However, it is difficult to directly obtain liver specimens by surgery or biopsy because these procedures are highly invasive, especially for patients with hemophilia. Patient-derived iPSCs possess full-length patient genes but do not express coagulation factor mRNA or protein in the undifferentiated state. However, we found that even undifferentiated iPSCs can express full-length F8 mRNA if a promoter is inserted. This technique allowed us to identify the pathogenic variant in introns through the detection of abnormal mRNA sequences. Additionally, we directly demonstrated the restoration of F8 mRNA and protein expression by genome editing.
One of the most important advantages of our method is the potential to obtain full-length F8 mRNA. iPSCs can reproduce patient genes, including unidentified pathological variants. Many groups have reported the detection of aberrant sequences at the in silico predicted site in F8 mRNA expressed ectopically in white blood cells.12-16 On the other hand, the application of iPSCs allows for the evaluation of full-length F8 mRNA with long-read RNA-seq analysis, such as PacBio sequencing. Using these techniques, it is possible to detect not only splice abnormalities at the predicted sites but also unidentified variants. We can also assess the recovery of full-length F8 mRNA by genome editing of the candidate site. This approach will also lead to the confirmation of the actual therapeutic efficacy of future individual genome editing treatments.
Furthermore, the application of iPSCs allowed us to evaluate nonsense-mediated mRNA decay. Nonsense-mediated RNA decay targets aberrant mRNAs with premature terminal codons, resulting in a reduction in mRNA for translation.30 It has been reported that premature terminal codons occur in 1 × 108 sites of the translated region in the human genome, and approximately half of these can cause nonsense-mediated RNA decay.31 However, it is difficult to assess nonsense-mediated RNA decay by the conventional transfection procedure to reconstitute the target gene,32 and it is impossible to artificially reconstitute cells to express a full-length patient-derived F8 gene because it is a 186 kb gene consisting of 26 exons.33 Indeed, the decrease in F8 mRNA expression by the variant could be observed in the experiment with the patient-derived iPSCs but not in HEK293 cells expressing the F8 minigene containing the variant. Hence, the plasma FVIII levels of the patient may be determined not only by abnormal F8 mRNA expression but also by nonsense-mediated RNA decay due to aberrant splicing.
The strategies used in this study could be applied to the diagnosis of other genetic diseases. Advances in NGS technologies have reportedly led to the identification of more than 180 deep intronic variants with possible etiologies in at least 70 different genes.34 Our method can evaluate mRNA sequence and expression without inducing differentiation by inserting a ubiquitous promoter. In addition, it is not necessary to induce the differentiation of iPSCs to obtain mRNA to diagnose diseases of somatic cells, in which it is difficult to induce differentiation from iPSCs.
Genome editing treatment for splice abnormalities resulting from intron variants seems to be much easier than that for variants in exons. Conventional genome editing is essentially mediated by site-specific DSBs generated by genome editing tools, including Cas9.35 DSBs are repaired by 2 DNA repair pathways, namely, HDR and nonhomologous end-joining (NHEJ).3,35 For the correction of the exon variant, DSBs should be repaired by HDR. However, HDR requires the introduction of a donor sequence and has extremely poor efficiency compared with NHEJ.36 In contrast, splice abnormalities can be resolved by NHEJ, disrupting the abnormal splice acceptor sites with DSBs. Indeed, in this study, splice abnormalities could be ameliorated by the introduction of only Cas9 and guide RNA. This approach has already been applied in human clinical trials for the treatment of Leber congenital amaurosis 10 (splice defect in the CEP290 gene).37
There are several limitations to this study that need to be discussed. First, we used undifferentiated iPSCs for the analysis to easily assess mRNA expression without differentiation. The result might not reflect the actual native splicing that occurs in FVIII-producing cells in the patients. Furthermore, we could not obtain FVIII protein in the supernatant even with the induction of a ubiquitous promoter. Hence, the efficient differentiation of iPSCs into sinusoidal endothelial cells is an ideal strategy. Endothelial cells derived from iPSCs have been reported to improve the bleeding tendency of hemophilia A mice to produce functional FVIII after transplantation.38,39 However, we could not obtain F8 mRNA through the differentiation of iPSCs into the endothelial lineage (data not shown). A highly efficient method for differentiating iPSCs into mature sinusoidal endothelial cells to efficiently produce FVIII remains to be established.40 The improvement of novel technologies, including 3-dimensional culture and organoid formation,41 may also lead to the improvement of iPSC-based diagnosis for hemophilia. Second, we could not completely explain the severity of hemophilia caused by the variant. A patient with this variant described in the previous literature reportedly exhibited mild hemophilia,14 but both of our patients had moderate hemophilia. At this stage, it is difficult to explain the discrepancy in severity. Coagulation factor activity in each case may be measured by different reagents in different laboratories, and the measurement has not been standardized. Hence, it is necessary to verify the actual coagulation factor levels using the same method. In addition, we cannot rule out the possibility that a small amount of FVIII in the patient's blood may be derived from normal transcripts because we observe low levels of normal transcripts in our experiments. Finally, we analyzed only 1 family in this study. Our procedure would be relatively time-consuming and complicated to apply to the evaluation of multiple patients. An alternative approach to enhance transcription would be to use CRISPR activation (CRISPRa).42,43 The introduction of catalytically dead Cas conjugated with transcriptional activators can increase the transcription of a target gene.42,43 We have recently developed a CRISPRa strategy involving an all-in-one AAV vector using the AsCas12f-based compact genome editing tool.44 CRISPRa may allow us to construct a more convenient assay that can be applied to many patients. We would like to analyze other cases in the future to verify the usefulness of this approach.
In conclusion, we showed that the deep intronic variant in intron 14 of F8 was pathogenic, leading to aberrant splicing and nonsense-mediated RNA decay using iPSCs derived from patients with hemophilia A. Furthermore, we confirmed that the repair of the variant by genome editing could restore mRNA and protein expression. The strategy used in this study may lead to a reliable diagnosis not only for patients with hemophilia but also for patients with a variety of genetic diseases. This strategy could also be applied to validate the therapeutic efficacy of individualized genome editing therapy targeting the candidate variant.
Acknowledgments
The authors thank Yuiko Ogiwara, Yaeko Suto, Mika Kishimoto, Tamaki Aoki, Hiromi Ozaki, Mai Hayashi, Tomoko Noguchi, Nagako Sekiya, and Hiroko Hayakawa (Jichi Medical University) for their technical assistance. The graphical abstract was created with BioRender.com.
This study was supported by Japan Agency for Medical Research and Development grants JP22fk0410037 (T.O.) and JP22am0401005 (O.N.), and Japan Society for the Promotion of Science KAKENHI grant 23H02819 (T.O.).
Authorship
Contribution: T.H. and T.O. designed the study, performed experiments, analyzed data, and wrote the manuscript; H.I., N.B., and Y.K. performed experiments, analyzed data, and revised the manuscript; M.H., N.K., and E.K. helped with data analysis and revised the manuscript; and H.N. and O.N. provided critical reagents, analyzed the data, and revised the manuscript.
Conflict-of-interest disclosure: H.I. received consultation fees from CSL Behring and Bayer; received speaker fees from Sanofi, Bayer, and CSL Behring; and is a member of an endowed chair funded by CSL Behring. E.K. received grants from Chugai Pharmaceutical and CSL Behring, and speaker fees from Chugai Pharmaceutical, Sanofi, Bayer, Takeda Pharmaceutical, Novo Nordisk, Fujimoto Pharmaceutical, and CSL Behring. T.O. received consultation fees from Chugai Pharmaceutical; grants from Chugai Pharmaceutical and Novo Nordisk; and speaker fees from Chugai Pharmaceutical, Sanofi, Pfizer, Bayer, Daiichi Sankyo, Takeda Pharmaceutical, Novo Nordisk, Fujimoto Pharmaceutical, LSI Medience, and CSL Behring. The remaining authors declare no competing financial interests.
Correspondence: Tsukasa Ohmori, Department of Biochemistry, Jichi Medical University School of Medicine, 3311-1 Yakushiji, Shimotsuke, Tochigi 329-0498, Japan; e-mail: tohmori@jichi.ac.jp.
References
Author notes
Presented in abstract form at the 84th annual meeting of the Japanese Society of Hematology, Fukuoka, Japan, 14 October 2022.
Long-read RNA-seq data of the full-length F8 transcript in iPSCs were deposited in the Gene Expression Omnibus database (accession number GSE229621).
Original data are available on reasonable request from the corresponding author, Tsukasa Ohmori (tohmori@jichi.ac.jp).
The full-text version of this article contains a data supplement.

![Inhibition of FVIII production by the introduction of the patient variant in HEK293 cells. HEK293 cells were transfected with the plasmid for the expression of BDD F8 cDNA containing mini-intron 14 derived without or with the patient variant (c.5220-8563A>G). (A) Schematic presentation of F8 cDNA with mini-intron 14. The gray boxes indicate the abnormal exon. (B) Identification of aberrant F8 transcripts between exons 14 and 15 by RT-PCR in HEK293 cells with the variant. The red arrows indicate the size of the amplicons. (C) Relative expression of F8 mRNA between exons 25 and 26. Values represent the mean ± standard deviation (n = 3). Values represent the mean ± standard deviation (n = 3). Statistical analysis was performed by two-tailed Student t test. (D) FVIII expression within the cells was evaluated by immunohistochemical staining and photographed by conformal microscopy (Leica, TCS SP8). Red, FVIII; blue, 4′,6-diamidino-2-phenylindole (DAPI). The scales in merged images represent 10 μm. (E-G) FVIII activity (FVIII:C) (E), FVIII antigen (FVIII:Ag) (F), and the ratio of FVIII:C to FVIII:Ag (G) derived from the cell supernatant. Values are the mean ± standard deviation (n = 3). Statistical analysis was performed using two-tailed Student t test (comparison between 2 groups) or one-way analysis of variance with post hoc Tukey multiple comparison test (comparison between 2 values among 3 groups). (H) Immunoblotting of FVIII protein in the supernatant. The red and blue arrows represent the heavy chain (87.5 kilodalton [kDa]) and the light chain (79 kDa), respectively. Variant, HEK293 cells expressing F8 with the patient’s intronic variant; wild-type, HEK293 cells expressing F8 without the variant; control, parental HEK293 cells.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/7/22/10.1182_bloodadvances.2023010838/3/m_blooda_adv-2023-010838-gr3.jpeg?Expires=1765884760&Signature=JwHxaFr-xNYolM6xuUkXiSxbbsFyLcOFleTGOlyB1~XI9CVxpfXluGLU3BBnkBhlhvh-U8ZShoc23wlqxg11tpkA7MsA~VXV8hwnn5jgB04jEzelgZVrLEBZDKaT8uzD58NMlp3kgUii-7NAsMPq~U4U9oan5h-UmNGaC-v2TWpEzEsP6zlEpKd8aYVW35dst7VR-ct1ln5GfF9q2EZXBS2g6Qg0V51PecD2I1LRzussD5nbyf93wn7MOUvfL4egl9G~G~JvARjCVL8Xvjic4JX8fEVaBui81d56Hp9ROROW6tC~wntnfL01~Lo505J9QqOgPGtdxwHOvCfLgI61xQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)