Key Points
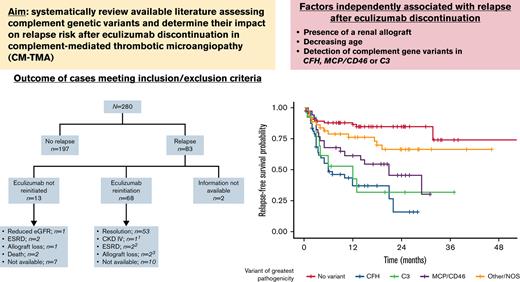
In CM-TMA, variants in specific genes/regions are associated with high risk of relapse after eculizumab is discontinued.
In CM-TMA, eculizumab discontinuation is appropriate in patients with a low risk of relapse.
Abstract
Eculizumab is effective for complement-mediated thrombotic microangiopathy (CM-TMA), also known as atypical hemolytic uremic syndrome. Although lifelong therapy had been suggested, discontinuation does not universally lead to relapse. Comprehensive data evaluating risk factors for recurrence following discontinuation are limited. Our aim was to systematically review available literature assessing the role of complement genetic variants in this setting. Reports on CM-TMA and eculizumab withdrawal published before 1 January 2021, were included. Key reasons for patient exclusion were no follow-up after drug withdrawal and patients lacking complement genetic testing. Two-hundred eighty patients from 40 publications were included. Median age was 28 years, and 25 patients had a known history of renal transplant. Complement genetic variants were identified in 60%, most commonly in CFH (n = 59) and MCP/CD46 (n = 38). Of patients with a complement gene variant, 51.3% had ≥1 likely pathogenic/pathogenic variant whereas the remaining had variants of uncertain significance (VUS). Overall relapse rate after therapy discontinuation was 29.6%. Relapse rate was highest among patients with CFH variants and MCP/CD46 variants in canonical splice regions. VUS (P < .001) and likely pathogenic/pathogenic variants (P < .001) were associated with increased relapse. Presence of a renal allograft (P = .009); decreasing age (P = .029); and detection of variants in CFH (P < .001), MCP/CD46 (P < .001), or C3 (P < .001) were all independently associated with relapse after eculizumab discontinuation. Eculizumab discontinuation is appropriate in specific patients with CM-TMA. Caution should be exerted when attempting such a strategy in patients with high risk of recurrence, including a subgroup of patients with MCP/CD46 variants.
Introduction
Thrombotic microangiopathy (TMA) syndromes encompass a diverse set of entities characterized by the presence of microangiopathic hemolytic anemia, thrombocytopenia, and end-organ injury.1 Complement-mediated TMA (CM-TMA), also known as atypical hemolytic uremic syndrome (aHUS), is a clinical entity characterized by uncontrolled activation of the alternative complement pathway in the setting of defects of complement regulation in genetically susceptible individuals.2,3
Prior to the approval of the anti-C5 monoclonal antibody eculizumab, up to 67% of patients diagnosed with CM-TMA required dialysis or died within 3 years of diagnosis.4 However, eculizumab use during the past decade has altered the natural history of CM-TMA and has resulted in long-term improvements of hematologic parameters, renal function, quality of life, and overall survival.5-7
Although the improvement of clinical outcomes with terminal complement inhibition is not a matter of debate, the markedly high costs of therapy, increased risk of potentially fatal meningococcal infections during therapy, and demonstration of a lack of universal relapse upon therapy withdrawal8,9 have resulted in attempts to discontinue therapy across multiple study groups after achievement of a clinical response.9-13
Results from these reports have shed light on the multifactorial nature of relapse after treatment withdrawal. Of particular relevance, the detection of complement genetic variants as well as the type and number of these genetic alterations have been associated with differing risks of relapse in different cohorts, with CFH variants being associated with a risk of relapse >70% in some studies.10,14,15
Although these findings have certainly helped guide clinical practice, the available literature has mostly consisted of single-center experiences with a restricted sample size, which has limited broad applicability of results. Taking this into consideration, our aim was to provide a comprehensive summary of the current evidence assessing the impact of complement genetic variants on CM-TMA relapse rates after eculizumab discontinuation.
Patients and methods
In this systematic review, search protocols were developed for the Ovid and PubMed databases to identify existing reports on CM-TMA and eculizumab withdrawal published before 1 January 2021. Searches were conducted using the following medical subject heading terms in combination with a keyword search: “thrombotic microangiopathies,” “atypical h(a)emolytic ur(a)emic syndrome,” “eculizumab,” “complement inactivator proteins.” Articles were reviewed first by title and then abstract for appropriateness of inclusion. Because there were few interventional studies, observational cohort studies, case series, and case reports were included.
Cases were cross-referenced and information evaluated at the patient level to eliminate duplicates. Inclusion criteria included patients diagnosed with CM-TMA or aHUS who had undergone eculizumab discontinuation after an initial response to ≥2 doses of C5 inhibitor therapy. Discontinuation was defined as withdrawal of complement inhibition therapy for a minimum of 15 days with the intent of therapy discontinuation, if specified by authors in the original publication. Using this definition, we eliminated patients who would have discontinued due to nonadherence. Otherwise, the only cohort in which eculizumab was discontinued with no plan to resume treatment in the foreseeable future but without the specific intent of therapy discontinuation was that published by Neto et al in which discontinuation was unavoidable due to a national eculizumab drug shortage. Patients undergoing eculizumab dose extension, but not discontinuation, were not included.
Exclusion criteria included (1) patients lacking complement genetic testing, (2) subsequent evidence leading to reclassification of diagnosis into another primary TMA syndrome, (3) no follow-up after therapy discontinuation, and (4) inability to determine case-specific information from published data. Of note, patients with clinical follow-up after eculizumab discontinuation but without mention of specific time of follow-up were included in the study but were not included in the cohort from which the time-to-event-analysis of predictors was carried out.
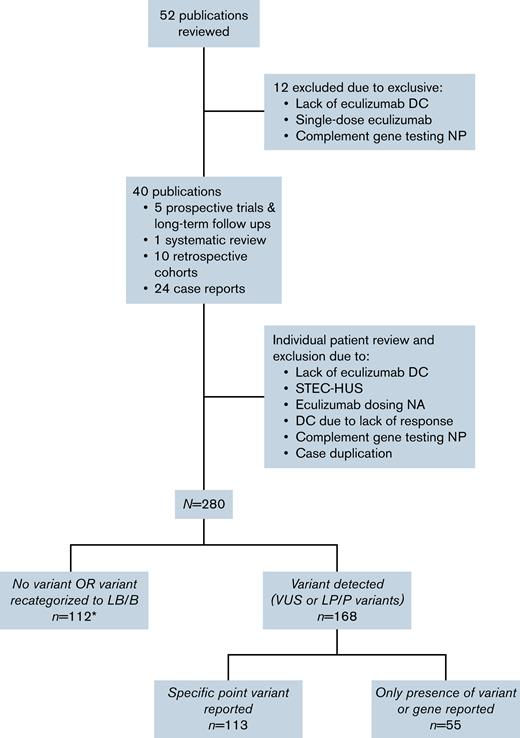
Fifty-two separate publications were reviewed for identification of patients meeting inclusion criteria, and patients from 12 publications were excluded due to exclusion criteria.13,16-26 The 40 remaining publications were reviewed extensively.3,5-7,9-12,14,15,27-56 Individual cases were excluded due to lack of eculizumab discontinuation, subsequent evidence of Shiga toxin-producing Escherichia coli, lack of information on eculizumab dosing, no complement genetic testing, case duplication, and eculizumab discontinuation in the setting of lack of a hematologic response.
Apart from patients from the study by Neto et al48 (n = 19) in which eculizumab discontinuation occurred due to eculizumab drug shortage, all patients in our final cohort had a return of hematogic parameters to baseline and an absence of laboratory evidence of hemolysis. Due to the heterogenous reporting of renal function in the parent publication, renal response was more difficult to classify. However, included publications, with the exception of author cases reported in Macia et al,27 at a minimum commented on renal response to eculizumab as stabilization or improvement of renal function after therapy initiation. Although the Neto study did not specifically comment on complete hematologic response before eculizumab discontinuation, based on the definition of relapse used in the study, it was assumed that patients had achieved at least a hematologic response and, as such, were included in our final cohort.
Variant analysis
For individuals found to have complement variants, analysis using Alamut Visual (version 2.11) software, including nucleotide and amino acid conservation, in silico prediction tools for amino acid changes (Align GVGD, SIFT, MutationTaster, and PolyPhen-2), and in silico prediction tools for potential splicing impact (SpliceSiteFinder-like, MaxEntScan, NNSPLICE, and GeneSplicer), was performed. Variant frequency was reviewed using data in the Genome Aggregation Database.57 To evaluate the frequency of variants in aHUS cases as compared with controls, the database of complement gene variants was also reviewed.58 The Combined Annotation Dependent Depletion score59 was calculated for each variant and a mutation significance cutoff at the 99% confidence interval (CI) was applied. The constraint metrics of Z score and observed/expected for missense variants was determined. The Rare Exome Variant Ensemble Learner precomputed scores were obtained for reported variants.60 Using these tools as well as literature reports, variants were classified per American College of Medical Genetics and Genomics and Association for Molecular Pathology guidelines for variant interpretation61 as benign/likely benign, variant of uncertain significance (VUS), likely pathogenic, and pathogenic.
Statistical analysis
For purposes of this analysis, patients with variants that were thought to be benign/likely benign were grouped with patients without variants. If a patient had >1 variant, the variant with the highest pathogenicity was used for classification. In addition, patients with documented factor H antibodies with no complement genetic variant of higher pathogenicity were classified as VUS.
Categorical variables are presented as n (%) and continuous variables as median (Q1-Q3). Association of demographic and genetic variant characteristics with risk of relapse was assessed using logistic regression. As a sensitivity analysis, Cox proportional hazards analysis was also performed on patients who had follow-up data available, considering relapse as a time-dependent event, from time of discontinuation of eculizumab until relapse or last relapse-free follow-up (supplemental Tables 1 and 2). All statistical analysis was performed using R, version 3.6.2.
Results
Demographics of study population
After assessment of individual publications, a total of 280 patients with a history of eculizumab discontinuation and complement genetic testing were included in the final cohort (Figure 1). There was significant heterogeneity in the report of baseline characteristics and follow-up across publications (Table 1). Among patients with available information, median age at time of the CM-TMA event for which eculizumab was initiated was 28 years (IQR 14.25-41 years). Twenty-five patients in our cohort had a history of renal transplant prior to initiation of eculizumab. Of those with available information (n = 86), 55.8% underwent dialysis at one point during their clinical course.
CONSORT diagram of publication and patient workflow for inclusion in the final cohort. Includes 7 patients with variants recategorized as LB/B (included in supplemental Table 1). DC, discontinuation; LB/B, likely benign or benign; LP/P, likely pathogenic or pathogenic; NA, not available; NP, not performed; STEC-HUS, Shiga toxin-producing Escherichia coli hemolytic uremic syndrome; VUS, variant of uncertain significance.
CONSORT diagram of publication and patient workflow for inclusion in the final cohort. Includes 7 patients with variants recategorized as LB/B (included in supplemental Table 1). DC, discontinuation; LB/B, likely benign or benign; LP/P, likely pathogenic or pathogenic; NA, not available; NP, not performed; STEC-HUS, Shiga toxin-producing Escherichia coli hemolytic uremic syndrome; VUS, variant of uncertain significance.
Baseline demographics of patients undergoing eculizumab discontinuation
| . | N = 280 . |
|---|---|
| Age at time of eculizumab initiation | |
| Median (Q1-Q3) | 28 (14.25-41.0) |
| N/A | 54 |
| Sex, n (%) | |
| Male | 85 (40.1) |
| Female | 127 (59.9) |
| N/A | 68 |
| Prior TMA event∗, n (%) | |
| Yes | 94 (79.7) |
| No | 24 (20.3) |
| N/A | 162 |
| Kidney status, n (%) | |
| Native | 201 (88.9) |
| Allograft | 25 (11.1) |
| N/A | 54 |
| Dialysis during CM-TMA event, n (%) | |
| Yes | 48 (55.8) |
| No | 38 (44.2) |
| N/A | 194 |
| Complement gene variant, n (%) | |
| Yes | 168 (60) |
| No variant | 112 (40) |
| Gene variant number†, n (%) | |
| Single variant | 148 (88.1) |
| >1 concomitant variant | 20 (11.9) |
| Single-gene variant type, n (%) | |
| CFH | 47 (31.8) |
| MCP/CD46 | 35 (23.6) |
| C3 | 11 (7.4) |
| CFI | 15 (10.1) |
| Time on eculizumab (mo) | |
| Median (Q1-Q3) | 8 (4.5-14.0) |
| N/A | 67 |
| Dialysis at eculizumab d/c, n (%) | |
| Yes | 12 (6) |
| No | 187 (94) |
| N/A | 81 |
| Follow up after eculizumab d/c (mo) | |
| Median (95% CI) | 23 (20-24) |
| N/A | 62 |
| . | N = 280 . |
|---|---|
| Age at time of eculizumab initiation | |
| Median (Q1-Q3) | 28 (14.25-41.0) |
| N/A | 54 |
| Sex, n (%) | |
| Male | 85 (40.1) |
| Female | 127 (59.9) |
| N/A | 68 |
| Prior TMA event∗, n (%) | |
| Yes | 94 (79.7) |
| No | 24 (20.3) |
| N/A | 162 |
| Kidney status, n (%) | |
| Native | 201 (88.9) |
| Allograft | 25 (11.1) |
| N/A | 54 |
| Dialysis during CM-TMA event, n (%) | |
| Yes | 48 (55.8) |
| No | 38 (44.2) |
| N/A | 194 |
| Complement gene variant, n (%) | |
| Yes | 168 (60) |
| No variant | 112 (40) |
| Gene variant number†, n (%) | |
| Single variant | 148 (88.1) |
| >1 concomitant variant | 20 (11.9) |
| Single-gene variant type, n (%) | |
| CFH | 47 (31.8) |
| MCP/CD46 | 35 (23.6) |
| C3 | 11 (7.4) |
| CFI | 15 (10.1) |
| Time on eculizumab (mo) | |
| Median (Q1-Q3) | 8 (4.5-14.0) |
| N/A | 67 |
| Dialysis at eculizumab d/c, n (%) | |
| Yes | 12 (6) |
| No | 187 (94) |
| N/A | 81 |
| Follow up after eculizumab d/c (mo) | |
| Median (95% CI) | 23 (20-24) |
| N/A | 62 |
N/A, not available.
Prior TMA event: authors of original publication indicated that episodes for which eculizumab was started was not their first TMA episode; however, the episode described in the current cohort was the first TMA episode for which eculizumab had been used as therapy.
Patients with only likely benign or benign variants were not included in this count.
Complement variants
One or more complement genetic variants were identified in 168 patients (60%). Seven additional patients presented with only benign/likely benign variants and were classified as having no variants for the purposes of subsequent analysis. The most common single variants included those in CFH (n = 47), MCP/CD46 (n = 35), CFI (n = 15), and the presence of anti-CFH antibodies with or without concomitant CFHR1 deletions (n = 12). Multiple concomitant variants were reported in 20 patients, in 12 of whom this included ≥1 in CFH variant (Table 2). Six patients were reported to have CFH or MCP/CD46 risk haplotypes, including 2 with no variants, 2 with an MCP/CD46 variant, 1 with a CFH variant, and 1 with a C3 variant.
Relapse rates according to complement gene variants and complement antibodies
| . | N . | n . | Relapse . | Relapse rate, % . |
|---|---|---|---|---|
| Overall | 280 | — | 83 | 29.6 |
| No variant detected | 112 | — | 12 | 10.7 |
| Variant detected | 168 | — | 71 | 42.3 |
| >1 concomitant variants | 20 | — | 10 | 50.0 |
| CFH-inclusive | — | 12 | 6 | 50.0 |
| Non CFH-inclusive | — | 8 | 4 | 50.0 |
| Single-gene variants | 148 | — | 61 | 41.2 |
| CFH | 47 | — | 27 | 57.4 |
| Exon 22 | — | 11 | 10 | 90.9 |
| NOS | — | 19 | 7 | 36.8 |
| Other | — | 17 | 10 | 58.8 |
| MCP/CD46 | 35 | — | 18 | 51.4 |
| Splice regions | — | 12 | 10 | 83.3 |
| NOS | — | 9 | 3 | 33.3 |
| Other | — | 14 | 5 | 35.7 |
| C3 | 11 | — | 5 | 45.5 |
| CFI | 15 | — | 5 | 33.3 |
| Anti-CFH antibodies | 12 | — | 2 | 16.7 |
| Other single variants | 13∗ | — | 4 | 30.8 |
| Variant NOS | 15 | — | 0 | 0.00 |
| CFHvariant type† | 28 | — | — | — |
| Missense variants | — | 18 | 14 | 77.8 |
| Non-missense variants | — | 10 | 6 | 60.0 |
| MCP/CD46variant type† | 26 | — | — | — |
| Non-splice region | — | 14 | 5 | 35.7 |
| Canonical splice region | — | 12 | 10 | 83.3 |
| . | N . | n . | Relapse . | Relapse rate, % . |
|---|---|---|---|---|
| Overall | 280 | — | 83 | 29.6 |
| No variant detected | 112 | — | 12 | 10.7 |
| Variant detected | 168 | — | 71 | 42.3 |
| >1 concomitant variants | 20 | — | 10 | 50.0 |
| CFH-inclusive | — | 12 | 6 | 50.0 |
| Non CFH-inclusive | — | 8 | 4 | 50.0 |
| Single-gene variants | 148 | — | 61 | 41.2 |
| CFH | 47 | — | 27 | 57.4 |
| Exon 22 | — | 11 | 10 | 90.9 |
| NOS | — | 19 | 7 | 36.8 |
| Other | — | 17 | 10 | 58.8 |
| MCP/CD46 | 35 | — | 18 | 51.4 |
| Splice regions | — | 12 | 10 | 83.3 |
| NOS | — | 9 | 3 | 33.3 |
| Other | — | 14 | 5 | 35.7 |
| C3 | 11 | — | 5 | 45.5 |
| CFI | 15 | — | 5 | 33.3 |
| Anti-CFH antibodies | 12 | — | 2 | 16.7 |
| Other single variants | 13∗ | — | 4 | 30.8 |
| Variant NOS | 15 | — | 0 | 0.00 |
| CFHvariant type† | 28 | — | — | — |
| Missense variants | — | 18 | 14 | 77.8 |
| Non-missense variants | — | 10 | 6 | 60.0 |
| MCP/CD46variant type† | 26 | — | — | — |
| Non-splice region | — | 14 | 5 | 35.7 |
| Canonical splice region | — | 12 | 10 | 83.3 |
Other single variants are CFHR1-5 deletions without autoantibodies (n = 8) and variants in ADAMTS13 (n = 2), CFB (n = 2), and DGKE (n = 1).
Only includes patients with listed variant as a single variant.
Of patients with reports of a variant, 120 had detailed information that allowed for variant evaluation and reclassification. One hundred thirteen of the 120 patients had a variant or Factor H antibody that was classified as a VUS or higher. A total of 58 of these patients had ≥1 likely pathogenic or pathogenic variants whereas the remaining (n = 55) had 1 or multiple VUS (supplemental Table 5).
Eculizumab discontinuation and relapse
Eculizumab was discontinued after a median of 8 months (IQR 4.5-14 months) on therapy. At time of withdrawal, all patients had hematologic remission, and all but 12 patients (7.2%) had achieved a stable renal function not requiring renal replacement therapy. Of the 8 patients with a known anti-CFH titer and mention of titer status at time of therapy discontinuation, 5 anti-CFH titers had been reduced with the use of additional therapies, and the remaining 3 had detectable antibody titers between 500 and 1500 AU/mL.
Overall relapse rate of CM-TMA after therapy discontinuation was 29.6% (n = 83). Among patients with information available regarding time of follow-up after eculizumab discontinuation (n = 218), median follow up was 23 months (95% CI, 20-24). Median time to relapse was 3.60 months (range, 0.5-31.75). Relapse rates according to complement genetic variant assessment are shown in Table 2. The highest relapse rates observed in the cohort were among patients with single CFH variants (57.4%), particularly if affecting exon 22 (90.9%); patients with single MCP/CD46 variants (51.4%); and patients with multiple concomitant genetic variants (50%). Of patients with single MCP/CD46 variants, those involving splice regions were more likely to relapse than those not involving splice regions (83.3% vs 35.7%; P = .021).
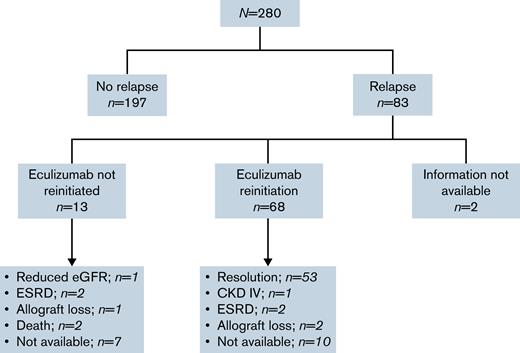
Clinical outcomes after relapse are presented in Figure 2. Anti–complement therapy was not reinitiated after confirmation of CM-TMA relapse in 13 cases (16%), 9 of which did not do so due to a drug shortage in the original authors’ region. Of the 6 patients with description of postrelapse clinical outcomes within this subgroup, 1 patient maintained a stable but reduced renal function on plasma exchange; 2 progressed to end-stage renal disease that required dialysis; 1 lost renal allograft function; and 2 patients died. One additional death was observed throughout the cohort follow-up in a patient without relapse, due to an unrelated medical comorbidity.
Flow of clinical outcomes of patients after eculizumab discontinuation. Slowly progressive proteinuria and worsening renal function over 3 months prior to reinitiation of eculizumab in setting of a patient with baseline CKD KDIGO IIIA and a CFH variant that stabilized at an eGFR of 29 mL/min at last follow-up.2 Progressive worsening renal function and long-term dialysis need27 and progressive worsening of kidney function despite eculizumab reinitiation in setting of an MCP variant and progression to CKD KDIGO V requiring renal transplantation.3 Postinfectious TMA relapse on a kidney allograft with initial stabilization of renal function after eculizumab reinitiation subsequently developed overlapping acute tubular necrosis due to a second insult leading to progressive renal function loss over 6 months and, ultimately, allograft loss29 and posttransplant recurrence of TMA in patient with CFI NOS variant with no response to 3 months of eculizumab retrial and subsequent allograft loss.47 CKD, chronic kidney disease; eGFR, estimated glomerular filtration rate; ESRD, end-stage renal disease.
Flow of clinical outcomes of patients after eculizumab discontinuation. Slowly progressive proteinuria and worsening renal function over 3 months prior to reinitiation of eculizumab in setting of a patient with baseline CKD KDIGO IIIA and a CFH variant that stabilized at an eGFR of 29 mL/min at last follow-up.2 Progressive worsening renal function and long-term dialysis need27 and progressive worsening of kidney function despite eculizumab reinitiation in setting of an MCP variant and progression to CKD KDIGO V requiring renal transplantation.3 Postinfectious TMA relapse on a kidney allograft with initial stabilization of renal function after eculizumab reinitiation subsequently developed overlapping acute tubular necrosis due to a second insult leading to progressive renal function loss over 6 months and, ultimately, allograft loss29 and posttransplant recurrence of TMA in patient with CFI NOS variant with no response to 3 months of eculizumab retrial and subsequent allograft loss.47 CKD, chronic kidney disease; eGFR, estimated glomerular filtration rate; ESRD, end-stage renal disease.
Among patients receiving eculizumab after relapse (n = 68), 10 patients had no information available regarding eculizumab response. Of the remaining 58 patients, a return to previously achieved hematologic and renal parameters was observed in 91.4% (n = 53) of patients. Five patients did not achieve response; a complement genetic variant was reported in 3 of the 5 cases not reaching previous response (CFH, MCP/CD46, and CFI). All 5 patients experienced a decline in renal function leading to stable KDIGO grade IV chronic kidney disease in 1 case, 2 progressions to end-stage renal disease, and 2 renal allograft losses.
Of 53 patients responding to reinitiaton of eculizumab, 36 remained on eculizumab, and discontinuation had not been retrialed at last follow-up of their respective reports. Of the 7 patients with reported reattempts at discontinuation, eculizumab, 2 had a new relapse event leading to reinitiation of eculizumab at last follow-up. No follow up data were available for 10 patients.
Predictors of CM-TMA relapse
Detailed univariate and multivariable analyses are presented in Tables 3 and 4. The baseline demographic characteristic associated with a significant increase in risk of relapse was decreasing age (odds ratio [OR] per 10-year decrease 1.312; 95% CI, 1.120-1.553; P = .001), whereas a significant impact with CM-TMA in the setting of a renal allograft was not observed (P = .610) with the univariate logistic regression model. A trend toward association between time on eculizumab prior to discontinuation and relapse was noted (P = .076).
Univariable logistic regression analysis of predictors for CM-TMA recurrence after eculizumab discontinuation
| . | Odds ratio (95% CI) . | Level P value . | Overall P value . |
|---|---|---|---|
| Nonvariant factors | |||
| Age, per 10-y decrease | 1.312 (1.120-1.553) | — | < .001 |
| Male (vs female) | 0.814 (0.466-1.467) | — | .497 |
| Transplant (vs native) | 1.248 (0.518-2.895) | — | .610 |
| Time on Eculizumab prior to d/c (mo) | 1.023 (0.997-1.049) | — | .076 |
| Variant-specific factors | |||
| One or more variants detected (vs none) | 6.100 (3.215-12.466) | — | < .001 |
| Number of variants | — | — | < .001 |
| Single variant (vs none) | 7.060 (3.654-14.644) | < .001 | — |
| Multiple variants (vs none) | 8.333 (2.896-24.685) | < .001 | — |
| CFH-inclusive variant (vs other variants + none) | 4.341 (2.386-7.997) | — | < .001 |
| Type of variant - CFH | — | — | < .001 |
| Non-CFH inclusive variant (vs none) | 4.460 (2.235-9.468) | < .001 | — |
| CFH inclusive variant (vs none) | 10.577 (4.928-24.085) | < .001 | — |
| Variant of greatest pathogenicity | — | < .001 | |
| CFH (vs none) | 10.667 (4.939-24.431) | < .001 | — |
| C3 (vs none) | 9.524 (2.947-32.050) | < .001 | — |
| MCP/CD46 (vs none) | 7.895 (3.326-19.534) | < .001 | — |
| Other/variant not specified (vs none) | 2.355 (0.995-5.629) | .051 | — |
| Variant pathogenicity | — | — | < .001 |
| VUS (vs none) | 4.398 (1.967-10.196) | < .001 | — |
| Pathogenic/likely pathogenic variants (vs none) | 14.683 (6.764-33.991) | < .001 | — |
| . | Odds ratio (95% CI) . | Level P value . | Overall P value . |
|---|---|---|---|
| Nonvariant factors | |||
| Age, per 10-y decrease | 1.312 (1.120-1.553) | — | < .001 |
| Male (vs female) | 0.814 (0.466-1.467) | — | .497 |
| Transplant (vs native) | 1.248 (0.518-2.895) | — | .610 |
| Time on Eculizumab prior to d/c (mo) | 1.023 (0.997-1.049) | — | .076 |
| Variant-specific factors | |||
| One or more variants detected (vs none) | 6.100 (3.215-12.466) | — | < .001 |
| Number of variants | — | — | < .001 |
| Single variant (vs none) | 7.060 (3.654-14.644) | < .001 | — |
| Multiple variants (vs none) | 8.333 (2.896-24.685) | < .001 | — |
| CFH-inclusive variant (vs other variants + none) | 4.341 (2.386-7.997) | — | < .001 |
| Type of variant - CFH | — | — | < .001 |
| Non-CFH inclusive variant (vs none) | 4.460 (2.235-9.468) | < .001 | — |
| CFH inclusive variant (vs none) | 10.577 (4.928-24.085) | < .001 | — |
| Variant of greatest pathogenicity | — | < .001 | |
| CFH (vs none) | 10.667 (4.939-24.431) | < .001 | — |
| C3 (vs none) | 9.524 (2.947-32.050) | < .001 | — |
| MCP/CD46 (vs none) | 7.895 (3.326-19.534) | < .001 | — |
| Other/variant not specified (vs none) | 2.355 (0.995-5.629) | .051 | — |
| Variant pathogenicity | — | — | < .001 |
| VUS (vs none) | 4.398 (1.967-10.196) | < .001 | — |
| Pathogenic/likely pathogenic variants (vs none) | 14.683 (6.764-33.991) | < .001 | — |
Multivariable logistic regression analysis of predictors for CM-TMA recurrence after eculizumab discontinuation
| . | Odds ratio (95% CI) . | Level P value . | Overall P value . |
|---|---|---|---|
| Age, per 10-y decrease | 1.248 (1.024-1.523) | — | .029 |
| Transplant (vs native) | 4.287 (1.428-12.866) | — | .009 |
| Variant of highest pathogenicity | < .001 | ||
| CFH (vs none) | 14.561 (5.659-37.469) | < .001 | |
| C3 (vs none) | 9.385 (2.586-34.069) | < .001 | |
| MCP/CD46 (vs none) | 6.413 (2.413-17.047) | < .001 | |
| Other/variant not specified (vs none) | 2.002 (0.753-5.318) | .164 |
| . | Odds ratio (95% CI) . | Level P value . | Overall P value . |
|---|---|---|---|
| Age, per 10-y decrease | 1.248 (1.024-1.523) | — | .029 |
| Transplant (vs native) | 4.287 (1.428-12.866) | — | .009 |
| Variant of highest pathogenicity | < .001 | ||
| CFH (vs none) | 14.561 (5.659-37.469) | < .001 | |
| C3 (vs none) | 9.385 (2.586-34.069) | < .001 | |
| MCP/CD46 (vs none) | 6.413 (2.413-17.047) | < .001 | |
| Other/variant not specified (vs none) | 2.002 (0.753-5.318) | .164 |
Regarding complement genetic variants, detection of ≥1 variants was associated with a >6-fold increase in the risk of relapse after eculizumab discontinuation (OR 6.10; 95% CI, 3.215-12.466; P < .001). Presence of multiple concomitant genetic variants and detection of a single-gene variant were associated with an increase in risk of relapse (P < .001), but presence of multiple variants did not significantly increase the risk of relapse as compared with a single variant (P = .73).
On further analysis of the specific complement gene impacted by variants, CFH-inclusive variants (P < .001), MCP/CD46-inclusive variants (P < .001), and C3-inclusive variants (P < .001) increased the likelihood of relapse after eculizumab discontinuation compared with patients without evidence of complement gene variants. Additionally, CFH-inclusive variants showed a significant increase in risk of relapse when compared with other non–CFH-inclusive variants (P = .009).
Compared with those with no variants or variants thought to be benign or likely benign, VUS were associated with >4-fold increase in relapse (P < .001) whereas likely pathogenic/pathogenic variants had an almost 15-fold increase (P < .001). Difference in risk of relapse between these 2 categories was statistically significant (P < .001) (Table 3).
Younger age at time of eculizumab withdrawal (P = .029), presence of renal allograft (P = .009), presence of CFH variants (P < .001), MCP/CD46 variants (P < .001), and C3 variants (P < .001) were all independently associated with relapse after discontinuation of complement inhibition therapy on multivariable analysis. This model had a c-statistic of 0.796.
To assess relapse as a time-dependent event, survival analysis (supplemental Figures 1-3) was also performed on patients who had follow-up data available (n = 218). Univariate (supplemental Table 1) and multivariable (supplemental Table 2) results were comparable to results from logistic analysis, with decreasing age, presence of renal allograft, and presence of CFH, MCP/CD46, or C3 variants independently associated with higher risk of relapse.
In addition, to determine whether the same conclusions regarding decreasing age and presence of complement variants would persist when posttransplant recurrences were excluded, analysis was performed to exclude patients with a history of renal transplant (supplemental Tables 6-8). Decreasing age and detection of variants in CFH, MCP/CD46, or C3 continued to be independent risks for relapse after eculizumab discontinuation (supplemental Table 8).
Discussion
As experience with the use of eculizumab as standard of care in CM-TMA has expanded, so has the realization that all patients with CM-TMA do not require lifelong eculizumab, particularly when guided by complement genetic variant detection. Whereas recent literature has impacted daily clinical practice and will continue to help tailor management strategies for CM-TMA, the rarity of the disorder in addition to the constant evolution of the knowledge of genetics surrounding CM-TMA continue to result in important challenges when determining the best treatment approach for individual patients.
In our systematic review of the literature, detection of complement genetic variants was associated with an almost 3-fold increase in risk of relapse after therapy withdrawal. This effect was similarly maintained in individuals with reports of either single or multiple concomitant variants. Importantly, no significant differences in relapse were observed in individuals with multiple variants as compared with the single variant group, even when tested only among those with pathogenic variants.
As anticipated, variants in CFH and C3 conferred the highest risk of relapse among those reported, which was particularly evident for variants in CFH exon 22. Unexpectedly, a marked and significant increase in relapse risk was also observed among the population with variants within the MCP/CD46 gene. Relapse in this subgroup was primarily driven by patients with variants within canonical splice sites; however, interpretation should also consider a possible influence of a decreased exposure to complement inhibition before therapy discontinuation (median time on eculizumab 5 months; IQR 2.6-11 months). MCP/CD46 variants have historically been thought to have a more benign course, which may have led to earlier discontinuation of eculizumab due to association of these variants with more favorable outcomes in the pre-eculizumab era.61 Another hypothesis is that because variant classification has not been standardized historically and MCP/CD46 has some rather common “risk” alleles, it is possible that MCP/CD46 has been associated with a more benign course because the common risk alleles were being counted the same as pathogenic variants. Further evaluation of MCP/CD46 variants and risks of eculizumab discontinuation and characteristics of relapse (ie, whether these patients have more subacute relapse) needs to be conducted.
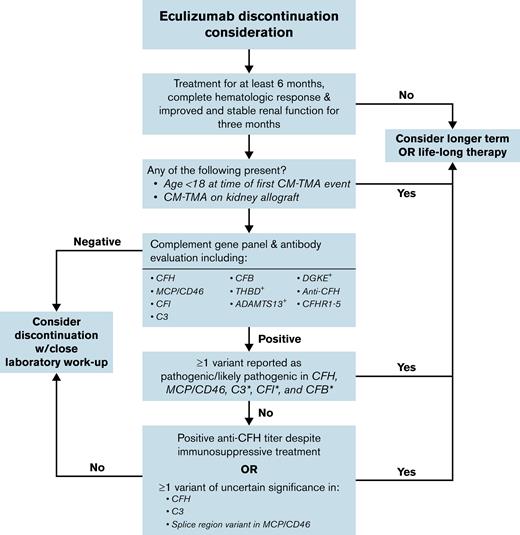
An important addition of our study to the current working knowledge of TMA-associated variants was the reevaluation of each of the published nucleotide changes and their restratification into benign/likely benign (not included in analysis), VUS, or pathogenic/likely pathogenic variants according to contemporary clinical laboratory standards.61 In addition to using clinical standards, we incorporated data that may not have been available at the time of initial case presentation, including recently published in silico meta-predictors60 and CFH functional assays.62 Our results demonstrate that although the highest risks in the cohort were associated with pathogenic/likely pathogenic variants, a nonomissible increase in relapse was also present in patients with VUS. This is likely because the VUS category includes variants for which there is insufficient information for a definitive classification; thus, some are truly pathogenic whereas others are benign. In situations in which eculizumab discontinuation is being considered for patients with VUS, in silico tools to aid in modeling likelihood of pathogenicity may be useful. For some patients, until these genetic alterations are better characterized, continuation of eculizumab may be warranted given higher risk of relapse among patients whose variant is truly pathogenic. Using the data summarized, a framework for how complement genetics can be used to guide discussion regarding eculizumab discontinuation is provided (Figure 3). Note that in addition to complement genetics and clinical equipoise, other patient-specific factors, including age, baseline renal function, and presence or absence of prior kidney transplant, should be incorporated consideration of eculizumab discontinuation. The decision to discontinue eculizumab should incorporate a patient’s ability to tolerate a recurrent TMA event (ie, patients with borderline renal function on therapy should not attempt discontinuation of eculizumab). Given the importance of initiating eculizumab as early as possible if a relapse is detected, ensuring appropriate laboratory and clinical follow-up is also imperative.
Suggested framework for consideration of eculizumab discontinuation in CM-TMA.+The role of complement inhibitor therapy and discontinuation is less clear in the setting of these “noncomplement” variants. ∗Note that the current study had small numbers of patients with variants in these genes, which should be considered when making decisions regarding complement inhibitor duration.
Suggested framework for consideration of eculizumab discontinuation in CM-TMA.+The role of complement inhibitor therapy and discontinuation is less clear in the setting of these “noncomplement” variants. ∗Note that the current study had small numbers of patients with variants in these genes, which should be considered when making decisions regarding complement inhibitor duration.
The main limitations of our study include its retrospective nature. The presented population pulls predominantly from retrospective cohorts, case series, and individual case reports, some of which were published when experience regarding use and discontinuation of terminal complement inhibitors was minimal, leading to heterogeneous treatment of CM-TMA events. Only 1 clinical trial designed to evaluate the predictive role of factors, including genetic variants in CM-TMA, has been published to date.12 Other prospective trials included in this study5-7 were not designed or powered to evaluate the possible discontinuation of complement inhibition therapy in CM-TMA but rather to assess the efficacy of eculizumab in this disorder and, as such, are limited with regards to the level of detail when reporting complement gene variants. Similarly, in the setting of this single prospective trial12 designed to investigate risk of relapse after eculizumab discontinuation, the current cohort does not provide insight on other laboratory assessments (ie, complement serology, urine protein/creatinine ratio) that may prove to be useful for relapse risk stratification after eculizumab discontinuation.
Due to the retrospective nature of the study, relevant demographic and follow-up data are not available for a subset of the cohort. Furthermore, presence of undetected variants in patients cannot be entirely ruled out given the lack of details of the specific genes included in the complement panel assessed in various publications. Although specifically queried in the original publications, presence of additional genetic risk factors such as the presence of CFH and MCP/CD46 haplotypes was rarely reported, and additional research is required to develop more precise estimates of their effect on relapse rates.
Despite the limitations, this study represents the most comprehensive evidence to date assessing the impact of complement gene variants on CM-TMA relapse after eculizumab withdrawal. Our results affirm that attempts at eculizumab discontinuation are not only feasible but appropriate in the right patient population. Our study also highlights the value of complement genetics in the decision-making algorithm when considering complement inhibitor discontinuation and underscores the need for robust variant pathogenicity prediction models. Presence of certain complement genetic variants, particularly pathogenic CFH, C3, or variants in canonical splice sites of MCP/CD46, should discourage attempts at complement inhibition discontinuation. Patients and clinicians should be aware that the success of such strategies is highly nuanced and should only be attempted after thorough consideration of personalized risks and benefits in patients who can be closely monitored for relapse, recognizing that although not frequent, eculizumab reinitiation will not always achieve prewithdrawal responses.
Acknowledgment
M.S. acknowledges the Mayo Clinic Department of Medicine Catalyst for Advancing in Academics grant for providing protected time to support the writing of this manuscript.
Authorship
Contribution: A.A.A.-M., R.S.G., and M.S. designed the research study; A.A.A.-M., M.S., A.M.M., and G.M.S. collected and analyzed the data and wrote the manuscript; G.M.S. and S.C.B. performed statistical analysis; and R.S.G., M.A.V.W., F.C.F., N.L., C.B., J.L.W., G.M.S., and S.C.B. provided critical review of data analysis and the manuscript.
Conflict-of-interest disclosure: M.S. received consulting fees from Alexion. The remaining authors declare no competing financial interests.
A complete list of the members of the Mayo Clinic Complement Alternative Pathway-Thrombotic Microangiopathy (CAP-TMA) Disease-Oriented Group appears in “Appendix.”
Correspondence: Meera Sridharan, Division of Hematology, Mayo Clinic, 200 1st St SW, Rochester, MN 55905; e-mail: sridharan.meera@mayo.edu.
Appendix
The members of the Mayo Clinic Complement Alternative Pathway-Thrombotic Microangiopathy (CAP-TMA) Disease-Oriented Group are: Hatem Amer, Fernando Fervenza, Nelson Leung, Cheryl Tran, Ladan Zand, Hassan Alkhateeb, Ronald Go, William Hogan, Christopher Hook, Rajiv Pruthi, Meera Sridharan, Jill Adamski, David Murray, Jeffrey Winters, Mariam Alexander, Dong Chen, Ann Moyer, Sanjeev Sethi, Maria Willrich, and Nahla Heikal.
References
Author notes
Data will be provided upon direct request to the authors, Aldo A. Acosta-Medina (acostamedina.aldo@mayo.edu) or Meera Sridharan (sridharan.meera@mayo.edu).
The full-text version of this article contains a data supplement.